Characterization of the Phenanthrene-Degrading Sphingobium yanoikuyae SJTF8 in Heavy Metal Co-Existing Liquid Medium and Analysis of Its Metabolic Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultures and Chemicals
2.2. Enrichment and Isolation of Phenanthrene-Degrading Strains
2.3. The Physiological and Evolutionary Analysis of Strain SJTF8
2.4. Utilization Detection of Phenanthrene and Other Aromatics by Strain SJTF8
2.5. Quantification and Metabolites Detection of Phenanthrene Degraded by Strain SJTF8
2.6. Effect of Heavy Metals and Other Stress Conditions on the Cell Growth and the Phenanthrene-Degrading Efficiency of Strain SJTF8
2.7. Plasmid Elimination of Strain SJTF8
2.8. Whole Genome Sequencing and Genome Mining of Strain SJTF8
3. Results
3.1. Isolation and Identification of Sphingobium yanoikuyae SJTF8
3.2. S. yanoikuyae SJTF8 Could Degrade Phenanthrene and Several Other Typical Aromatics Efficiently
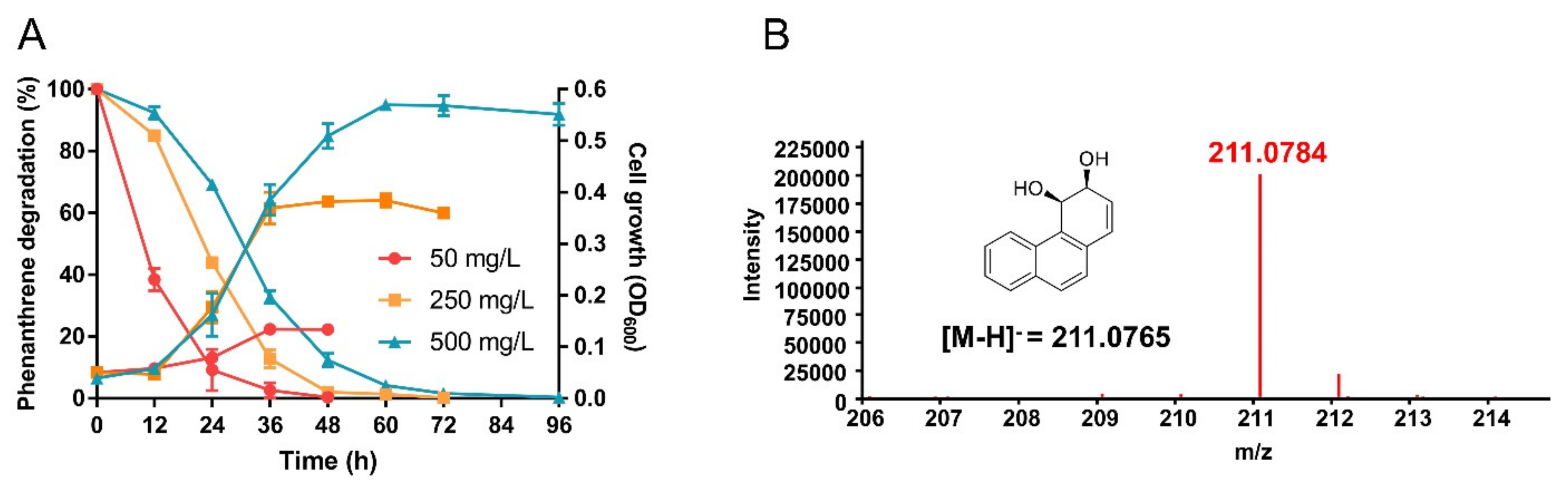
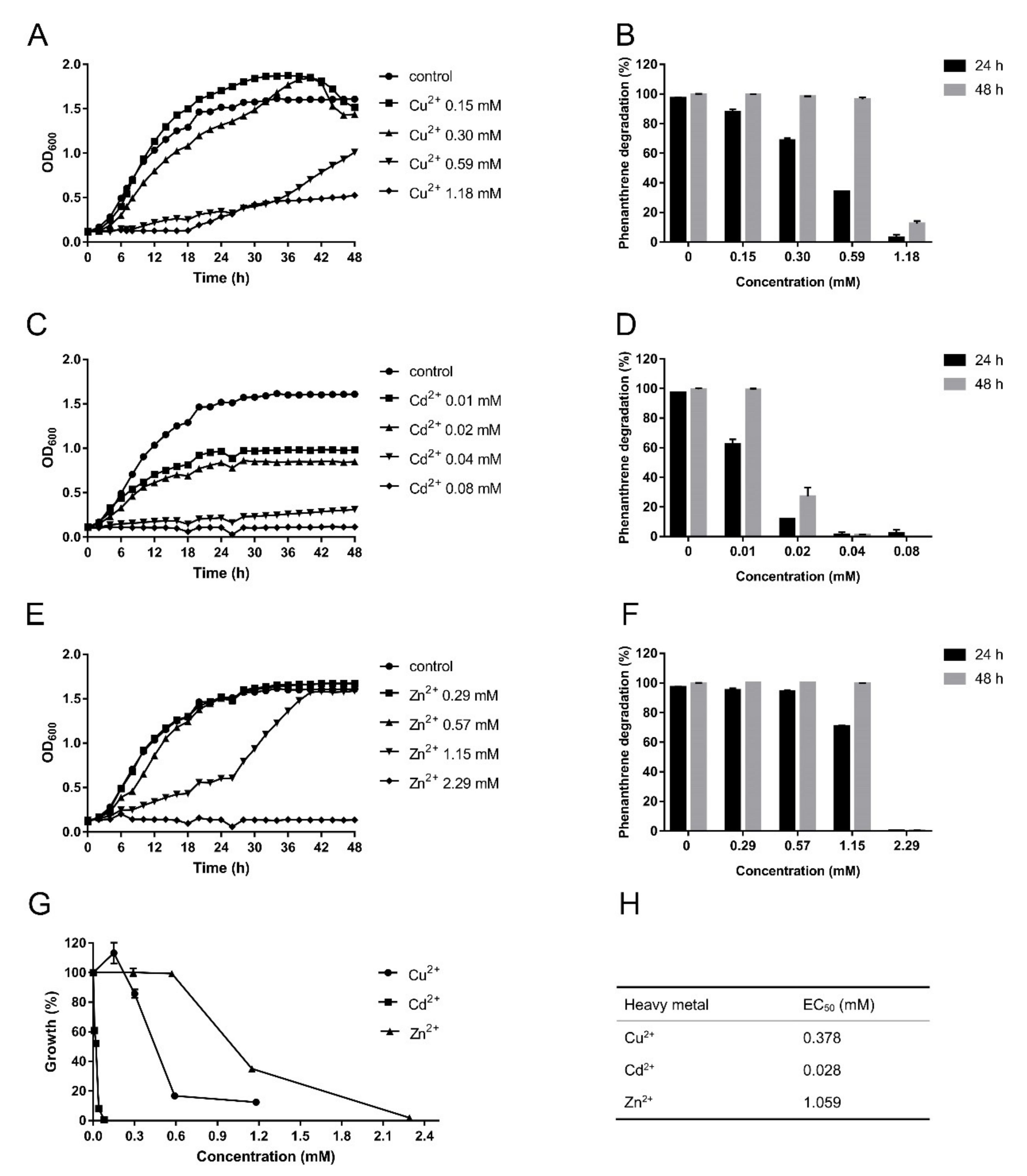
3.3. S. yanoikuyae SJTF8 Could Tolerate High Concentration of Heavy Metals and Degrade Phenanthrene Efficiently
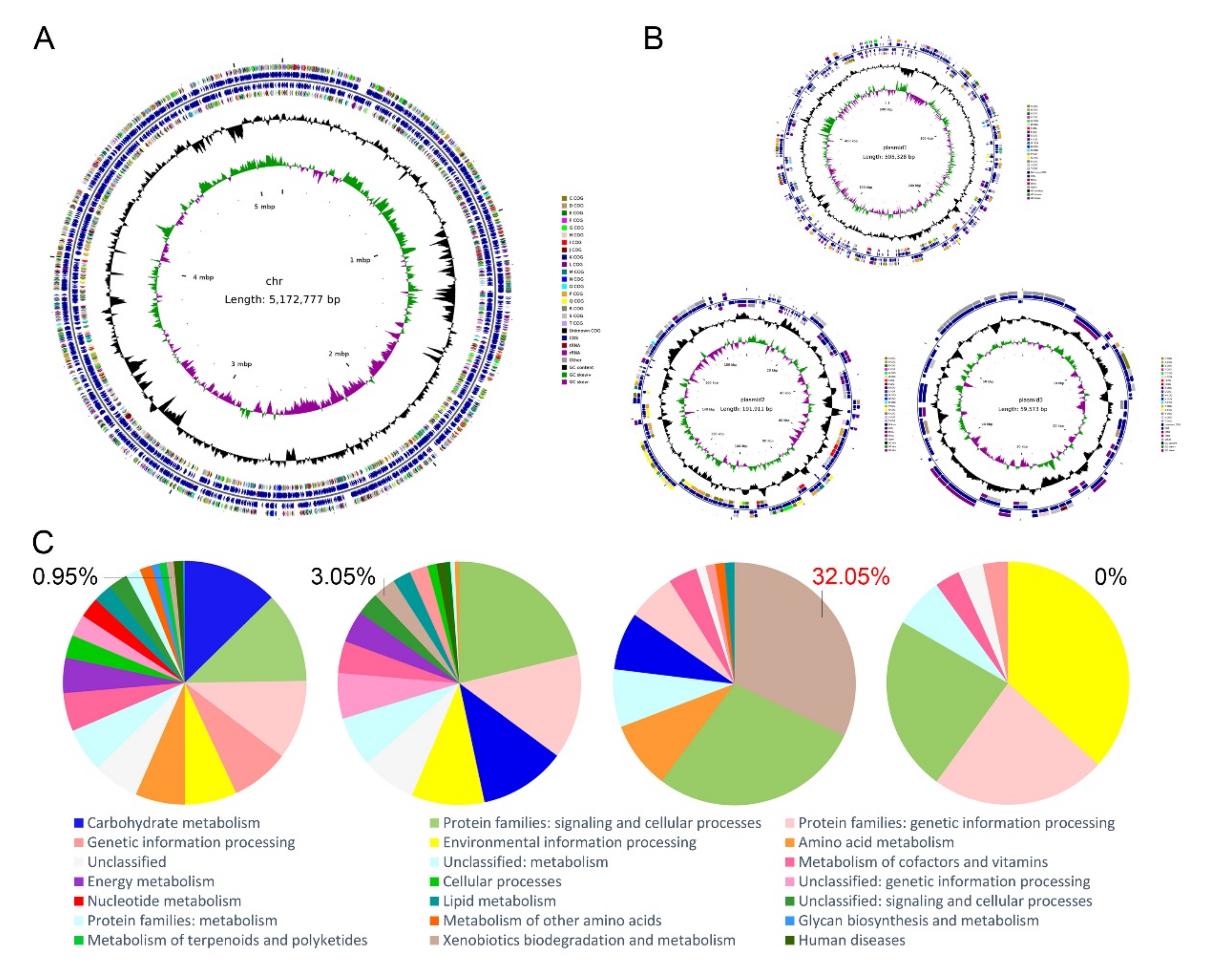
3.4. Whole Genome Sequence Analysis of S. yanoikuyae SJTF8
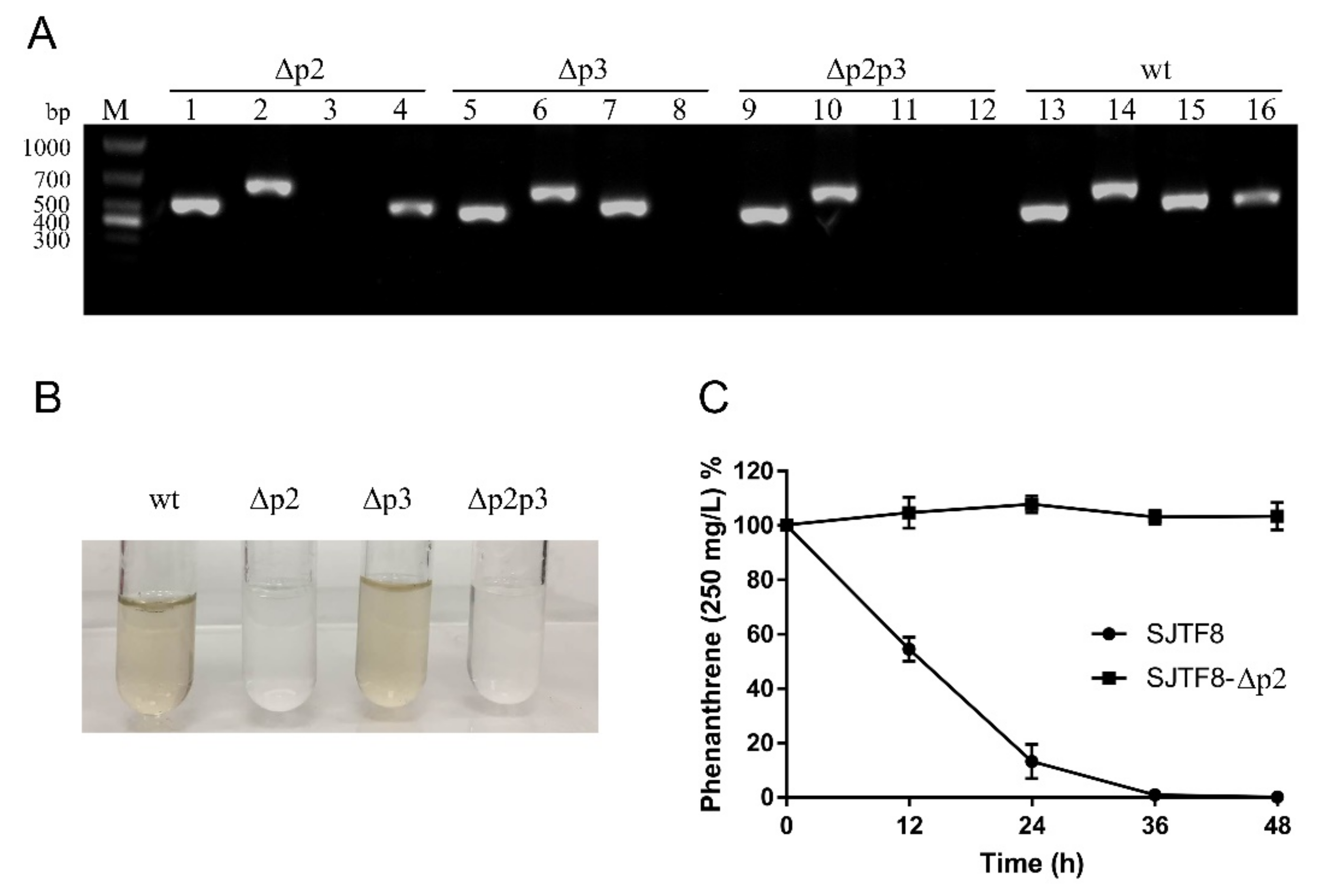
3.5. Genes in Plasmid 2 Were Responsible for PAH Degradation
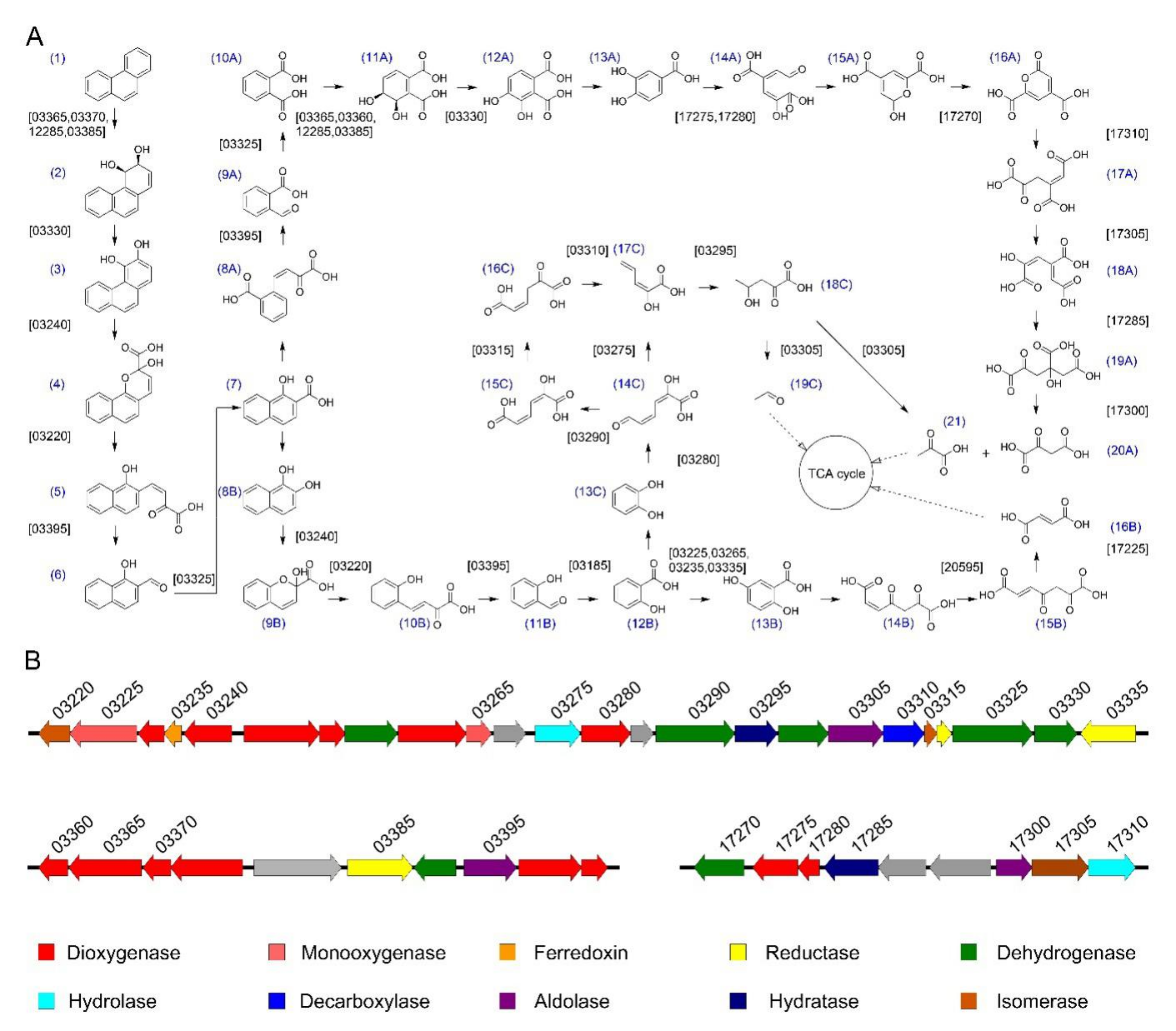
3.6. The Phenanthrene Catabolic Pathway in S. yanoikuyae SJTF8
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alegbeleye, O.O.; Opeolu, B.O.; Jackson, V.A. Polycyclic aromatic hydrocarbons: A critical review of environmental occurrence and bioremediation. Environ. Manage. 2017, 60, 758–783. [Google Scholar] [CrossRef]
- Gao, P.; da Silva, E.; Hou, L.; Denslow, N.D.; Xiang, P.; Ma, L.Q. Human exposure to polycyclic aromatic hydrocarbons: Metabolomics perspective. Environ. Int. 2018, 119, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, J. Different partition of polycyclic aromatic hydrocarbon on environmental particulates in freshwater: Microplastics in comparison to natural sediment. Ecotoxicol. Environ. Saf. 2018, 147, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, M.; Trably, E.; Bernet, N.; Patureau, D. Biodegradation of polycyclic aromatic hydrocarbons: Using microbial bioelectrochemical systems to overcome an impasse. Environ. Pollut. 2017, 231, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Hennessee, C.T.; Li, Q.X. Effects of Polycyclic aromatic hydrocarbon mixtures on degradation, gene expression, and metabolite production in four Mycobacterium species. Appl. Environ. Microbiol. 2016, 82, 3357–3369. [Google Scholar] [CrossRef] [PubMed]
- Hernández Blanco, F.J.; García de Llasera, M.P. Monitoring dihydrodiol polyaromatic hydrocarbon metabolites produced by the freshwater microalgae Selenastrum capricornutum. Chemosphere 2016, 158, 80–90. [Google Scholar] [CrossRef]
- Pozdnyakova, N.; Dubrovskaya, E.; Chernyshova, M.; Makarov, O.; Golubev, S.; Balandina, S.; Turkovskaya, O. The degradation of three-ringed polycyclic aromatic hydrocarbons by wood-inhabiting fungus Pleurotus ostreatus and soil-inhabiting fungus Agaricus bisporus. Fungal Biol. 2018, 122, 363–372. [Google Scholar] [CrossRef]
- Lu, X.Y.; Zhang, T.; Fang, H.H. Bacteria-mediated PAH degradation in soil and sediment. Appl. Microbiol. Biotechnol. 2011, 89, 1357–1371. [Google Scholar] [CrossRef]
- Aylward, F.O.; McDonald, B.R.; Adams, S.M.; Valenzuela, A.; Schmidt, R.A.; Goodwin, L.A.; Woyke, T.; Currie, C.R.; Suen, G.; Poulsen, M. Comparison of 26 sphingomonad genomes reveals diverse environmental adaptations and biodegradative capabilities. Appl. Environ. Microbiol. 2013, 79, 3724–3733. [Google Scholar] [CrossRef]
- Willison, J.C. Isolation and characterization of a novel sphingomonad capable of growth with chrysene as sole carbon and energy source. FEMS Microbiol. Lett. 2004, 241, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Kunihiro, M.; Ozeki, Y.; Nogi, Y.; Hamamura, N.; Kanaly, R.A. Benz[a]anthracene biotransformation and production of ring fission products by Sphingobium sp. strain KK22. Appl. Environ. Microbiol. 2013, 79, 4410–4420. [Google Scholar] [CrossRef] [PubMed]
- Chadhain, S.M.; Moritz, E.M.; Kim, E.; Zylstra, G.J. Identification, cloning, and characterization of a multicomponent biphenyl dioxygenase from Sphingobium yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 2007, 34, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.H.; Xiong, A.S.; Xue, Y.; Fu, X.Y.; Gao, F.; Zhao, W.; Tian, Y.S.; Yao, Q.H. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008, 32, 927–955. [Google Scholar] [CrossRef] [PubMed]
- Habe, H.; Omori, T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 2003, 67, 225–243. [Google Scholar] [CrossRef] [PubMed]
- Demanèche, S.; Meyer, C.; Micoud, J.; Louwagie, M.; Willison, J.C.; Jouanneau, Y. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2004, 70, 6714–6725. [Google Scholar] [CrossRef]
- Waigi, M.G.; Kang, F.; Goikavi, C.; Ling, W.; Gao, Y. Phenanthrene biodegradation by sphingomonads and its application in the contaminated soils and sediments: A review. Int. Biodeter. Biodegr. 2015, 104, 333–349. [Google Scholar] [CrossRef]
- Jouanneau, Y.; Meyer, C.; Jakoncic, J.; Stojanoff, V.; Gaillard, J. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 2006, 45, 12380–12391. [Google Scholar] [CrossRef]
- Jouanneau, Y.; Micoud, J.; Meyer, C. Purification and characterization of a three-component salicylate 1-hydroxylase from Sphingomonas sp. strain CHY-1. Appl. Environ. Microbiol. 2007, 73, 7515–7521. [Google Scholar] [CrossRef]
- Jouanneau, Y.; Meyer, C. Purification and characterization of an arene cis-dihydrodiol dehydrogenase endowed with broad substrate specificity toward polycyclic aromatic hydrocarbon dihydrodiols. Appl. Environ. Microbiol. 2006, 72, 4726–4734. [Google Scholar] [CrossRef]
- Jakoncic, J.; Jouanneau, Y.; Meyer, C.; Stojanoff, V. The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. FEBS J. 2007, 274, 2470–2481. [Google Scholar] [CrossRef] [PubMed]
- Librando, V.; Pappalardo, M. Computational study on the interaction of a ring-hydroxylating dioxygenase from Sphingomonas CHY-1 with PAHs. J. Mol. Graph. Model. 2011, 29, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, D.J.; Brown, E.N.; Yu, C.L.; Parales, R.E.; Gibson, D.T.; Ramaswamy, S. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct. Biol. 2007, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Haarstad, K.; Bavor, H.J.; Mæhlum, T. Organic and metallic pollutants in water treatment and natural wetlands: A review. Water Sci. Technol. 2012, 65, 76–99. [Google Scholar] [CrossRef]
- Ma, X.K.; Li, T.T.; Fam, H.; Charles Peterson, E.; Zhao, W.W.; Guo, W.; Zhou, B. The influence of heavy metals on the bioremediation of polycyclic aromatic hydrocarbons in aquatic system by a bacterial-fungal consortium. Environ. Technol. 2018, 39, 2128–2137. [Google Scholar] [CrossRef]
- Deng, G.; Yang, W.; Zhou, G.; Li, Y.; Liu, S. Heavy metals and polycyclic aromatic hydrocarbons in sediments from the Shenzhen River, South China. Environ. Sci. Pollut. Res. Int. 2014, 21, 10594–10600. [Google Scholar] [CrossRef]
- Liu, S.H.; Zeng, G.M.; Niu, Q.Y.; Liu, Y.; Zhou, L.; Jiang, L.H.; Tan, X.F.; Xu, P.; Zhang, C.; Cheng, M. Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: A mini review. Bioresour. Technol. 2017, 224, 25–33. [Google Scholar] [CrossRef]
- Malakul, P.; Srinivasan, K.R.; Wang, H.Y. Metal toxicity reduction in naphthalene biodegradation by use of metal-chelating adsorbents. Appl. Environ. Microbiol. 1998, 64, 4610–4613. [Google Scholar] [CrossRef]
- Kuo, C.; Genthner, B. Effect of added heavy metal ions on biotransformation and biodegradation of 2-chlorophenol and 3-chlorobenzoate in anaerobic bacterial consortia. Appl. Environ. Microbiol. 1996, 62, 2317–2323. [Google Scholar] [CrossRef]
- Chen, C.; Lei, W.; Lu, M.; Zhang, J.; Zhang, Z.; Luo, C.; Chen, Y.; Hong, Q.; Shen, Z. Characterization of Cu(II) and Cd(II) resistance mechanisms in Sphingobium sp. PHE-SPH and Ochrobactrum sp. PHE-OCH and their potential application in the bioremediation of heavy metal-phenanthrene co-contaminated sites. Environ. Sci. Pollut. Res. Int. 2016, 23, 6861–6872. [Google Scholar] [CrossRef]
- Gran-Scheuch, A.; Fuentes, E.; Bravo, D.M.; Jimenez, J.C.; Perez-Donoso, J.M. Isolation and characterization of phenanthrene degrading bacteria from diesel fuel-contaminated antarctic soils. Front. Microbiol. 2017, 8, 1634. [Google Scholar] [CrossRef] [PubMed]
- Breed, R.S.; Murray, E.G.; Hitchens, A.P. The Outline classification used in the Bergey Manual of Determinative Bacteriology. Bacteriol. Rev. 1944, 8, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, A.; Chmelnitsky, I.; Ofek, I.; Carmeli, Y.; Navon-Venezia, S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 2010, 65, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Tritt, A.; Eisen, J.A.; Facciotti, M.T.; Darling, A.E. An integrated pipeline for de novo assembly of microbial genomes. PLoS ONE 2012, 7, e42304. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Delcher, A.L.; Kasif, S.; Fleischmann, R.D.; Peterson, J.; White, O.; Salzberg, S.L. Alignment of whole genomes. Nucleic Acids Res. 1999, 27, 2369–2376. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.; Forslund, K.; Szklarczyk, D.; Trachana, K.; Roth, A.; Huerta-Cepas, J.; Gabaldon, T.; Rattei, T.; Creevey, C.; Kuhn, M.; et al. eggNOG v4.0: Nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014, 42, D231–D239. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Zhao, H.P.; Wu, Q.S.; Wang, L.; Zhao, X.T.; Gao, H.W. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. J. Hazard. Mater. 2009, 164, 863–869. [Google Scholar] [CrossRef]
- Pagnout, C.; Frache, G.; Poupin, P.; Maunit, B.; Muller, J.F.; Ferard, J.F. Isolation and characterization of a gene cluster involved in PAH degradation in Mycobacterium sp. strain SNP11: Expression in Mycobacterium smegmatis mc2155. Res. Microbiol. 2007, 158, 175–186. [Google Scholar] [CrossRef]
- Subashchandrabose, S.R.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. Interaction effects of polycyclic aromatic hydrocarbons and heavy metals on a soil microalga, Chlorococcum sp. MM11. Environ. Sci. Pollut. R. Int. 2015, 22, 8876–8889. [Google Scholar] [CrossRef]
- Karl, H.; Kammann, U.; Aust, M.O.; Manthey-Karl, M.; Luth, A.; Kanisch, G. Large scale distribution of dioxins, PCBs, heavy metals, PAH-metabolites and radionuclides in cod (Gadus morhua) from the North Atlantic and its adjacent seas. Chemosphere 2016, 149, 294–303. [Google Scholar] [CrossRef]
- Khatoon, K.; Malik, A. Screening of polycyclic aromatic hydrocarbon degrading bacterial isolates from oil refinery wastewater and detection of conjugative plasmids in polycyclic aromatic hydrocarbon tolerant and multi-metal resistant bacteria. Heliyon 2019, 5, e02742. [Google Scholar] [CrossRef] [PubMed]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Sa. 2000, 45, 198–207. [Google Scholar] [CrossRef]
- Fu, B.; Li, Q.X.; Xu, T.; Cui, Z.L.; Sun, Y.; Li, J. Sphingobium sp. FB3 degrades a mixture of polycyclic aromatic hydrocarbons. Int. Biodeter. Biodegr. 2014, 87, 44–51. [Google Scholar] [CrossRef]
- Balciunas, E.M.; Kappelmeyer, U.; Harms, H.; Heipieper, H.J. Increasing ibuprofen degradation in constructed wetlands by bioaugmentation with gravel containing biofilms of an ibuprofen-degrading Sphingobium yanoikuyae. Eng. Life Sci. 2020, 20, 160–167. [Google Scholar] [CrossRef]
- Tao, X.Q.; Lu, G.-N.; Dang, Z.; Yang, C.; Yi, X.Y. A phenanthrene-degrading strain Sphingomonas sp. GY2B isolated from contaminated soils. Process. Biochem. 2007, 42, 401–408. [Google Scholar] [CrossRef]
- Pepi, M.; Lobianco, A.; Renzi, M.; Perra, G.; Bernardini, E.; Marvasi, M.; Gasperini, S.; Volterrani, M.; Franchi, E.; Heipieper, H.J.; et al. Two naphthalene degrading bacteria belonging to the genera Paenibacillus and Pseudomonas isolated from a highly polluted lagoon perform different sensitivities to the organic and heavy metal contaminants. Extremophiles 2009, 13, 839–848. [Google Scholar] [CrossRef]
- Chen, S.; Yin, H.; Ye, J.; Peng, H.; Zhang, N.; He, B. Effect of copper(II) on biodegradation of benzo[a]pyrene by Stenotrophomonas maltophilia. Chemosphere 2013, 90, 1811–1820. [Google Scholar] [CrossRef]
- Obuekwe, I.S.; Semple, K.T. Impact of Zn and Cu on the development of phenanthrene catabolism in soil. Environ. Monit. Assess. 2013, 185, 10039–10047. [Google Scholar] [CrossRef]
- Li, W.; Shi, J.; Wang, X.; Han, Y.; Tong, W.; Ma, L.; Liu, B.; Cai, B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 2004, 336, 231–240. [Google Scholar] [CrossRef]
- Romine, M.F.; Stillwell, L.C.; Wong, K.K.; Thurston, S.J.; Sisk, E.C.; Sensen, C.; Gaasterland, T.; Fredrickson, J.K.; Saffer, J.D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 1999, 181, 1585–1602. [Google Scholar] [CrossRef]
- Zylstra, G.J.; Kim, E. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biot. 1997, 19, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Pinyakong, O.; Habe, H.; Yoshida, T.; Nojiri, H.; Omori, T. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation of Sphingobium sp. strain P2. Biochem. Bioph. Res. Co. 2003, 301, 350–357. [Google Scholar] [CrossRef]
| Aromatic Chemicals | Degradation Capability a | Degradation Efficiency (%) b | Medium Color |
|---|---|---|---|
| Naphthalene | + | > 99 | persistent brown |
| Dibenzothiophene | + | 70 | temporary red |
| 9-Bromophenanthrene | + | 54 | persistent brown |
| Anthracene | + | 33 | temporary pink |
| Benz[a]anthracene | – | – | – |
| Benz[b]anthracene | – | – | – |
| Pyrene | – | – | – |
| Catechol | + | > 99 | persistent brown |
| Salicylic acid | + | > 99 | colorless |
| Phthalic acid | – | – | – |
| Protocatechuate | + | > 99 | colorless |
| Xylene | – | – | – |
| Biphenyl | + | > 99 | persistent brown |
| Genome Features | Chromosome | Plasmid 1 | Plasmid 2 | Plasmid 3 |
|---|---|---|---|---|
| Size (bp) | 5,172,777 | 505,328 | 191,911 | 59,573 |
| GC content (%) | 64.31% | 62.37% | 62.65% | 61.34% |
| Total genes | 4,896 | 547 | 188 | 72 |
| Coding genes | 4781 | 545 | 186 | 72 |
| rRNA | 12 | 0 | 0 | 0 |
| tRNA | 62 | 0 | 0 | 0 |
| Other ncRNA | 25 | 2 | 2 | 0 |
| COG Categories | Categories Function | ORF Number | Value % |
|---|---|---|---|
| A | RNA processing and modification | 0 | 0 |
| B | Chromatin structure and dynamics | 0 | 0 |
| C | Energy production and conversion | 257 | 5.249 |
| D | Cell cycle control, cell division, chromosome partitioning | 31 | 0.633 |
| E | Amino acid transport and metabolism | 258 | 5.270 |
| F | Nucleotide transport and metabolism | 66 | 1.348 |
| G | Carbohydrate transport and metabolism | 241 | 4.922 |
| H | Coenzyme transport and metabolism | 117 | 2.390 |
| I | Lipid transport and metabolism | 165 | 3.370 |
| J | Translation, ribosomal structure and biogenesis | 159 | 3.248 |
| K | Transcription | 248 | 5.065 |
| L | Replication, recombination and repair | 280 | 5.719 |
| M | Cell wall/membrane/envelope biogenesis | 250 | 5.106 |
| N | Cell motility | 32 | 0.654 |
| O | Posttranslational modification, protein turnover, chaperones | 156 | 3.186 |
| P | Inorganic ion transport and metabolism | 249 | 5.086 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 99 | 2.022 |
| R | General function prediction only | 0 | 0 |
| S | Function unknown | 1065 | 21.752 |
| T | Signal transduction mechanisms | 193 | 3.942 |
| U | Intracellular trafficking, secretion, and vesicular transport | 86 | 1.757 |
| V | Defense mechanisms | 69 | 1.409 |
| W | Extracellular structures | 0 | 0 |
| Y | Nuclear structure | 0 | 0 |
| Z | Cytoskeleton | 0 | 0 |
| – | Not in eggNOG | 875 | 17.872 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Xiong, W.; Qiu, H.; Peng, W.; Deng, Z.; Lin, S.; Liang, R. Characterization of the Phenanthrene-Degrading Sphingobium yanoikuyae SJTF8 in Heavy Metal Co-Existing Liquid Medium and Analysis of Its Metabolic Pathway. Microorganisms 2020, 8, 946. https://doi.org/10.3390/microorganisms8060946
Yin C, Xiong W, Qiu H, Peng W, Deng Z, Lin S, Liang R. Characterization of the Phenanthrene-Degrading Sphingobium yanoikuyae SJTF8 in Heavy Metal Co-Existing Liquid Medium and Analysis of Its Metabolic Pathway. Microorganisms. 2020; 8(6):946. https://doi.org/10.3390/microorganisms8060946
Chicago/Turabian StyleYin, Chong, Weiliang Xiong, Hua Qiu, Wanli Peng, Zixin Deng, Shuangjun Lin, and Rubing Liang. 2020. "Characterization of the Phenanthrene-Degrading Sphingobium yanoikuyae SJTF8 in Heavy Metal Co-Existing Liquid Medium and Analysis of Its Metabolic Pathway" Microorganisms 8, no. 6: 946. https://doi.org/10.3390/microorganisms8060946
APA StyleYin, C., Xiong, W., Qiu, H., Peng, W., Deng, Z., Lin, S., & Liang, R. (2020). Characterization of the Phenanthrene-Degrading Sphingobium yanoikuyae SJTF8 in Heavy Metal Co-Existing Liquid Medium and Analysis of Its Metabolic Pathway. Microorganisms, 8(6), 946. https://doi.org/10.3390/microorganisms8060946







