NetMet: A Network-Based Tool for Predicting Metabolic Capacities of Microbial Species and their Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of the Expansion Algorithm and its Implementation in the NetMet Tool
2.2. Single-Species Mode
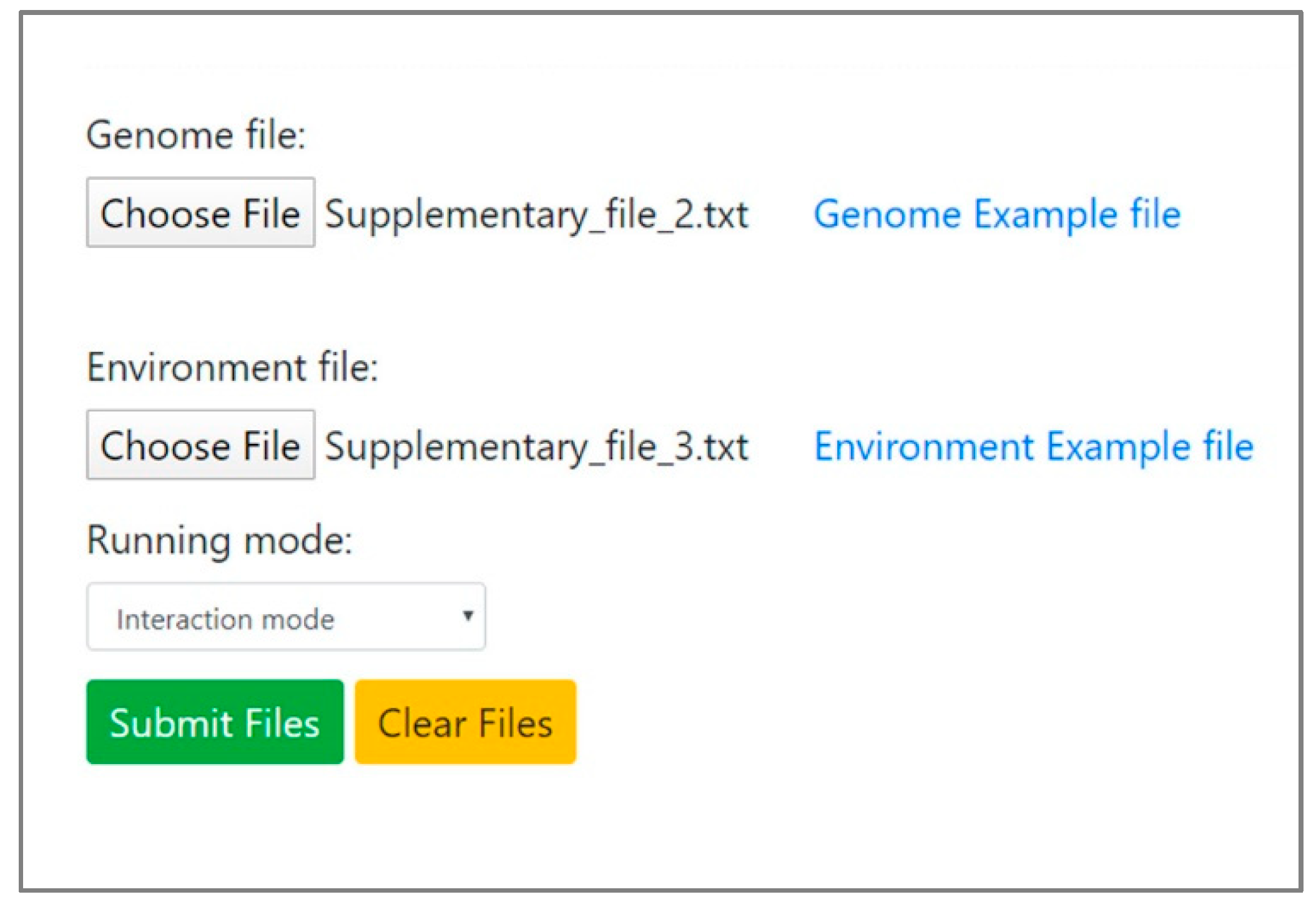
2.3. Interaction Mode
3. Results
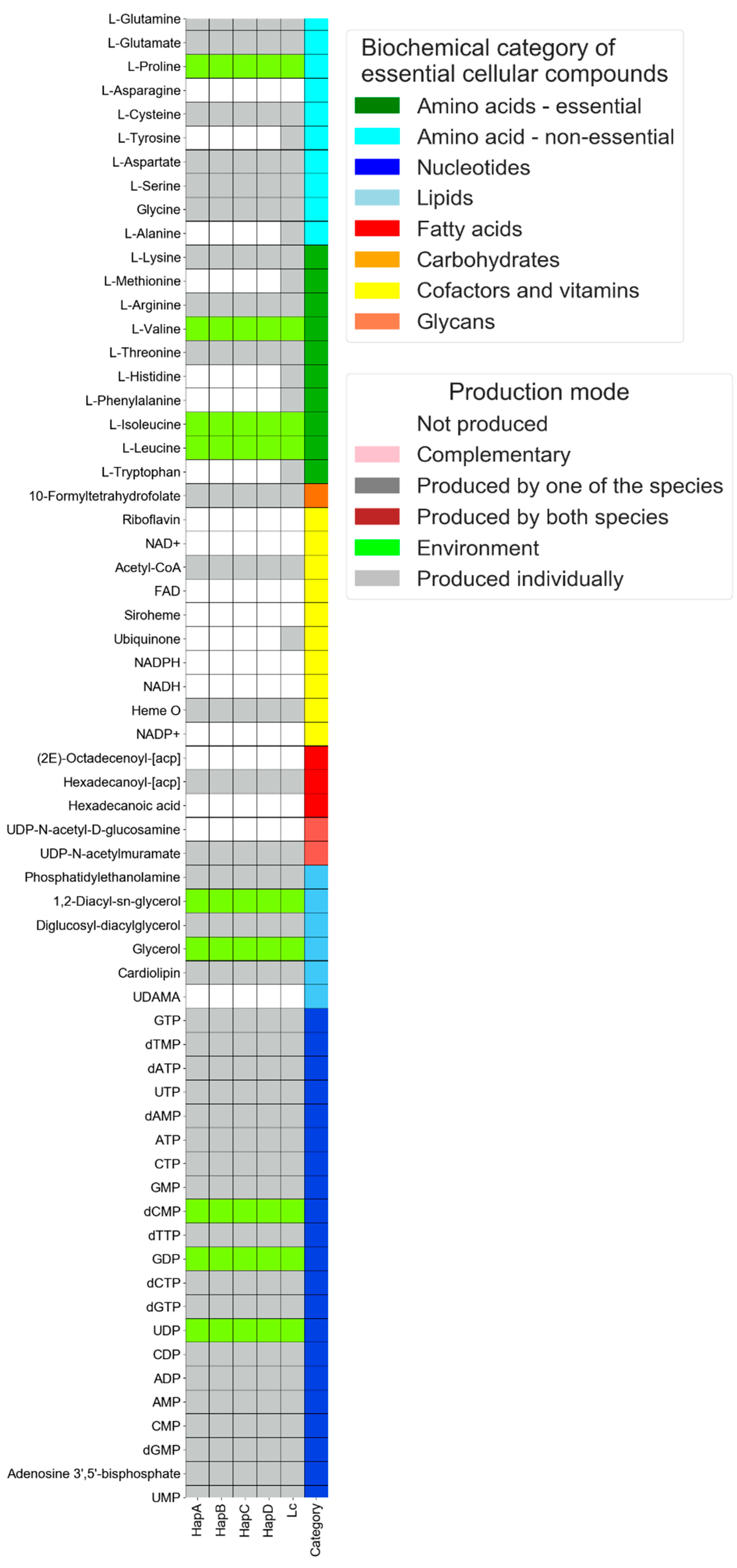
3.1. Single-Species Mode: Comparative Analysis of the Metabolic Capacities of Liberibacter Species That Have Undergone Genomic Reduction
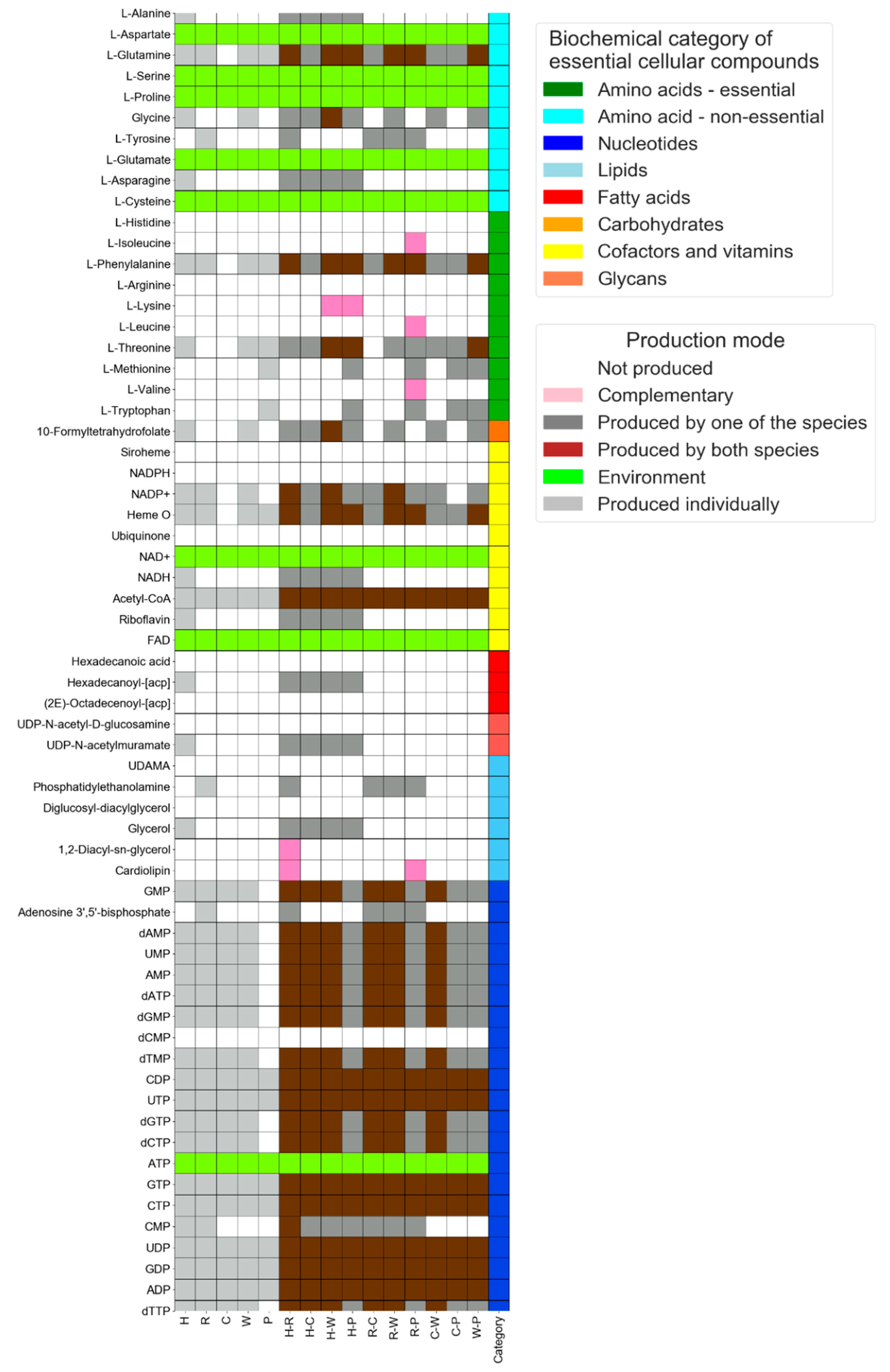
3.2. Interaction Mode: Delineating Exchange Interactions between Endosymbionts of Phloem-Feeding Whitefly Bemisia tabaci
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muller, E.E.L.; Faust, K.; Widder, S.; Herold, M.; Martínez Arbas, S.; Wilmes, P. Using metabolic networks to resolve ecological properties of microbiomes. Curr. Op. Syst. Biol. 2018, 8, 73–80. [Google Scholar] [CrossRef]
- Kruse, K.A.I.; Ebenhöh, O. Comparing flux balance analysis to network expansion: Producibility, sustainability and the scope of compounds. Genome Inform. 2008, 20, 91–101. [Google Scholar]
- Ebenhöh, O.; Handorf, T.; Heinrich, R. Structural analysis of expanding metabolic networks. Genome Inform. 2004, 15, 35–45. [Google Scholar]
- Ebenhöh, O.; Handorf, T.; Heinrich, R. A Cross Species Comparison of Metabolic Network Functions. Genome Inform. 2005, 16, 203–213. [Google Scholar]
- Katsir, L.; Zhepu, R.; Santos Garcia, D.; Piasezky, A.; Jiang, J.; Sela, N.; Freilich, S.; Bahar, O. Genome analysis of haplotype D of Candidatus Liberibacter Solanacearum. Front. Microbiol. 2018, 9, 2933. [Google Scholar] [CrossRef] [PubMed]
- Freilich, S.; Kreimer, A.; Borenstein, E.; Yosef, N.; Sharan, R.; Gophna, U.; Ruppin, E. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 2009, 10, R61. [Google Scholar] [CrossRef] [PubMed]
- Freilich, S.; Kreimer, A.; Borenstein, E.; Gophna, U.; Sharan, R.; Ruppin, E. Decoupling Environment-Dependent and Independent Genetic Robustness across Bacterial Species. PLoS Comput. Biol. 2010, 6, e1000690. [Google Scholar] [CrossRef] [PubMed]
- Handorf, T.; Ebenhöh, O.; Heinrich, R. Expanding Metabolic Networks: Scopes of Compounds, Robustness, and Evolution. J. Mol. Evol. 2005, 61, 498–512. [Google Scholar] [CrossRef] [PubMed]
- Zomorrodi, A.R.; Segrè, D. Synthetic ecology of microbes: Mathematical models and applications. J. Mol. Biol. 2016, 428, 837–861. [Google Scholar] [CrossRef]
- Klitgord, N.; Segrè, D. Ecosystems biology of microbial metabolism. Curr. Opin. Biotech. 2011, 22, 541–546. [Google Scholar] [CrossRef]
- Widder, S.; Allen, R.J.; Pfeiffer, T.; Curtis, T.P.; Wiuf, C.; Sloan, W.T.; Cordero, O.X.; Brown, S.P.; Momeni, B.; Shou, W.; et al. Challenges in microbial ecology: Building predictive understanding of community function and dynamics. ISME J. 2016, 10, 2557–2568. [Google Scholar] [CrossRef]
- Borenstein, E.; Feldman, M.W. Topological signatures of species interactions in metabolic networks. J. Comput. Biol. 2009, 16, 191–200. [Google Scholar] [CrossRef]
- Christian, N.; Handorf, T.; Ebenhöh, O. Metabolic synergy: Increasing biosynthetic capabilities by network cooperation. Genome Inform. 2007, 18, 320–329. [Google Scholar]
- Duan, G.; Christian, N.; Schwachtje, J.; Walther, D.; Ebenhöh, O. The metabolic interplay between plants and phytopathogens. Metabolites 2013, 3, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Freilich, S.; Kreimer, A.; Meilijson, I.; Gophna, U.; Sharan, R.; Ruppin, E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010, 38, 3857–3868. [Google Scholar] [CrossRef]
- Levy, R.; Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 2013, 110, 12804–12809. [Google Scholar] [CrossRef] [PubMed]
- Ofaim, S.; Ofek-Lalzar, M.; Sela, N.; Jinag, J.; Kashi, Y.; Minz, D.; Freilich, S. Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front. Microbiol. 2017, 8, 1606. [Google Scholar] [CrossRef] [PubMed]
- Opatovsky, I.; Santos-Garcia, D.; Ruan, Z.; Lahav, T.; Ofaim, S.; Mouton, L.; Barbe, V.; Jiang, J.; Zchori-Fein, E.; Freilich, S. Modeling trophic dependencies and exchanges among insects’ bacterial symbionts in a host-simulated environment. BMC Genom. 2018, 19, 402. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, A.; Doron-Faigenboim, A.; Borenstein, E.; Freilich, S. NetCmpt: A network-based tool for calculating the metabolic competition between bacterial species. Bioinformatics 2012, 28, 2195–2197. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.; Carr, R.; Kreimer, A.; Freilich, S.; Borenstein, E. NetCooperate: A network-based tool for inferring host-microbe and microbe-microbe cooperation. BMC Bioinform. 2015, 16, 164. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2015, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Woyke, T.; Huntemann, M.; et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2013, 42, D560–D567. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Blimkie, T.; Lee, A.H.-Y.; Hancock, R.E.W. MetaBridge: An Integrative Multi-Omics Tool for Metabolite-Enzyme Mapping. Curr. Protoc. Bioinform. 2020, 70, e98. [Google Scholar] [CrossRef]
- Burton, E.G.; Sakami, W. The formation of methionine from the monoglutamate form of methyltetrahydrofolate by higher plants. Biochem. Biophys. Res. Commun. 1969, 36, 228–234. [Google Scholar] [CrossRef]
- Guest, J.R.; Friedman, S.; Foster, M.A.; Tejerina, G.; Woods, D.D. Transfer of the methyl group from N5-methyltetrahydrofolates to homocysteine in Escherichia coli. Biochem. J. 1964, 92, 497–504. [Google Scholar] [CrossRef]
- Lin, H.; Lou, B.; Glynn, J.M.; Doddapaneni, H.; Civerolo, E.L.; Chen, C.; Duan, Y.; Zhou, L.; Vahling, C.M. The complete genome sequence of ‘Candidatus Liberibacter solanacearum’, the bacterium associated with potato zebra chip disease. PLoS ONE 2011, 6, e19135. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.M.; Johnson, C.P.; Lu, A.Y.; Frampton, R.A.; Sullivan, K.L.; Fiers, M.W.E.J.; Crowhurst, R.N.; Pitman, A.R.; Scott, I.A.W.; Wen, A.; et al. Genomes of ‘Candidatus Liberibacter solanacearum’ haplotype A from New Zealand and the United States suggest significant genome plasticity in the species. Phytopathology 2015, 105, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Haapalainen, M.; Schott, T.; Thompson, S.M.; Smith, G.R.; Nissinen, A.I.; Pirhonen, M. Genomic sequence of ‘Candidatus Liberibacter solanacearum’ haplotype C and its comparison with haplotype A and B genomes. PLoS ONE 2017, 12, e0171531. [Google Scholar] [CrossRef] [PubMed]
- Hartung, J.S.; Shao, J.; Kuykendall, L.D. Comparison of the ‘Ca. Liberibacter asiaticus’ genome adapted for an intracellular lifestyle with other members of the rhizobiales. PLoS ONE 2011, 6, e23289. [Google Scholar] [CrossRef]
- Bennett, G.M.; Moran, N.A. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 2015, 112, 10169–10176. [Google Scholar] [CrossRef]
- Fagen, J.R.; Leonard, M.T.; Coyle, J.F.; McCullough, C.M.; Davis-Richardson, A.G.; Davis, M.J.; Triplett, E.W. Liberibacter crescens gen. nov., sp. nov., the first cultured member of the genus Liberibacter. Int. J. Syst. Evol. Microbiol 2014, 64, 2461–2466. [Google Scholar] [CrossRef]
- Carr, R.; Borenstein, E. NetSeed: A network-based reverse-ecology tool for calculating the metabolic interface of an organism with its environment. Bioinformatics 2012, 28, 734–735. [Google Scholar] [CrossRef]
- Ankrah, N.Y.D.; Luan, J.; Douglas, A.E. Cooperative metabolism in a three-partner insect-bacterial symbiosis revealed by metabolic modeling. J. Bacteriol. 2017, 199, e00872-00816. [Google Scholar] [CrossRef]
- Luan, J.-B.; Chen, W.; Hasegawa, D.K.; Simmons, A.M.; Wintermantel, W.M.; Ling, K.-S.; Fei, Z.; Liu, S.-S.; Douglas, A.E. Metabolic Coevolution in the Bacterial Symbiosis of Whiteflies and Related Plant Sap-Feeding Insects. Genome Biol. Evol. 2015, 7, 2635–2647. [Google Scholar] [CrossRef]
- Santos-Garcia, D.; Juravel, K.; Freilich, S.; Zchori-Fein, E.; Latorre, A.; Moya, A.; Morin, S.; Silva, F.J. To B or Not to B: Comparative Genomics Suggests Arsenophonus as a Source of B Vitamins in Whiteflies. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Zchori-Fein, E.; Lahav, T.; Freilich, S. Variations in the identity and complexity of endosymbiont combinations in whitefly hosts. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Heinken, A.; Thiele, I. Systems biology of host–microbe metabolomics. WIREs Syst. Biol. Med. 2015, 7, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Charitou, T.; Bryan, K.; Lynn, D.J. Using biological networks to integrate, visualize and analyze genomics data. Genet. Sel. Evol. 2016, 48, 27. [Google Scholar] [CrossRef]
- Xu, X.; Zarecki, R.; Medina, S.; Ofaim, S.; Liu, X.; Chen, C.; Hu, S.; Brom, D.; Gat, D.; Porob, S.; et al. Modeling microbial communities from atrazine contaminated soils promotes the development of biostimulation solutions. ISME J. 2019, 13, 494–508. [Google Scholar] [CrossRef] [PubMed]
- Faust, K. Microbial consortium design benefits from metabolic modeling. Trends Biotechnol. 2019, 37, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Freilich, S.; Zarecki, R.; Eilam, O.; Segal, E.S.; Henry, C.S.; Kupiec, M.; Gophna, U.; Sharan, R.; Ruppin, E. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2011, 2, 589. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tal, O.; Selvaraj, G.; Medina, S.; Ofaim, S.; Freilich, S. NetMet: A Network-Based Tool for Predicting Metabolic Capacities of Microbial Species and their Interactions. Microorganisms 2020, 8, 840. https://doi.org/10.3390/microorganisms8060840
Tal O, Selvaraj G, Medina S, Ofaim S, Freilich S. NetMet: A Network-Based Tool for Predicting Metabolic Capacities of Microbial Species and their Interactions. Microorganisms. 2020; 8(6):840. https://doi.org/10.3390/microorganisms8060840
Chicago/Turabian StyleTal, Ofir, Gopinath Selvaraj, Shlomit Medina, Shany Ofaim, and Shiri Freilich. 2020. "NetMet: A Network-Based Tool for Predicting Metabolic Capacities of Microbial Species and their Interactions" Microorganisms 8, no. 6: 840. https://doi.org/10.3390/microorganisms8060840
APA StyleTal, O., Selvaraj, G., Medina, S., Ofaim, S., & Freilich, S. (2020). NetMet: A Network-Based Tool for Predicting Metabolic Capacities of Microbial Species and their Interactions. Microorganisms, 8(6), 840. https://doi.org/10.3390/microorganisms8060840





