Characterization of Indigenous Lactic Acid Bacteria in Cow Milk of the Maltese Islands: A Geographical and Seasonal Assessment
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Raw Milk Samples
2.2. Quantification and Isolation of Bacteria from Raw Cow’s Milk
2.3. DNA Extraction and Strain Differentiation
2.4. Species Identification
2.5. Statistical Analysis
3. Results
3.1. Microbial Population of Raw Maltese Cow Milk
3.2. Microbial Diversity of Raw Maltese Cow Milk
4. Discussion
4.1. Prevalence of Bacteria in Raw Cow’s Milk
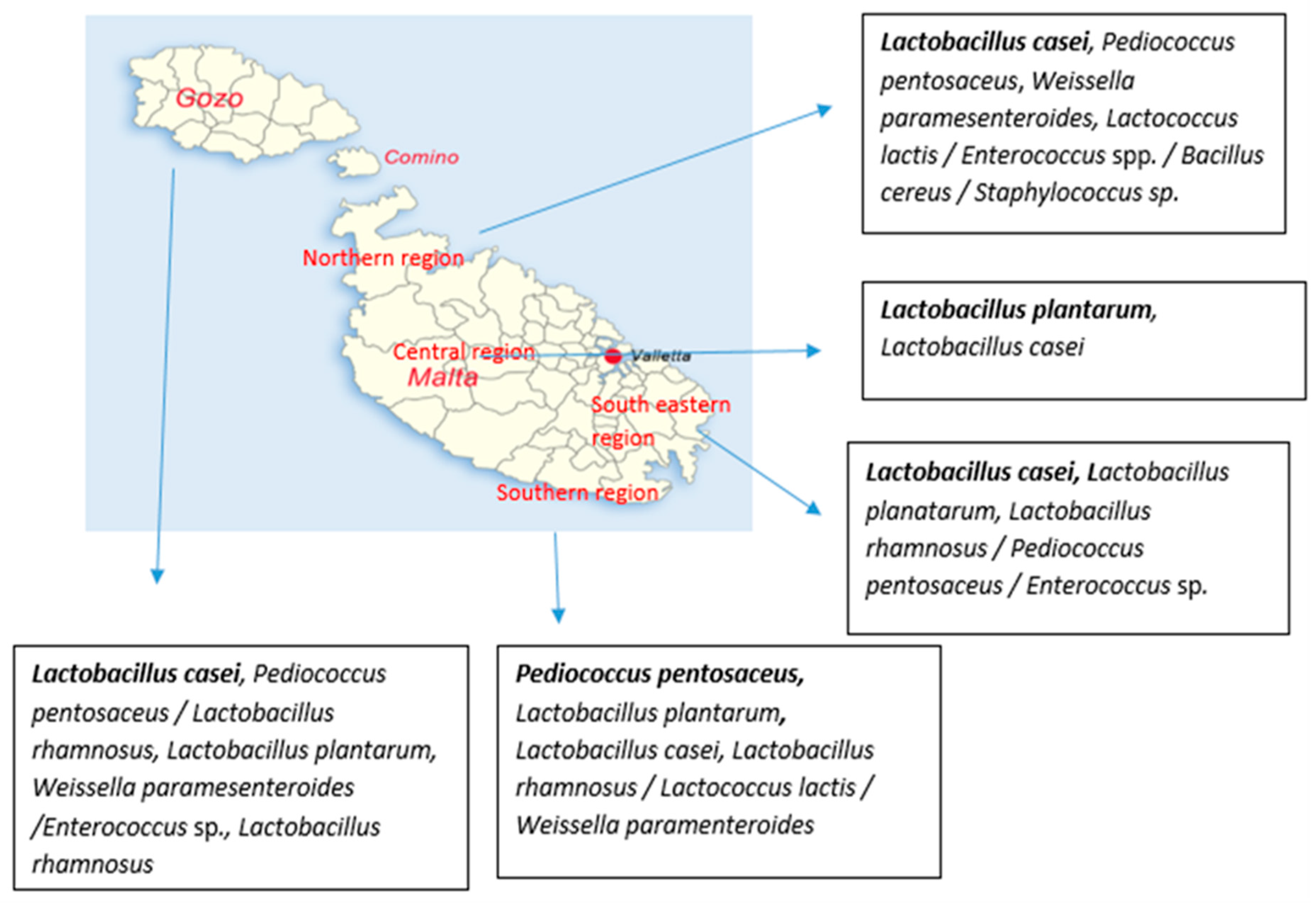
4.2. Seasonal and Geographical Prevalence of Bacteria in Raw Cow’s Milk
4.3. Effect of Seasonality and Geographical Area on Distribution of Different Species Isolated from Raw Cow’s Milk
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- László, V. Microbiological quality of commercial dairy products. In Communicating Current Research and Educational Topics and Trends in Applied Microbiology; Mendez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2007; pp. 487–494. ISBN 8461194225. [Google Scholar]
- Fernández García, L.; Riera Rodríguez, F.A. Combination of microfiltration and heat treatment for ESL milk production: Impact on shelf life. J. Food Eng. 2014, 128, 1–9. [Google Scholar] [CrossRef]
- Public Health Service/Food and Drug Administration. Grade “A” Pasteurized Milk Ordinance, Including Provisions from the Grade “A” Condensed and Dry Milk Products and Condensed and Dry Whey—Supplement I to the Grade “A” Pasteurized Milk Ordinance. 2017. Available online: https://www.fda.gov/media/114169/download (accessed on 30 March 2020).
- White, C.H. Rapid methods for estimation and prediction of shelf-life of milk and dairy products. J. Dairy Sci. 1993, 76, 2126–3132. [Google Scholar] [CrossRef]
- Marth, E.; Steele, J. Applied Dairy Microbiology, 2nd ed.; Marcel Dekker, Inc.: New York, NY, USA, 2001; ISBN 9780824705367. [Google Scholar]
- Spiteri, R.; Attard, E. Determination of major and minor elements in maltese sheep, goat and cow milk using microwave plasma-atomic emission spectrophotometry. J. Agric. Sci. 2017, 9, 43–50. [Google Scholar] [CrossRef][Green Version]
- Spiteri, R.; Attard, E. Determination of Physicochemical characteristics of maltese ovine, caprine and bovine milk. HSOA J. Dairy Res. Technol. 2019, 2. [Google Scholar] [CrossRef]
- Buehner, K.P.; Anand, S.; Garcia, A. Prevalence of thermoduric bacteria and spores on 10 Midwest dairy farms. J. Dairy Sci. 2014, 97, 6777–67784. [Google Scholar] [CrossRef] [PubMed]
- De Garnica, M.L.; Linage, B.; Carriedo, J.A.; De La Fuente, L.F.; García-Jimeno, M.C.; Santos, J.A.; Gonzalo, C. Relationship among specific bacterial counts and total bacterial and somatic cell counts and factors influencing their variation in ovine bulk tank milk. J. Dairy Sci. 2013, 96, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Carafa, I.; Navarro, I.C.; Bittante, G.; Tagliapietra, F.; Gallo, L.; Tuohy, K.; Franciosi, E. Shift in the cow milk microbiota during alpine pasture as analyzed by culture dependent and high-throughput sequencing techniques. Food Microbiol. 2020, 91, 103504. [Google Scholar] [CrossRef]
- Pidcock, K.; Heard, G.M.; Henriksson, A. Application of non-traditional meat starter cultures in production of Hungarian salami. J. Food Microbiol. 2002, 76, 75–81. [Google Scholar] [CrossRef]
- Pantoja, J.C.F.; Reinemann, D.J.; Ruegg, P.L. Associations among milk quality indicators in raw bulk milk. J. Dairy Sci. 2009, 92, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Salman, A.A.; Elnasri, H.A. Somatic cell count, total bacterial count and acidity properties of milk in Khartoum State, Sudan. J. Cell Anim. Biol. 2011, 5, 223–230. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Ercolini, D.; Villani, F.; Nychas, G.-J.E. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012, 157, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Kacem, M.; Zadi-Karam, H.; Karam, N. Identification of lactic acid bacteria isolated from milk and fermented olive oil in western Algeria. Rev. Maroc. Sci. Agron. Vét. 2004, 23, 135–141. [Google Scholar]
- Ercolini, D.; Russo, F.; Ferrocino, I.; Villani, F. Molecular identification of mesophilic and psychrotrophic bacteria from raw cow’s milk. Food Microbiol. 2009, 26, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Tormo, H.; Ali Haimoud Lekhal, D.; Roques, C. Phenotypic and genotypic characterization of lactic acid bacteria isolated from raw goat milk and effect of farming practices on the dominant species of lactic acid bacteria. J. Food Microbiol. 2015, 20, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Fguiri, I.; Ziadi, M.; Atigui, M.; Ayeb, N.; Arroum, S.; Assadi, M.; Khorchan, T. Isolation and characterisation of lactic acid bacteria strains from raw camel milk for potential use in the production of fermented Tunisian dairy products. Int. J. Dairy Technol. 2016, 69, 103–113. [Google Scholar] [CrossRef]
- Abdullah, S.A.; Osman, M.M. Isolation and identification of lactic acid bacteria from raw cow milk, white cheese and rob in Sudan. Pak. J. Nutr. 2010, 9, 50–65. [Google Scholar] [CrossRef]
- Mallet, A.; Guéguen, M.; Kauffmann, F.; Chesneau, C.; Sesboué, A.; Desmasures, N. Quantitative and qualitative microbial analysis of raw milk reveals substantial diversity influenced by herd management practices. Int. Dairy J. 2012, 27, 13–21. [Google Scholar] [CrossRef]
- Nalepa, B.; Olszewska, M.A.; Markiewicz, L.H. Seasonal variances in bacterial microbiota and volatile organic compounds in raw milk. Int. J. Food Microbiol. 2018, 267, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. Impacts of seasonal housing and teat preparation on raw milk microbiota: A high-throughput sequencing study. Appl. Environ. Microbiol. 2017, 83, e02694-16. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Amiranashvili, L.; Chitchyane, K.; Ghazanchyane, N.; Darbinyane, K.; Gagelidzed, N.; Sadunishvilid, T.; Goginyane, V.; Kvesitadzed, G.; Torokf, T.; et al. Microbial biogeography of the transnational fermented milk matsoni. Food Microbiol. 2015, 50, 12–19. [Google Scholar] [CrossRef] [PubMed]




| Winter Samples | Summer Samples | ||||
|---|---|---|---|---|---|
| Region | Farmer Code | Average TVC (log CFU/mL) | Average Total Counts of LAB (log CFU/mL) | Average TVC (log CFU/mL) | Average Total Counts of LAB (log CFU/mL) |
| Central | 8 | 5.48 ± 0.10 | 1.93 ± 0.19 | 7.17 ± 0.12 | 3.81 ± 0.03 |
| Gozo | 20 | 5.58 ± 0.07 | 2.49 ± 0.06 | 4.79 ± 0.03 | 4.06 ± 0.01 |
| 21 | 5.00 ± 0.03 | 1.71 ± 0.04 | 4.68 ± 0.02 | 4.00 ± 0.06 | |
| 22 | 4.69 ± 0.05 | 1.81 ± 0.02 | 4.69 ± 0.09 | 4.13 ± 0.01 | |
| 23 | 4.77 ± 0.12 | 1.77 ± 0.16 | 4.13 ± 0.07 | 3.29 ± 0.06 | |
| 24 | 4.98 ± 0.09 | 1.69 ± 0.04 | 4.62 ± 0.07 | 3.89 ± 0.09 | |
| 25 | 5.42 ± 0.02 | 1.50 ± 0.05 | 4.70 ± 0.05 | 3.99 ± 0.03 | |
| Northern | 15 | 6.42 ± 0.02 | 0.89 ± 0.05 | 4.08 ± 0.01 | 2.59 ± 0.11 |
| 16 | 5.13 ± 0.25 | 2.77 ± 0.01 | 4.53 ± 0.01 | 3.71 ± 0.01 | |
| 17 | 4.63 ± 0.21 | 1.38 ± 0.01 | 4.26 ± 0.04 | 3.75 ± 0.15 | |
| South-Eastern | 2 | 5.35 ± 0.05 | 1.70 ± 0.05 | 4.89 ± 0.31 | 4.02 ± 0.02 |
| 3 | 5.73 ± 0.02 | 1.90 ± 0.11 | 5.02 ± 0.24 | 3.61 ± 0.04 | |
| 4 | 5.26 ± 0.09 | 2.73 ± 0.19 | 5.95 ± 0.23 | 4.33 ± 0.03 | |
| 6 | 4.40 ± 0.02 | 2.13 ± 0.01 | 5.12 ± 0.12 | 3.82 ± 0.02 | |
| 7 | 3.80 ± 0.01 | 0.88 ± 0.19 | 5.28 ± 0.01 | 3.75 ± 0.11 | |
| 10 | 4.34 ± 0.02 | 2.42 ± 0.21 | 3.98 ± 0.02 | 3.06 ± 0.01 | |
| 12 | 6.02 ± 0.04 | 2.02 ± 0.05 | 5.42 ± 0.01 | 3.50 ± 0.07 | |
| 14 | 5.03 ± 0.09 | 2.75 ± 0.24 | 4.22 ± 0.04 | 2.86 ± 0.01 | |
| Southern | 1 | 5.25 ± 0.07 | 1.58 ± 0.05 | 5.52 ± 0.28 | 3.82 ± 0.07 |
| 5 | 4.17 ± 0.08 | 1.33 ± 0.01 | 5.28 ± 0.05 | 3.29 ± 0.02 | |
| 9 | 4.43 ± 0.02 | 1.13 ± 0.18 | 4.70 ± 0.04 | 3.87 ± 0.01 | |
| 11 | 4.37 ± 0.01 | 1.27 ± 0.04 | 5.14 ± 0.02 | 3.29 ± 0.17 | |
| 13 | 3.21 ± 0.17 | 0.75 ± 0.15 | 3.90 ± 0.02 | 3.61 ± 0.01 | |
| 18 | 4.43 ± 0.49 | 1.13 ± 0.83 | 4.33 ± 0.07 | 3.46 ± 0.03 | |
| 19 | 6.21 ± 0.12 | 3.19 ± 0.14 | 4.56 ± 0.11 | 3.69 ± 0.08 | |
| Group | Species | Strain | Similar Pattern | Winter | Summer |
|---|---|---|---|---|---|
| I | Enterococcus sp. | MMS 10.4 | 11 | 2 | 9 |
| II | Enterococcus casseliflavus | MMS 22.2 | 7 | 0 | 7 |
| III | Enterococcus faecalis | MMS 8.5 | 19 | 0 | 19 |
| IV | Lactococcus lactis | MMS 2.9 | 16 | 0 | 16 |
| V | Lactobacillus rhamnosus | MM 19.1 | 14 | 12 | 2 |
| VI | Lactococcus garvieae | MMS 17.4 | 2 | 1 | 1 |
| VII | Lactobacillus plantarum | MM 12.8 | 11 | 11 | 0 |
| VIII | Enterococcus sp. | MMS 15.2 | 7 | 5 | 2 |
| IX | Lactobacillus plantarum | MM 8.2 | 12 | 12 | 0 |
| X | Lactobacillus casei | MM 12.11 | 12 | 12 | 0 |
| XI | Lactobacillus casei | MM 9.1 | 10 | 10 | 0 |
| XII | Lactobacillus casei | MM 20.13 | 8 | 8 | 0 |
| XIII | Lactobacillus casei | MM 23.6 | 11 | 11 | 0 |
| XIV | Weissella paramesenteroides | MM 27.2 | 22 | 7 | 15 |
| XV | Pediococcus pentosaceus | MM 23.2 | 10 | 10 | 0 |
| XVI | Pediococcus pentosaceus | MM 22.4 | 8 | 4 | 4 |
| XVII | Enterococcus sp. | MMS 10.1 | 7 | 0 | 7 |
| XVIII | Lactobacillus casei | MM 10.2 | 4 | 2 | 2 |
| XIX | Lactobacillus plantarum | MM 20.7 | 17 | 3 | 14 |
| XX | Enterococcus sp. | MMS 9.4 | 6 | 0 | 6 |
| XXI | Lactococcus lactis | MMS 16.2 | 3 | 1 | 2 |
| XXII | Lactococcus lactis | MMS 14.5 | 16 | 1 | 15 |
| XXIII | Pediococcus pentosaceus | MM 18.5 | 3 | 3 | 0 |
| XXIV | Pediococcus pentosaceus | MM 26.1 | 4 | 2 | 2 |
| XXV | Pediococcus pentosaceus | MM 19.5 | 10 | 8 | 2 |
| XXVI | Lactobacillus casei | MM 14.5 | 4 | 4 | 0 |
| XXVII | Weissella paramesenteroides | MM 27.1 | 2 | 2 | 0 |
| XXVIII | Enterococcus malodoratus | MMS 22.4 | 2 | 0 | 2 |
| XXIX | Streptococcus suis | MMS 19.1 | 1 | 0 | 1 |
| XXX | Bacillus cereus | MMS 1.4 | 1 | 0 | 1 |
| XXXI | Staphylococcus sp. | MM 16.9 | 1 | 1 | 0 |
| XXXII | Bacillus cereus | MM 16.2 | 1 | 1 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garroni, E.; Doulgeraki, A.I.; Pavli, F.; Spiteri, D.; Valdramidis, V.P. Characterization of Indigenous Lactic Acid Bacteria in Cow Milk of the Maltese Islands: A Geographical and Seasonal Assessment. Microorganisms 2020, 8, 812. https://doi.org/10.3390/microorganisms8060812
Garroni E, Doulgeraki AI, Pavli F, Spiteri D, Valdramidis VP. Characterization of Indigenous Lactic Acid Bacteria in Cow Milk of the Maltese Islands: A Geographical and Seasonal Assessment. Microorganisms. 2020; 8(6):812. https://doi.org/10.3390/microorganisms8060812
Chicago/Turabian StyleGarroni, Elisa, Agapi I. Doulgeraki, Foteini Pavli, David Spiteri, and Vasilis P. Valdramidis. 2020. "Characterization of Indigenous Lactic Acid Bacteria in Cow Milk of the Maltese Islands: A Geographical and Seasonal Assessment" Microorganisms 8, no. 6: 812. https://doi.org/10.3390/microorganisms8060812
APA StyleGarroni, E., Doulgeraki, A. I., Pavli, F., Spiteri, D., & Valdramidis, V. P. (2020). Characterization of Indigenous Lactic Acid Bacteria in Cow Milk of the Maltese Islands: A Geographical and Seasonal Assessment. Microorganisms, 8(6), 812. https://doi.org/10.3390/microorganisms8060812






