Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture
2.2. Assessment of Carbohydrate Utilization and Bacterial Growth
2.2.1. pH Measurements and Viable Cell Counts
2.2.2. SCFA Determinations
2.3. Cell Culture and Determination of Immunomodulatory Properties of Culture Supernatants
2.4. Animals and Experimental Design
2.4.1. Sample Preparation for Administration
2.4.2. DNA Extraction and Real-Time qPCR Amplification
2.4.3. 16S rRNA Gene Amplification and Sequence Analysis
2.4.4. Histological Analysis and RT-qPCR of the Colon Tissue
2.5. Statistical Analyses
3. Results
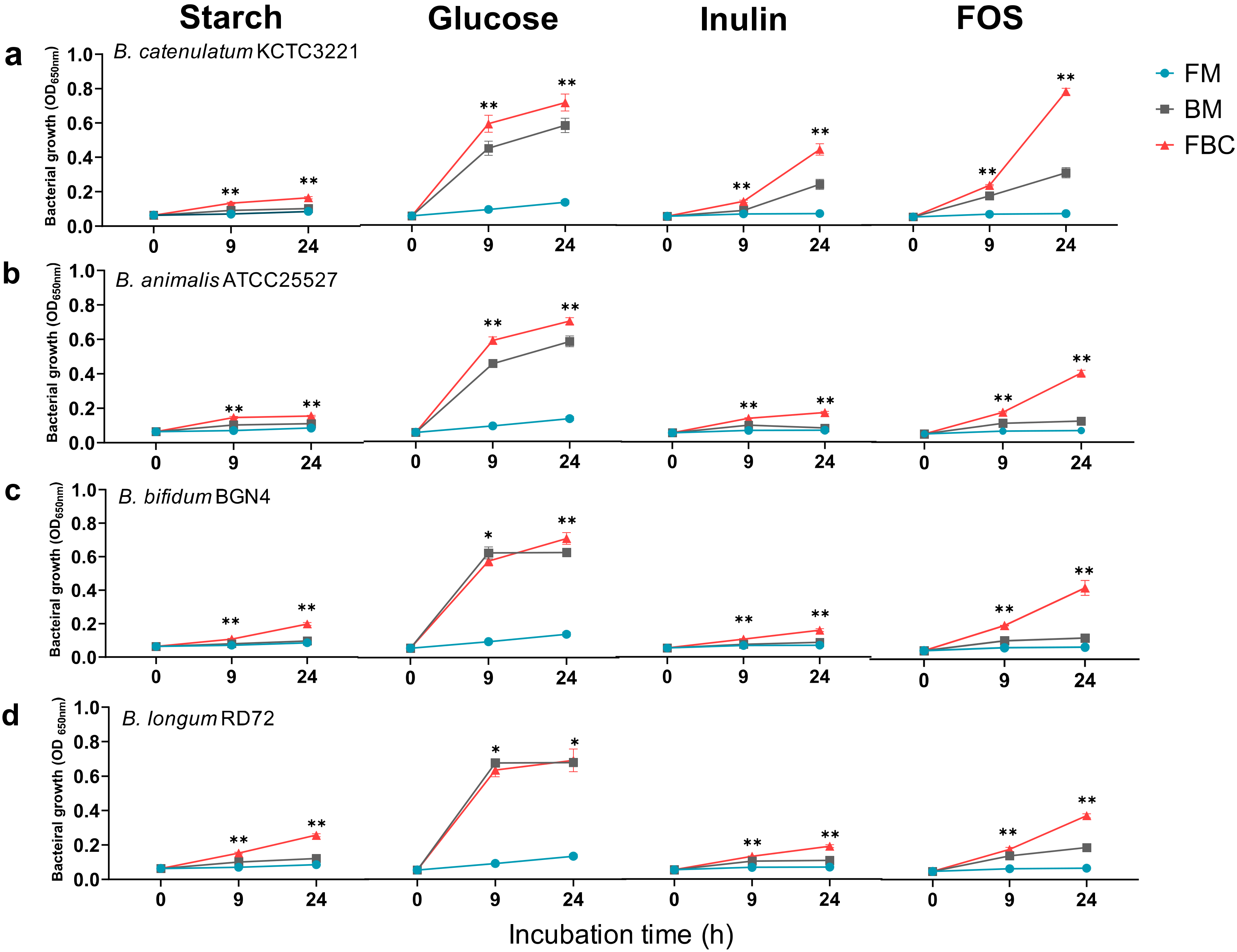
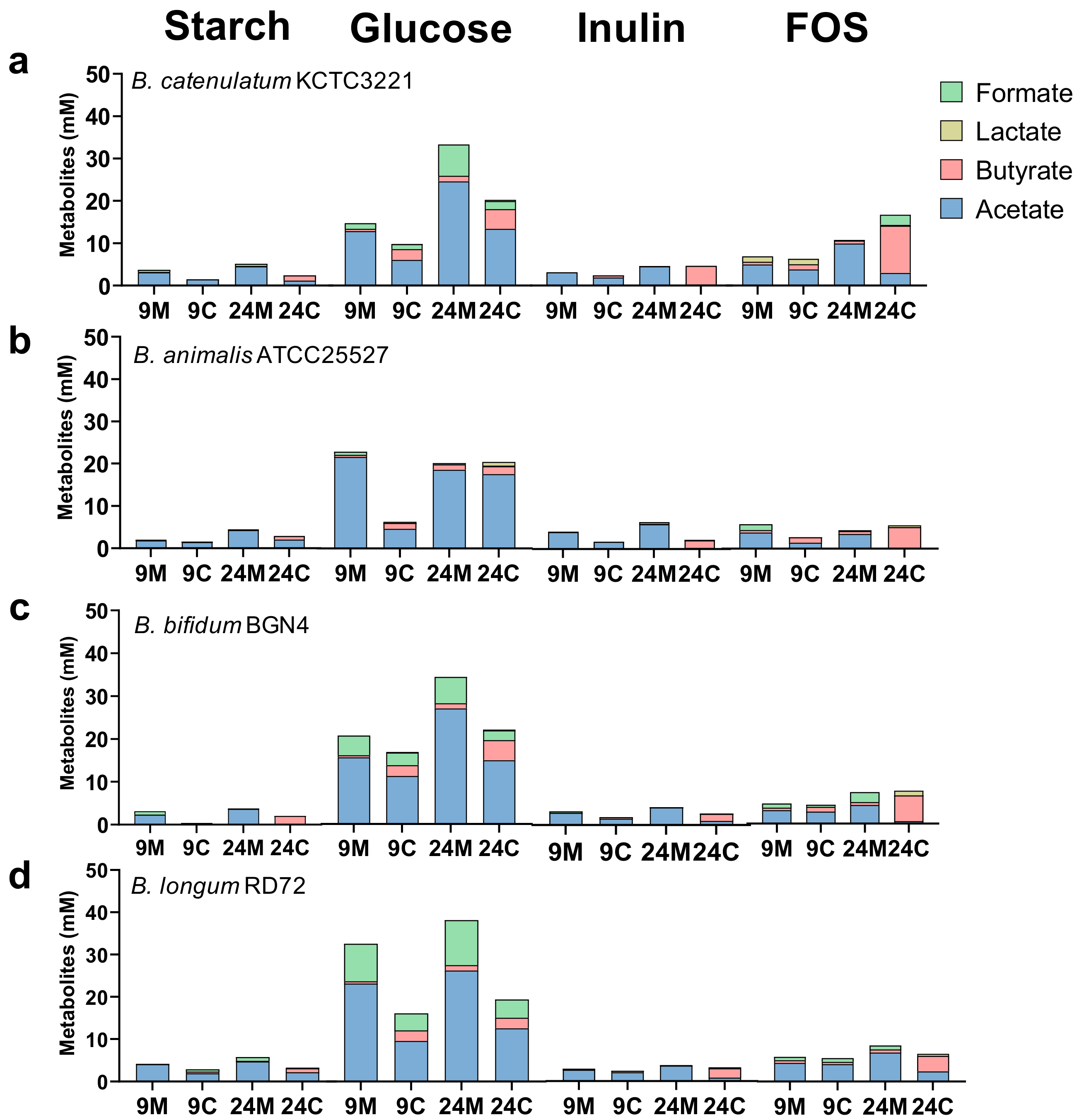
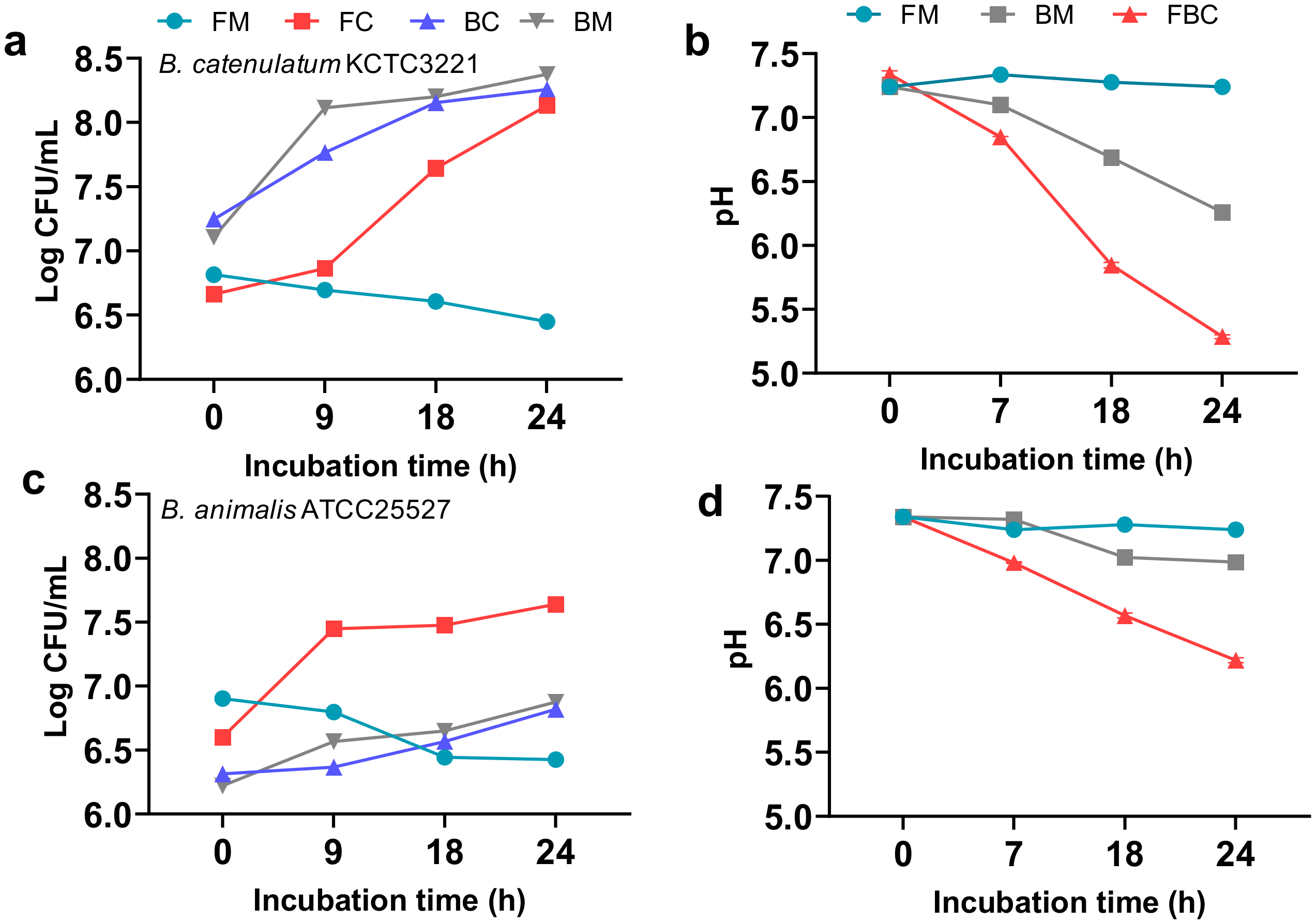
3.1. Effects of Mono- and Co-Culture, and Different Carbon Sources on Bacterial Growth, SCFA Production, Culture pH, and Viable Cell Counts
3.1.1. Growth of Bacteria in Mono- and Co-Cultures in the Presence of Different Carbon Sources
3.1.2. SCFA Production by Bacteria in Mono- and Co-Culture in the Presence of Different Carbon Sources
3.1.3. Effect of F. prausnitzii ATCC 27768 Co-Culture with B. catenulatum KCTC 3221 or B. animalis ATCC 25527 in a Medium Containing FOS on Culture pH and Viable Cell Counts
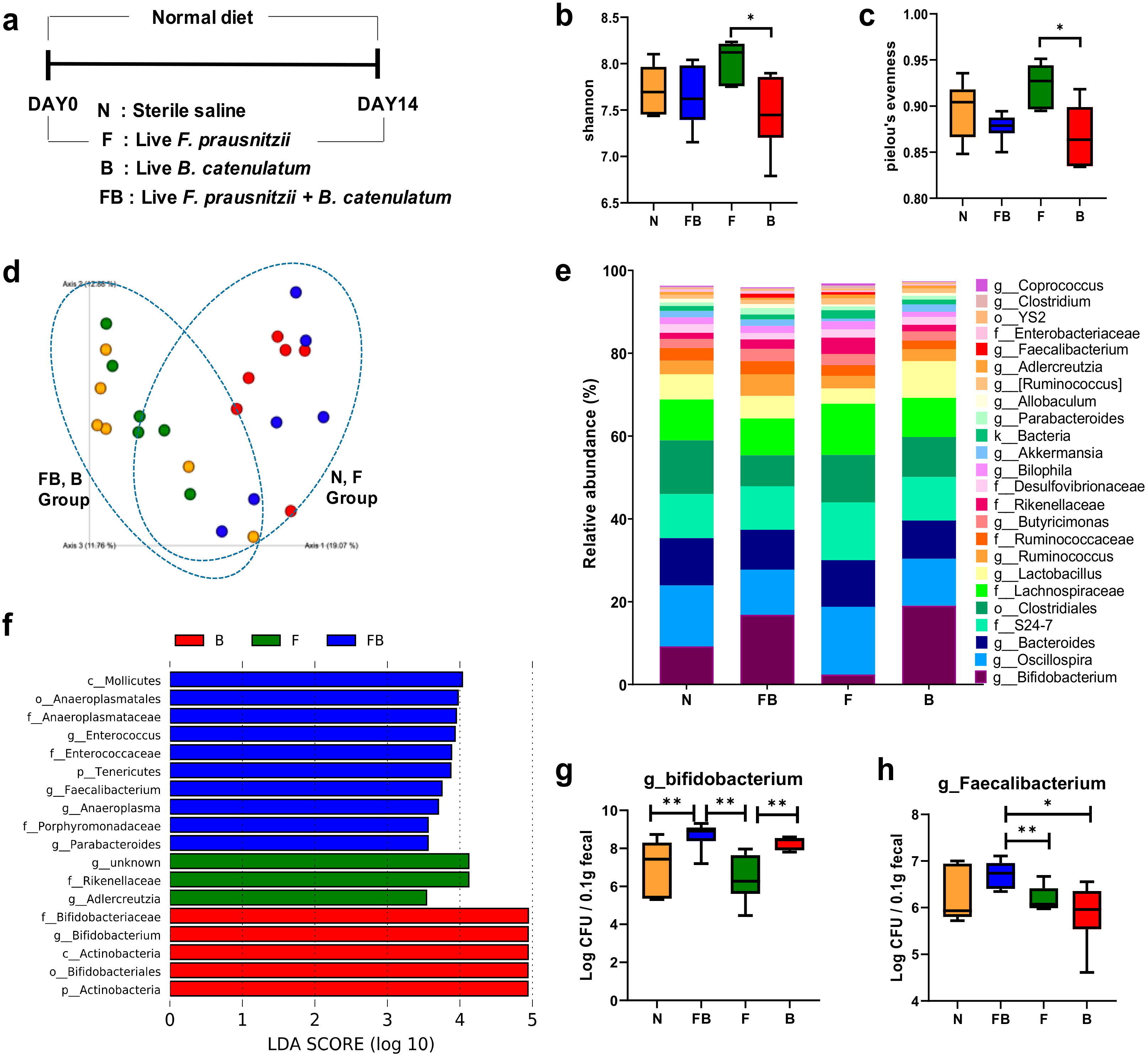
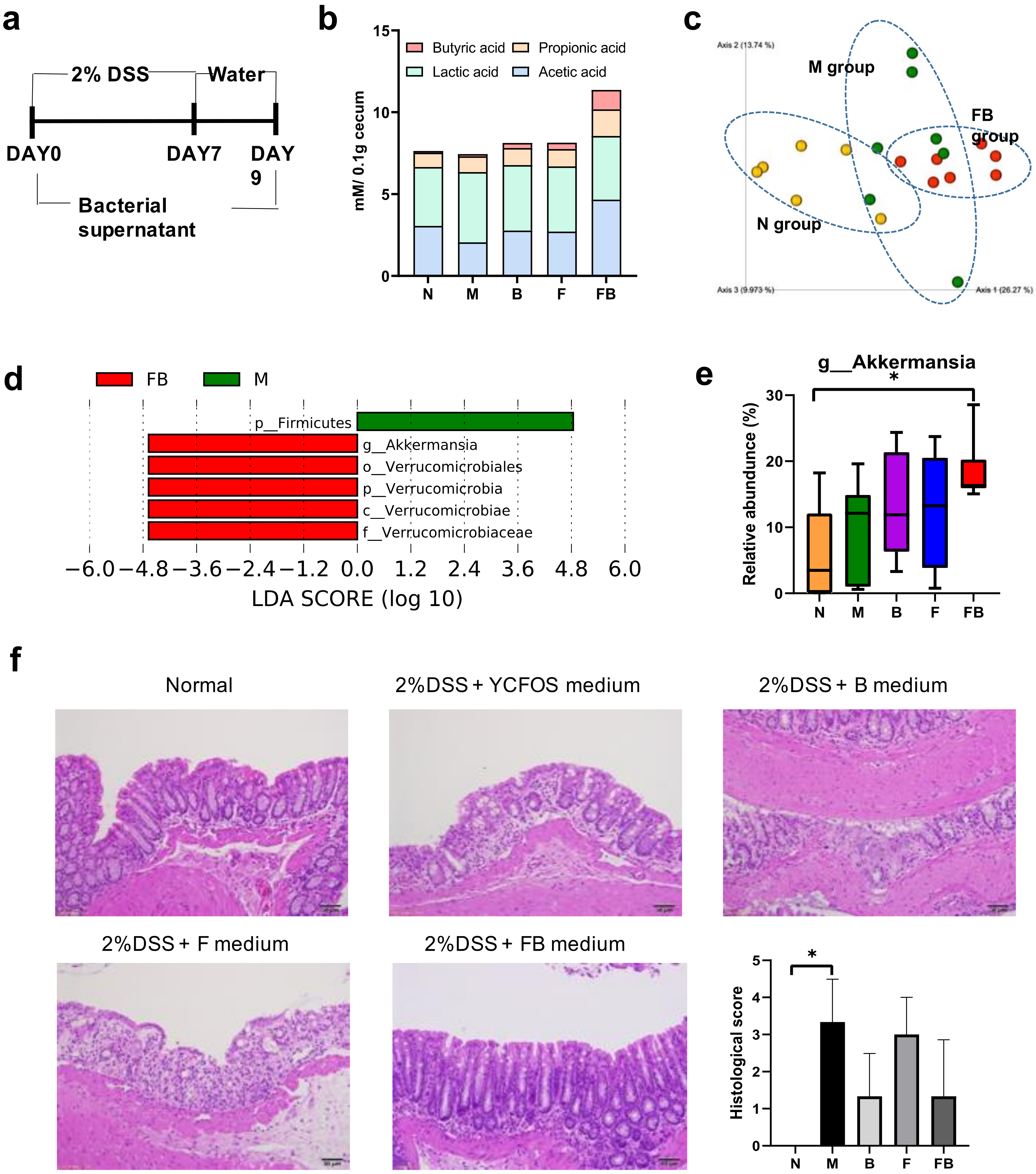
3.2. Comparison of the Fecal Microbiota of Normal Mice Fed F. prausnitzii ATCC 27768 and B. catenulatum KCTC 3221
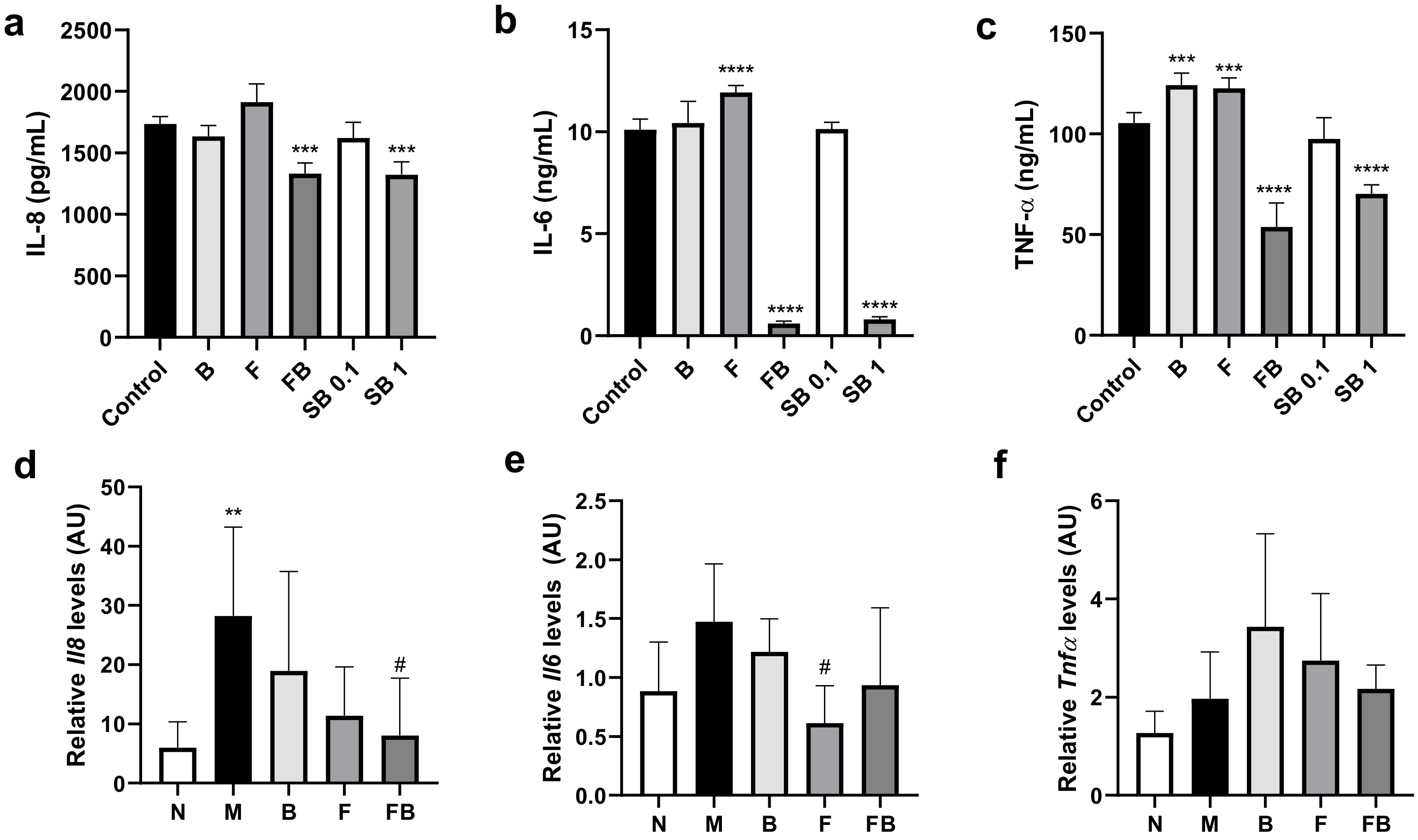
3.3. Effect of Bacterial Culture Supernatant on Proinflammatory Cytokine Production by HT-29 Cells and RAW 264.7 Cells, and in the Colon in the DSS-Induced Colitis Mouse Model
3.4. Effect of Co-Culture Bacterial Supernatant on DSS-Induced Colitis in Mouse
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kaplan, G.G.; Ng, S.C. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology 2017, 152, 313–321. [Google Scholar] [CrossRef] [PubMed]
- M’koma, A.E. Inflammatory bowel disease: An expanding global health problem. Clinical Medicine Insights: Gastroenterology 2013, 6, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Pithadia, A.B.; Jain, S. Treatment of inflammatory bowel disease (IBD). Pharmacol. Rep. 2011, 63, 629–642. [Google Scholar] [CrossRef]
- Rosen, M.J.; Dhawan, A.; Saeed, S.A. Inflammatory bowel disease in children and adolescents. JAMA Pediatr. 2015, 169, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Fakhoury, M.; Negrulj, R.; Mooranian, A.; Al-Salami, H. Inflammatory bowel disease: Clinical aspects and treatments. J. Inflamm. Res. 2014, 7, 113. [Google Scholar] [CrossRef]
- Martín, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M. Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: A step forward in the use of F. prausnitzii as a next-generation probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar]
- Martín, R.; Bermúdez-Humarán, L.G.; Langella, P. Searching for the bacterial effector: The example of the multi-skilled commensal bacterium Faecalibacterium prausnitzii. Front. Microbiol. 2018, 9, 346. [Google Scholar]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Carlsson, A.H.; Yakymenko, O.; Olivier, I.; Håkansson, F.; Postma, E.; Keita, Å.V.; Söderholm, J.D. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand. J. Gastroenterol. 2013, 48, 1136–1144. [Google Scholar] [CrossRef]
- Kawade, Y.; Sakai, M.; Okamori, M.; Morita, M.; Mizushima, K.; Ueda, T.; Takagi, T.; Naito, Y.; Itoh, Y.; Shimada, T. Administration of live, but not inactivated, Faecalibacterium prausnitzii has a preventive effect on dextran sodium sulfate-induced colitis in mice. Mol. Med. Rep. 2019, 20, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Miquel, S.; Chain, F.; Natividad, J.M.; Jury, J.; Lu, J.; Sokol, H.; Theodorou, V.; Bercik, P.; Verdu, E.F. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Rossi, O.; Khan, M.T.; Schwarzer, M.; Hudcovic, T.; Srutkova, D.; Duncan, S.H.; Stolte, E.H.; Kozakova, H.; Flint, H.J.; Samsom, J.N. Faecalibacterium prausnitzii strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. PLoS ONE 2015, 10, e0123013. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Venegas, D.P.; Marjorie, K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10. [Google Scholar]
- Zhang, M.; Zhou, L.; Wang, Y.; Dorfman, R.G.; Tang, D.; Xu, L.; Pan, Y.; Zhou, Q.; Li, Y.; Yin, Y. Faecalibacterium prausnitzii produces butyrate to decrease c-myc-related metabolism and th17 differentiation by inhibiting histone deacetylase 3. Int. Immunol. 2019, 31, 499–514. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.; Brummer, R.J. The role of butyrate on colonic function. Aliment. Pharm. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Gibson, G.R. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. In Gastrointestinal Microbiology; Springer: Boston, MA, USA; pp. 269–318.
- Wolin, M.J.; Miller, T.L.; Yerry, S.; Zhang, Y.; Bank, S.; Weaver, G.A. Changes of fermentation pathways of fecal microbial communities associated with a drug treatment that increases dietary starch in the human colon. Appl. Environ. Microbiol. 1999, 65, 2807–2812. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef]
- Duncan, S.H.; Holtrop, G.; Lobley, G.E.; Calder, A.G.; Stewart, C.S.; Flint, H.J. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 2004, 91, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Kurakawa, T.; Tsuji, H.; Takahashi, T.; Kawashima, K.; Nagata, S.; Nomoto, K.; Yamashiro, Y. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: A quantitative assessment. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Barcenilla, A.; Stewart, C.S.; Pryde, S.E.; Flint, H.J. Acetate utilization and butyryl coenzyme A (CoA): Acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 2002, 68, 5186–5190. [Google Scholar] [CrossRef] [PubMed]
- Rios-Covian, D.; Gueimonde, M.; Duncan, S.H.; Flint, H.J.; de los Reyes-Gavilan, C.G. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Foditsch, C.; Santos, T.M.; Teixeira, A.G.; Pereira, R.V.; Dias, J.M.; Gaeta, N.; Bicalho, R.C. Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS ONE 2014, 9, e116465. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Duncan, S.H.; Stams, A.J.; van Dijl, J.M.; Flint, H.J.; Harmsen, H.J. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012, 6, 1578–1585. [Google Scholar] [CrossRef]
- Miquel, S.; Martín, R.; Lashermes, A.; Gillet, M.; Meleine, M.; Gelot, A.; Eschalier, A.; Ardid, D.; Bermúdez-Humarán, L.; Sokol, H. Anti-nociceptive effect of Faecalibacterium prausnitzii in non-inflammatory IBS-like models. Sci. Rep. 2016, 6, 19399. [Google Scholar] [CrossRef]
- Kang, S.; You, H.J.; Lee, Y.-G.; Jeong, Y.; Johnston, T.V.; Baek, N.-I.; Ku, S.; Ji, G.E. Production, Structural Characterization, and In Vitro Assessment of the Prebiotic Potential of Butyl-Fructooligosaccharides. Int. J. Mol. Sci. 2020, 21, 445. [Google Scholar] [CrossRef]
- Brown, K.; Abbott, D.W.; Uwiera, R.R.; Inglis, G.D. Removal of the cecum affects intestinal fermentation, enteric bacterial community structure, and acute colitis in mice. Gut Microbes 2018, 9, 218–235. [Google Scholar] [CrossRef]
- Kang, S.; Denman, S.E.; Morrison, M.; Yu, Z.; Dore, J.; Leclerc, M.; McSweeney, C.S. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel Dis. 2010, 16, 2034–2042. [Google Scholar] [CrossRef]
- Fijan, S.; Šulc, D.; Steyer, A. Study of the in vitro antagonistic activity of various single-strain and multi-strain probiotics against Escherichia coli. Int. J. Env. Res. Public Health 2018, 15, 1539. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-K.; Lee, S.H.; Lee, S.-Y.; Kim, E.-K.; Kwon, J.-E.; Seo, H.-B.; Lee, H.H.; Lee, B.-I.; Park, S.-H.; Cho, M.-L. Grim19 attenuates DSS induced colitis in an animal model. PLoS ONE 2016, 11, e0155853. [Google Scholar] [CrossRef] [PubMed]
- Fachi, J.L.; Felipe, J.D.; Pral, L.P.; da Silva, B.K.; Corrêa, R.O.; de Andrade, M.C.P.; da Fonseca, D.M.; Basso, P.J.; Câmara, N.O.S.; Souza, É.L.D.S.E. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep. 2019, 27, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Oh, S.-H.; Jang, H.W.; Kwon, J.-H.; Lee, K.J.; Kim, C.H.; Park, S.J.; Hong, S.P.; Cheon, J.H.; Kim, T.I. Long-term effects of bone marrow-derived mesenchymal stem cells in dextran sulfate sodium-induced murine chronic colitis. Gut Liver 2016, 10, 412. [Google Scholar] [CrossRef]
- Bhatt, B.; Zeng, P.; Zhu, H.; Sivaprakasam, S.; Li, S.; Xiao, H.; Dong, L.; Shiao, P.; Kolhe, R.; Patel, N. Gpr109a limits microbiota-induced IL-23 production to constrain ILC3-mediated colonic inflammation. J. Immunol. 2018, 200, 2905–2914. [Google Scholar] [CrossRef]
- Smith, N.W.; Shorten, P.R.; Altermann, E.; Roy, N.C.; McNabb, W.C. The classification and evolution of bacterial cross-feeding. Front. Ecol. Evol. 2019, 7, 153. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; de Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Moens, F.; Rivière, A.; Selak, M.; de Vuyst, L. Inulin-type fructan degradation capacity of interesting butyrate-producing colon bacteria and cross-feeding interactions of Faecalibacterium prausnitzii DSM 17677 T with bifidobacteria. Arch. Public Health 2014, 72, O6. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Tanaka, R.; Fukuda, M.; Oyaizu, H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ.Microbol. 1999, 65, 4506–4512. [Google Scholar] [CrossRef]
- Fang, D.; Shi, D.; Lv, L.; Gu, S.; Wu, W.; Chen, Y.; Guo, J.; Li, A.; Hu, X.; Guo, F. Bifidobacterium pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10 attenuate D-galactosamine-induced liver injury by modifying the gut microbiota. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Xing, H.C.; Li, L.J.; Xu, K.J.; Shen, T.; Chen, Y.B.; Sheng, J.F.; Chen, Y.; Fu, S.Z.; Chen, C.L.; Wang, J.G. Protective role of supplement with foreign Bifidobacterium and Lactobacillus in experimental hepatic ischemia-reperfusion injury. J. Gastroenterol. Hepatol. 2006, 21, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.P.; Martin, J.C.; Duncan, S.H.; Flint, H.J. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, In Vitro. FEMS Microbiol. Lett. 2014, 87, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Foditsch, C.; Pereira, R.V.V.; Ganda, E.K.; Gomez, M.S.; Marques, E.C.; Santin, T.; Bicalho, R.C. Oral administration of Faecalibacterium prausnitzii decreased the incidence of severe diarrhea and related mortality rate and increased weight gain in preweaned dairy heifers. PLoS ONE 2015, 10, e0145485. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-C.; Liao, C.-C.; Hsu, M.-J.; Liao, Y.-T.; Lin, C.-C.; Sheu, J.-R.; Lin, C.-H. Peptidoglycan-induced IL-6 production in RAW 264.7 macrophages is mediated by cyclooxygenase-2, PGE2/PGE4 receptors, protein kinase A, IκB kinase, and NF-κB. J. Immunol. 2006, 177, 681–693. [Google Scholar] [CrossRef]
- Breyner, N.M.; Michon, C.; de Sousa, C.S.; Boas, P.B.V.; Chain, F.; Azevedo, V.A.; Langella, P.; Chatel, J.M. Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-κB pathway. Front. Microbiol. 2017, 8, 114. [Google Scholar] [CrossRef]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef]
- Huang, X.-L.; Zhang, X.; Fei, X.-Y.; Chen, Z.-G.; Hao, Y.-P.; Zhang, S.; Zhang, M.-M.; Yu, Y.-Q.; Yu, C.-G. Faecalibacterium prausnitzii supernatant ameliorates dextran sulfate sodium induced colitis by regulating Th17 cell differentiation. World J. Gastroenterol. 2016, 22, 5201. [Google Scholar] [CrossRef]
- Zhang, M.; Qiu, X.; Zhang, H.; Yang, X.; Hong, N.; Yang, Y.; Chen, H.; Yu, C. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS ONE 2014, 9, e109146. [Google Scholar] [CrossRef]
- Alrafas, H.R.D.; Busbee, B.P.; Nagarkatti, P.; Nagarkatti, M. Effect of Sodium Butyrate Supplementation on the Gut Microbiome during Colorectal Cancer. J. Immunol. 2019, 202 (Suppl. 1), 191.10. [Google Scholar]
- Roshanravan, N.; Mahdavi, R.; Jafarabadi, M.A.; Alizadeh, E.; Ghavami, A.; Saadat, Y.R.; Alamdari, N.M.; Dastouri, M.R.; Alipour, S.; Ostadrahimi, A. The effects of sodium butyrate and high-performance inulin supplementation on the promotion of gut bacterium Akkermansia muciniphila growth and alterations in miR-375 and KLF5 expression in type 2 diabetic patients: A randomized, double-blind, placebo-controlled trial. Eur. J. Integr. Med. 2018, 18, 1–7. [Google Scholar]
- Lopez-Siles, M.; Enrich-Capó, N.; Aldeguer, X.; Sabat-Mir, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Alterations in the abundance and co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the colonic mucosa of inflammatory bowel disease subjects. Front. Cell. Infect. Microbiol. 2018, 8, 281. [Google Scholar] [CrossRef] [PubMed]
- Lukovac, S.; Belzer, C.; Pellis, L.; Keijser, B.J.; de Vos, W.M.; Montijn, R.C.; Roeselers, G. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 2014, 5, e01438-14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, X.; Cao, S.; Cromie, M.; Shen, Y.; Feng, Y.; Yang, H.; Li, L. Chlorogenic acid ameliorates experimental colitis by promoting growth of Akkermansia in mice. Nutrients 2017, 9, 677. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Jeong, Y.; Kang, S.; You, H.J.; Ji, G.E. Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies. Microorganisms 2020, 8, 788. https://doi.org/10.3390/microorganisms8050788
Kim H, Jeong Y, Kang S, You HJ, Ji GE. Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies. Microorganisms. 2020; 8(5):788. https://doi.org/10.3390/microorganisms8050788
Chicago/Turabian StyleKim, Heejung, Yunju Jeong, Sini Kang, Hyun Ju You, and Geun Eog Ji. 2020. "Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies" Microorganisms 8, no. 5: 788. https://doi.org/10.3390/microorganisms8050788
APA StyleKim, H., Jeong, Y., Kang, S., You, H. J., & Ji, G. E. (2020). Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies. Microorganisms, 8(5), 788. https://doi.org/10.3390/microorganisms8050788





