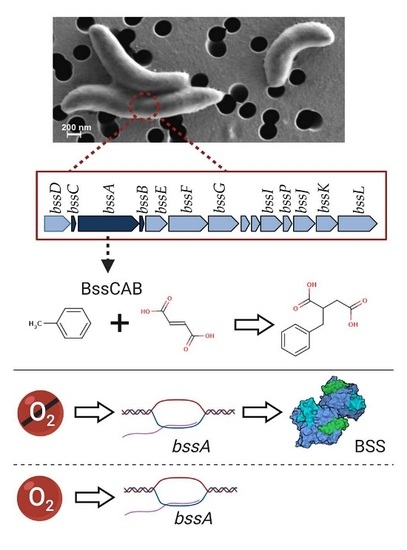
Benzylsuccinate Synthase is Post-Transcriptionally Regulated in the Toluene-Degrading Denitrifier Magnetospirillum sp. Strain 15-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Medium and Cultivation
2.2. RNA Extraction and cDNA Synthesis
2.3. Transcriptional Organization of bss, tdi, and bbs Gene Clusters by Reverse-Transcriptase PCR (RT-PCR)
2.4. Gene Expression Assessment by Quantitative RT- PCR (qRT-PCR)
2.5. Protein Extraction and Preparation
2.6. LC-MS/MS and Proteome Analysis
2.7. Data Availability
3. Results
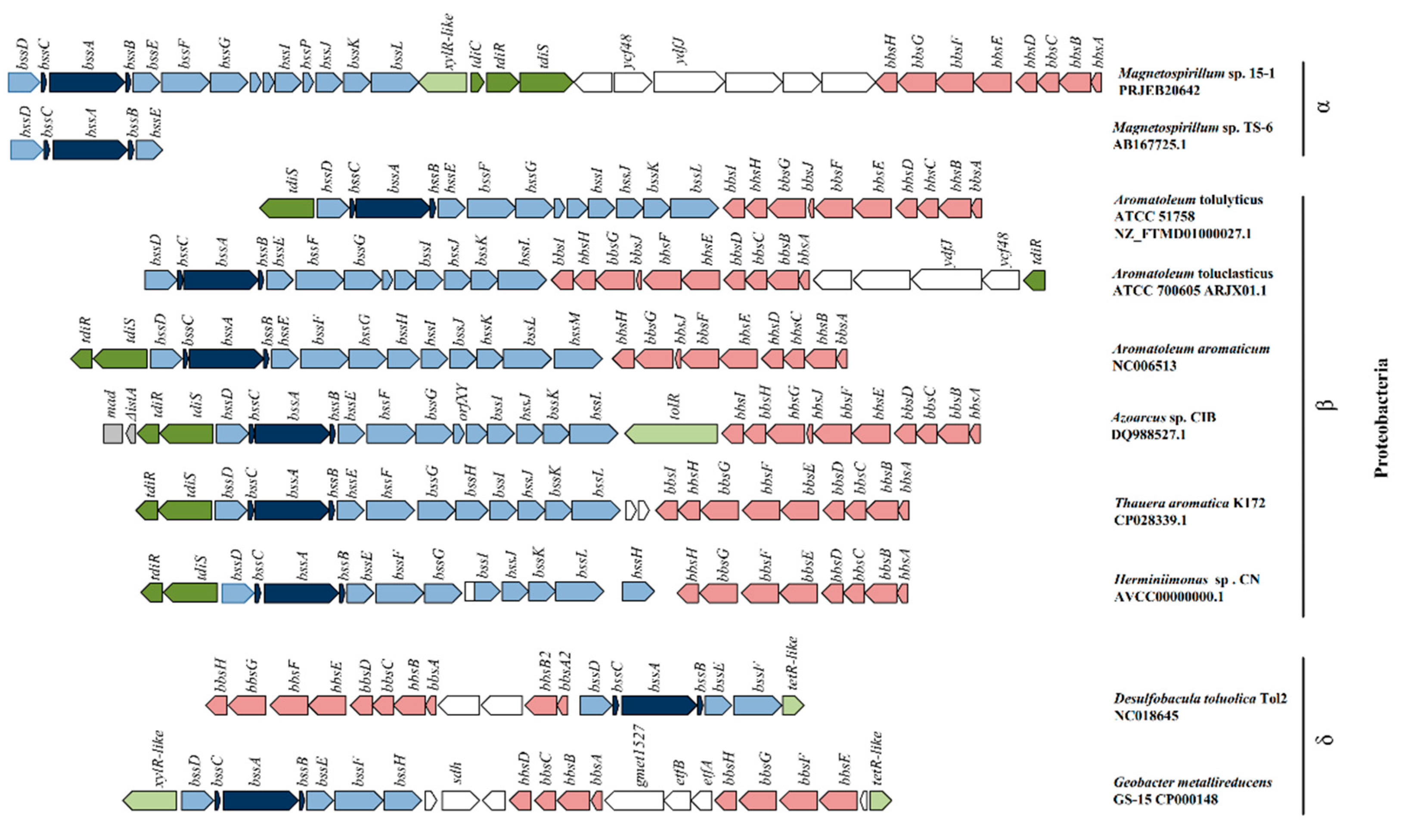
3.1. Genomic Organization of Genes Involved in Anaerobic Toluene Transformation
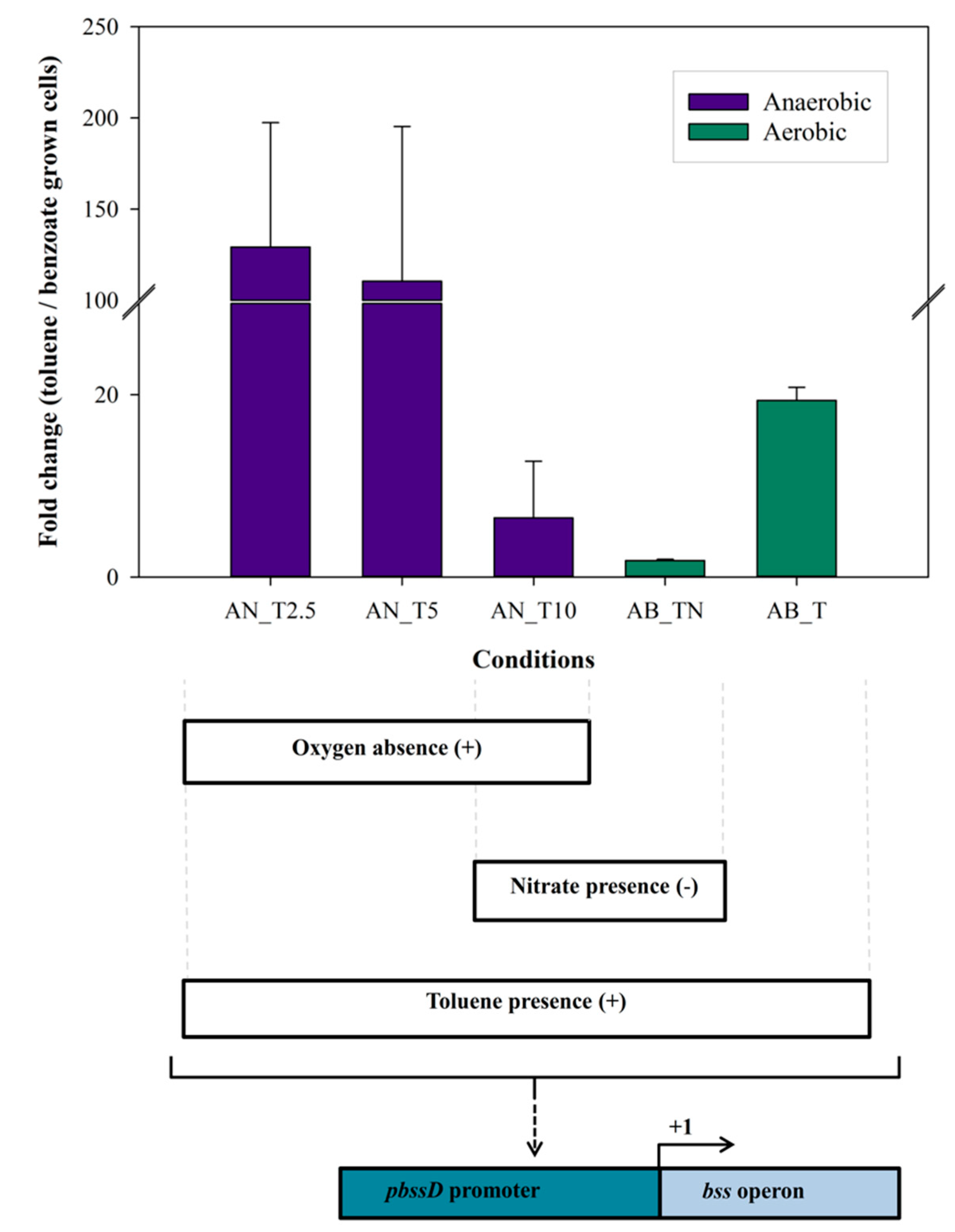
3.2. Regulation of bss Gene Expression in Magnetospirillum sp. Strain 15-1
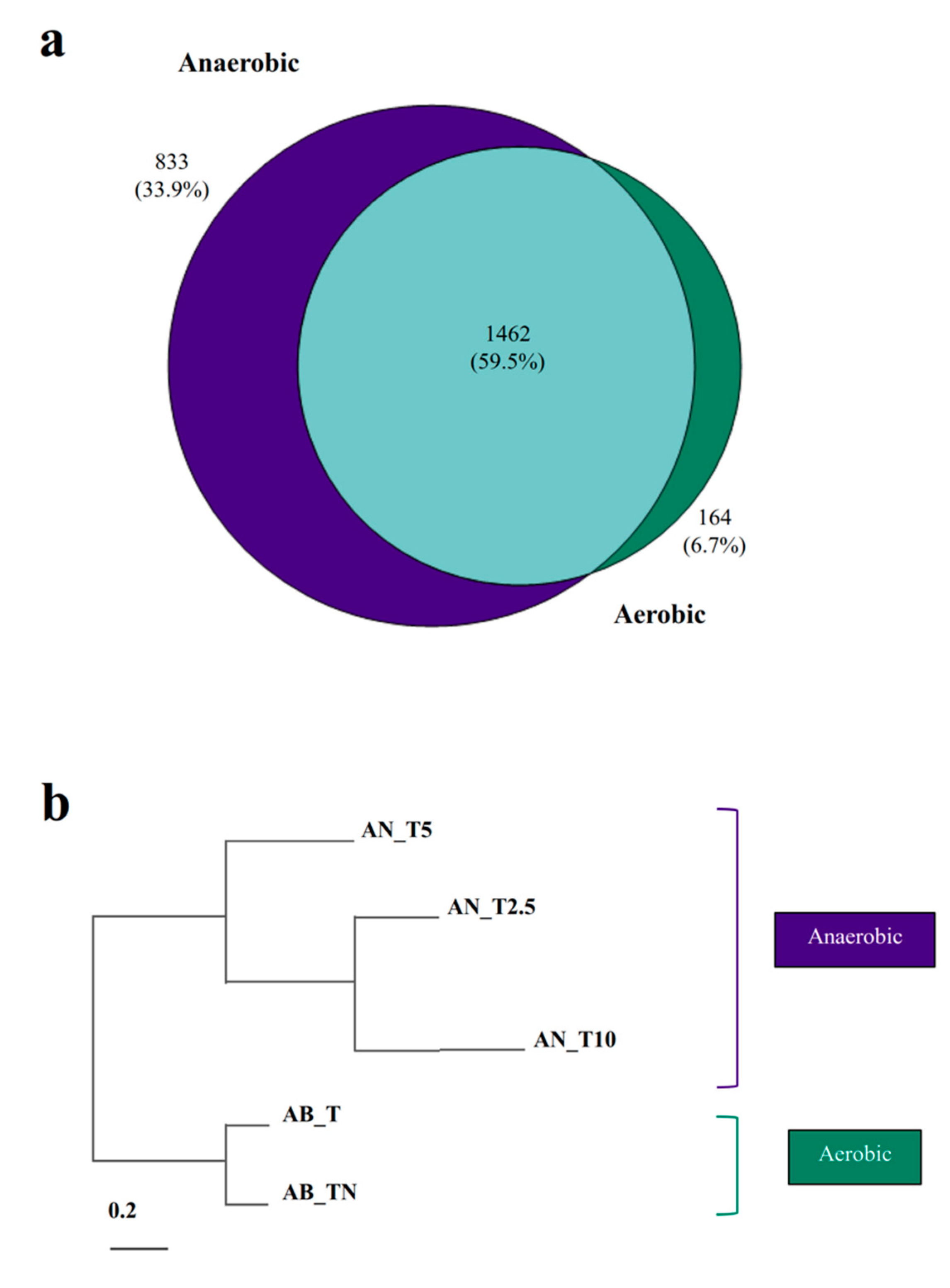
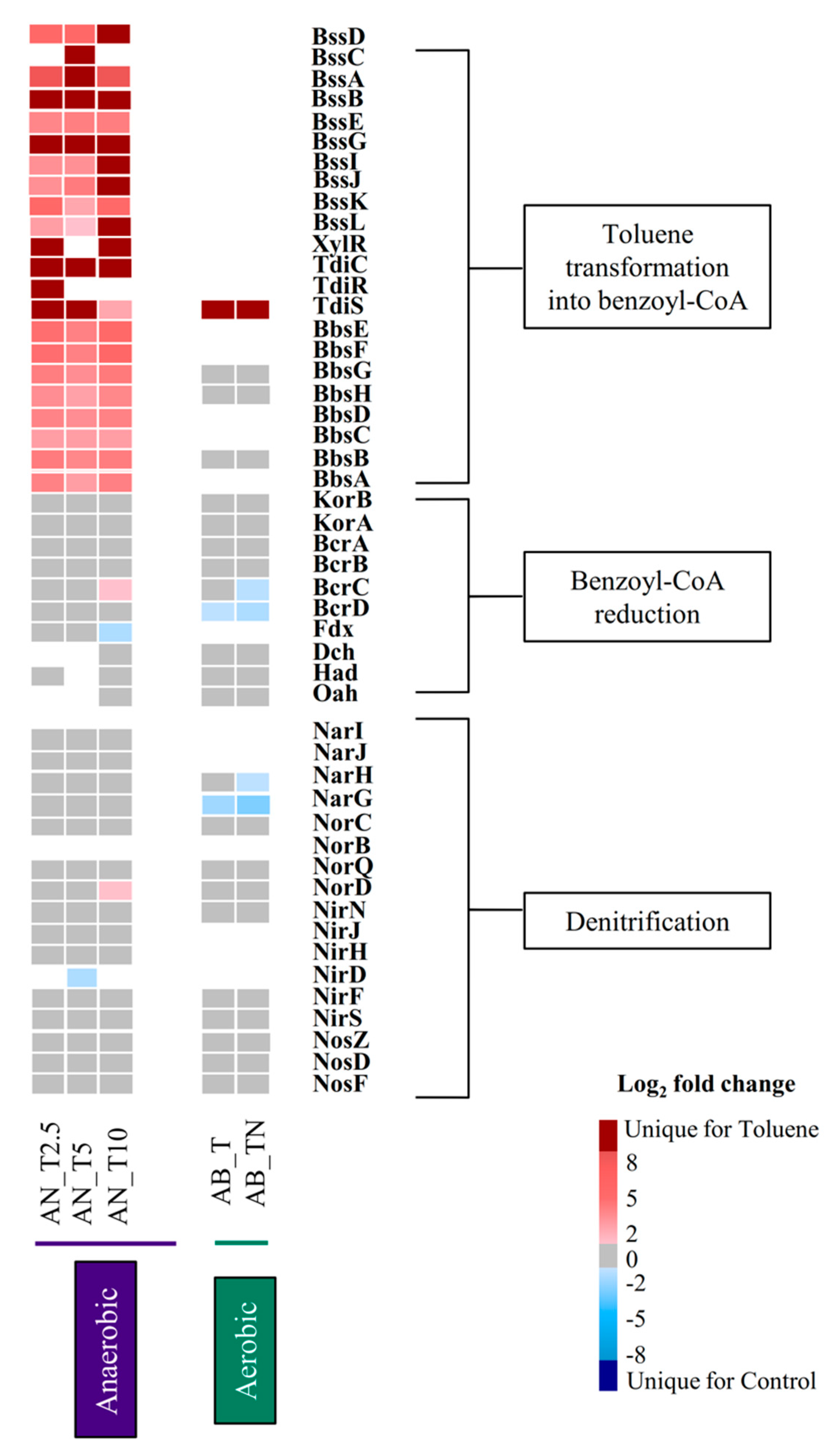
3.3. Proteins Involved in Anaerobic Toluene Degradation
3.4. Expression Pattern of the Transcriptional Regulatory Components TdiCRS and XylR
3.5. Proteins Involved in the Denitrification Process
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carmona, M.; Zamarro, M.T.; Blazquez, B.; Durante-Rodriguez, G.; Juarez, J.F.; Valderrama, J.A.; Barragan, M.J.; Garcia, J.L.; Diaz, E. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 2009, 73, 71–133. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 2008, 1125, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Parales, R.E.; Parales, J.V.; Pelletier, D.A.; Ditty, J.L. Diversity of microbial toluene degradation pathways; Academic Press: Cambridge, MA, USA, 2008; volume 64, pp. 1–73. [Google Scholar]
- Weelink, S.A.; van Eekert, M.H.A.; Stams, A.J.M. Degradation of BTEX by anaerobic bacteria: physiology and application. Rev. Environ. Sci. Biotechnol. 2010, 9, 359–385. [Google Scholar] [CrossRef]
- Funk, M.A.; Judd, E.T.; Marsh, E.N.G.; Elliott, S.J.; Drennan, C.L. Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity. Proc. Natl. Acad. Sci. USA 2014, 111, 10161–10166. [Google Scholar] [CrossRef]
- Funk, M.A.; Marsh, E.N.G.; Drennan, C.L. Substrate-bound structures of benzylsuccinate synthase reveal how toluene is activated in anaerobic hydrocarbon degradation. J. Biol. Chem. 2015, 290, 22398–22408. [Google Scholar] [CrossRef]
- Leuthner, B.; Heider, J. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of β oxidation of the intermediate benzylsuccinate. J. Bacteriol. 2000, 182, 272–277. [Google Scholar] [CrossRef]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds - from one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef]
- Kung, J.W.; Löffler, C.; Dörner, K.; Heintz, D.; Gallien, S.; Van Dorsselaer, A.; Friedrich, T.; Boll, M. Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc. Natl. Acad. Sci. USA 2009, 106, 17687–17692. [Google Scholar] [CrossRef]
- Boll, M.; Fuchs, G.; Heider, J. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 2002, 6, 604–611. [Google Scholar] [CrossRef]
- Widdel, F.; Rabus, R. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 2001, 12, 259–276. [Google Scholar] [CrossRef]
- Sawers, G. Biochemistry, physiology and molecular biology of glycyl radical enzymes. FEMS Microbiol. Rev. 1998, 22, 543–551. [Google Scholar] [CrossRef]
- Leuthner, B.; Leutwein, C.; Schulz, H.; Hörth, P.; Haehnel, W.; Schiltz, E.; Schägger, H.; Heider, J. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalyzing the first step in anaerobic toluene metabolism. Mol. Microbiol. 1998, 28, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Hilberg, M.; Pierik, A.J.; Bill, E.; Friedrich, T.; Lippert, M.; Heider, J. Identification of FeS clusters in the glycyl-radical enzyme benzylsuccinate synthase via EPR and Mössbauer spectroscopy. J. Biol. Inorg. Chem. 2012, 17, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Heider, J.; Szaleniec, M.; Martins, B.M.; Seyhan, D.; Buckel, W.; Golding, B.T. Structure and function of benzylsuccinate synthase and related fumarate-adding glycyl radical enzymes. J. Mol. Microbiol. Biotechnol. 2016, 26, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, B.; Carmona, M.; Díaz, E. Transcriptional regulation of the peripheral pathway for the anaerobic catabolism of toluene and m-xylene in Azoarcus sp. CIB. Front. Microbiol. 2018, 9, 506. [Google Scholar] [CrossRef] [PubMed]
- Achong, G.R.; Rodriguez, A.M.; Spormann, A.M. Benzylsuccinate synthase of Azoarcus sp. strain T: Cloning, sequencing, transcriptional organization and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 2001, 183, 6763–6770. [Google Scholar] [CrossRef]
- Kube, M.; Heider, J.; Amann, J.; Hufnagel, P.; Kühner, S.; Beck, A.; Reinhardt, R.; Rabus, R. Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 2004, 181, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Leuthner, B.; Heider, J. A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol. Lett. 1998, 166, 35–41. [Google Scholar] [CrossRef]
- Coschigano, P.W. Transcriptional analysis of the tutE tutFDGH gene cluster from Thauera aromatica strain T1. Appl. Environ. Microbiol. 2000, 66, 1147–1151. [Google Scholar] [CrossRef]
- Coschigano, P.W.; Bishop, B.J. Role of benzylsuccinate in the induction of the tutE tutFDGH gene complex of T. aromatica strain T1. FEMS Microbiol. Lett. 2004, 231, 261–266. [Google Scholar] [CrossRef]
- Kühner, S.; Wöhlbrand, L.; Fritz, I.; Wruck, W.; Hultschig, C.; Hufnagel, P.; Kube, M.; Reinhardt, R.; Rabus, R. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 2005, 187, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, Y.; Akagi, J.; Uchihashi, Y.; Hiraishi, A.; Yukawa, H.; Yurimoto, H.; Sakai, Y.; Kato, N. Anaerobic degradation of aromatic compounds by Magnetospirillum strains isolation and degradation genes. Biosci. Biotech. Bioch. 2005, 69, 1483–1491. [Google Scholar] [CrossRef]
- Shinoda, Y.; Sakai, Y.; Uenishi, H.; Uchihashi, Y.; Hiraishi, A.; Yukawa, H.; Yurimoto, H.; Kato, N. Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl. Environ. Microbiol. 2004, 70, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Brow, C.N.; O’Brien Johnson, R.; Johnson, R.L.; Simon, H.M. Assessment of anaerobic toluene biodegradation activity by bssA transcript/gene ratios. Appl. Environ. Microbiol. 2013, 79, 5338–5344. [Google Scholar] [CrossRef]
- Kappelmeyer, U.; Wiessner, A.; Kuschk, P.; Kästner, M. Operation of a universal test unit for planted soil filters- planted fixed bed reactor. Chem. Ing. Tech. 2002, 73, 1218. [Google Scholar] [CrossRef]
- Lünsmann, V.; Kappelmeyer, U.; Taubert, A.; Nijenhuis, I.; von Bergen, M.; Heipieper, H.J.; Müller, J.A.; Jehmlich, N. Aerobic toluene degraders in the rhizosphere of a constructed wetland model show diurnal polyhydroxyalkanoate metabolism. Appl. Environ. Microbiol. 2016, 8, 4126–4132. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.; Fiedler, S.; Müller, J.A.; Kappelmeyer, U.; Mäusezahl, I.; Heipieper, H.J. Draft genome sequence of Magnetospirillum sp. strain 15-1, a denitrifying toluene degrader isolated from a planted fixed-bed reactor. Genome Announc. 2017, 5, e00764-00717. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.; Martinez-Lavanchy, P.M.; Marin-Cevada, V.; Böhnke, S.; Harms, H.; Müller, J.A.; Heipieper, H.J. Isolation and characterization of Magnetospirillum sp. strain 15-1 as a representative anaerobic toluene-degrader from a constructed wetland model. PLoS ONE 2017, 12, e0174750. [Google Scholar] [CrossRef]
- Tschech, A.; Fuchs, G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 1987, 148, 213–217. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing; John Wiley and Sons, Ltd.: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Kuntze, K.; Vogt, C.; Richnow, H.H.; Boll, M. Combined application of PCR-based functional assays for the detection of aromatic-compound-degrading anaerobes. Appl. Environ. Microbiol. 2011, 77, 5056–5061. [Google Scholar] [CrossRef]
- He, F. Bradford protein assay. Bio Protoc. 2011, 1, e45. [Google Scholar] [CrossRef]
- Lünsmann, V.; Kappelmeyer, U.; Benndorf, R.; Martinez-Lavanchy, P.M.; Taubert, A.; Adrian, L.; Duarte, M.; Pieper, D.H.; von Bergen, M.; Müller, J.A.; et al. In situ protein-SIP highlights Burkholderiaceae as key players degrading toluene by para ring hydroxylation in a constructed wetland model. Environ. Microbiol. 2016, 18, 1176–1186. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–214. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923. [Google Scholar] [CrossRef] [PubMed]
- Chawade, A.; Alexandersson, E.; Levander, F. Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 2014, 13, 3114–3120. [Google Scholar] [CrossRef]
- Välikangas, T.; Suomi, T.; Elo, L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Littlefield, K. Venn Diagram Plotter, 1.5.5228.29250; Batelle Memorial Institute, Department of Energy (PNNL): Richland, WA, USA, 2004. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Letunic, I.; Yamada, T.; Kanehisa, M.; Bork, P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008, 33, 101–103. [Google Scholar] [CrossRef]
- Yamada, T.; Letunic, I.; Okuda, S.; Kanehisa, M.; Bork, P. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011, 39, W412–W415. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Singh, J. FigShare. J. Pharmacol. Pharmacother. 2011, 2, 138–139. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.; Gruhl, S.; Haange, S.B.; Lünsmann, V.; Jehmlich, N.; von Bergen, M.; Heipieper, H.J.; Müller, J. Data Set S1: qPCR raw data of bssA, bcrC and 16S rRNA from AN and AB cultures. Figshare 2020. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.; Gruhl, S.; Haange, S.B.; Lünsmann, V.; Jehmlich, N.; von Bergen, M.; Heipieper, H.J.; Müller, J. Data Set S2: Copy numbers of bssA, bcrC and 16S rRNA from AN and AB cultures. Figshare 2020. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.; Gruhl, S.; Haange, S.B.; Lünsmann, V.; Jehmlich, N.; von Bergen, M.; Heipieper, H.J.; Müller, J. Data Set S3: Proteins identification and abundances from AN and AB cultures. Figshare 2020. [Google Scholar] [CrossRef]
- Kim, S.J.; Park, S.J.; Jung, M.Y.; Kim, J.G.; Madsen, E.L.; Rhee, S.K. An uncultivated nitrate-reducing member of the genus Herminiimonas degrades toluene. Appl. Environ. Microbiol. 2014, 80, 3233–3243. [Google Scholar] [CrossRef]
- Koh, S.; Hwang, J.; Guchhait, K.; Lee, E.G.; Kim, S.Y.; Kim, S.J.; Lee, S.; Chung, J.M.; Jung, H.S.; Lee, S.J.; et al. Molecular Insights into toluene sensing in the TodS/TodT signal transduction system. J. Biol. Chem. 2016, 291, 8575–8590. [Google Scholar] [CrossRef]
- Ereño-Orbea, J.; Oyenarte, I.; Martinez-Cruz, L.A. CBS domains: Ligand binding sites and conformational variability. Arch. Biochem. Biophys. 2013, 540, 70–81. [Google Scholar] [CrossRef]
- Kümmel, S.; Kuntze, K.; Vogt, C.; Boll, M.; Heider, J.; Richnow, H.H. Evidence for benzylsuccinate synthase subtypes obtained by using stable isotope tools. J. Bacteriol. 2013, 195, 4660–4667. [Google Scholar] [CrossRef]
- Sawers, R.G.; Zenelein, E.; Böck, A. Two-dimensional gel electrophoretic analysis of Escherichia coli proteins: influence of various anaerobic growth conditions and the fnr gene product on cellular protein composition. Arch. Microbiol. 1988, 149, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, R.P.; Park, S.J. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res. Microbiol. 1994, 145, 437–450. [Google Scholar] [CrossRef]
- Chiang, R.C.; Cavicchioli, R.; Gunsalus, R.P. Identification and characterization of narQ, a second nitrate sensor for nitrate-dependent gene regulation in Escherichia coli. Mol. Microbiol. 1992, 6, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, R.P.; Kalman, L.V.; Stewart, R.R. Nucleotide sequence of the narL gene that is involved in global regulation of nitrate controlled respiratory genes of Escherichia coli. Nucleic Acids Res. 1989, 17, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kustu, S.; Stewart, V. In vitro Interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J. Mol. Biol. 1994, 241, 150–165. [Google Scholar] [CrossRef]
- Rabin, R.S.; Stewart, V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 1993, 175, 3259–3268. [Google Scholar] [CrossRef]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Stewart, V. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol. Microbiol. 1993, 9, 425–434. [Google Scholar] [CrossRef]
- Myers, K.S.; Yan, H.; Ong, I.M.; Chung, D.; Liang, K.; Tran, F.; Keleş, S.; Landick, R.; Kiley, P.J. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 2013, 9, e1003565. [Google Scholar] [CrossRef]
- Schmidt, A.; Kochanowski, K.; Vedelaar, S.; Ahrne, E.; Volkmer, B.; Callipo, L.; Knoops, K.; Bauer, M.; Aebersold, R.; Heinemann, M. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016, 34, 104–110. [Google Scholar] [CrossRef]
- Hino, T.; Matsumoto, Y.; Nagano, S.; Sugimoto, H.; Fukumori, Y.; Murata, T.; Iwata, S.; Shiro, Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 2010, 330, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Nnyepi, M.R.; Peng, Y.; Broderick, J.B. Inactivation of E. coli pyruvate formate-lyase: role of AdhE and small molecules. Arch. Biochem. Biophys. 2007, 459, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Toraya, T. Molecular architectures and functions of radical enzymes and their (re)activating proteins. J. Biochem. 2015, 158, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Hermuth, K.; Leuthner, B.; Heider, J. Operon structure and expression of the genes for benzylsuccinate synthase in Thauera aromatica strain K172. Arch. Microbiol. 2002, 177, 132–138. [Google Scholar] [CrossRef]
- Van Assche, E.; Van Puyvelde, S.; Vanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar] [CrossRef]
- Picard, F.; Dressaire, C.; Girbal, L.; Cocaign-Bousquet, M. Examination of post-transcriptional regulations in prokaryotes by integrative biology. C. R. Biol. 2009, 332, 958–973. [Google Scholar] [CrossRef]
- Potts, A.H.; Vakulskas, C.A.; Pannuri, A.; Yakhnin, H.; Babitzke, P.; Romeo, T. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 2017, 8, 1596. [Google Scholar] [CrossRef]
- Holmqvist, E.; Wright, P.R.; Li, L.; Bischler, T.; Barquist, L.; Reinhardt, R.; Backofen, R.; Vogel, J. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016, 35, 991–1011. [Google Scholar] [CrossRef]
- Kavanagh, K.L.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef]
- Baker, M.E.; Grundy, W.N.; Elkan, C.P. Spinach CSP41, an mRNA-binding protein and ribonuclease, is homologous to nucleotide-sugar epimerases and hydroxysteroid dehydrogenases. Biochem. Biophys. Res. Commun. 1998, 248, 250–254. [Google Scholar] [CrossRef]
- Stammers, D.K.; Ren, J.; Leslie, K.; Nichols, C.E.; Lamb, H.K.; Cocklin, S.; Dodds, A.; Hawkins, A.R. The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 2001, 20, 6619–6626. [Google Scholar] [CrossRef] [PubMed]
- LeGros, H.L.J.; Halim, A.B.; Geller, A.M.; Kotb, M. Cloning, expression, and functional characterization of the β regulatory subunit of human methionine adenosyltransferase (MAT II). J. Biol. Chem. 2000, 254, 2359–2366. [Google Scholar] [CrossRef]
- Duetz, W.A.; Marques, S.; Wind, B.; Ramos, J.L.; van Andel, J.G. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol. 1996, 62, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Lacal, J.; Silva-Jimenez, H.; Krell, T.; Ramos, J.L. Catabolite repression of the TodS/TodT two-component system and effector-dependent transphosphorylation of TodT as the basis for toluene dioxygenase catabolic pathway control. J. Bacteriol. 2010, 192, 4246–4250. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.A.; van Niel, E.W.; Torremans, R.A.; Kuenen, J.G. Simultaneous nitrification and denitrification in aerobic chemostat cultures of Thiosphaera pantotropha. Appl. Environ. Microbiol. 1988, 54, 2812–2818. [Google Scholar] [CrossRef]
- Körner, H.; Zumft, W.G. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 1989, 55, 1670–1676. [Google Scholar] [CrossRef] [PubMed]
- Durante-Rodríguez, G.; Gómez-Álvarez, H.; Nogales, J.; Carmona, M.; Díaz, E. One-component systems that regulate the expression of degradation pathways for aromatic compounds. In Cellular Ecophysiology of Microbe: Hydrocarbon and Lipid Interactions. Handbook of Hydrocarbon and Lipid Microbiology; Krell, T.E., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 137–175. [Google Scholar]
- Ebert, M.; Laaß, S.; Thürmer, A.; Roselius, L.; Eckweiler, D.; Daniel, R.; Härtig, E.; Jahn, D. FnrL and three Dnr regulators are used for the metabolic adaptation to low oxygen tension in Dinoroseobacter shibae. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
| Primers Used for PCR | ||||
| Target Sequence | Primer Name | Primer Sequence | Product Size (bp) | Source |
| 16S rRNA | 27F 1492R | AGAGTTTGATCMTGGCTCAG GGYTACCTTGTTACGACTT | 1465 | [31] |
| bssA | bssA_F1 bssA_R2 | GACGARTTCATCRTCGGCTACCACGC AGCAGRTTGSCYTTCTGRTTYTTCTG | 1546 | Junca, H. |
| bcrC | bcrC_F bcrC_R | CGHATYCCRCGSTCGACCATCG CGGATCGGCTGCATCTGGCC | 800 | [32] |
| bbsC region | bbsD_F bbsB_R | GGCGGGATGTTGTCCTATGG GCTTCGGCCCTATTTGCTTG | 1823 | this study |
| Primers Used for RT-qPCR | ||||
| Target Sequence | Primer Name | Primer Sequence | Product Size (bp) | Source |
| 16S rRNA | 16S_F 16S_R | TGATGAAGGCCTTAGGGTTG CCAGGGCTTTCACTTCTGAC | 170 | Marín, V. |
| bcrC | 2bcrC_F 2bcrC_R | CATGATCTTCCCGTT TCCTTCAGCTCCTTC | 159 | Marín, V. |
| bssA | bssA_F3 bssA_R3 | CGTCCTTCGCCTCGGGTTAC CATCGCCTGCCAGTTGTCAATC | 188 | Marín, V. |
| Primers Used for RT-PCR | ||||
| Target Sequence | Primer Name | Primer Sequence | Product Size (bp) | Source |
| Intergenic region bssD-bssA | bssD-bssA_F bssD-bssA_R | CGCATTCACATCCCGGTCATC CAGGACGTTGGCGGTCATATT | 575 | this study |
| Intergenic region bssA-bssE | bssA-bssE_F bssA-bssE_R | CTGAATTGCGACCTCTGAGC GCTTGCGCGAGATATTACGG | 587 | this study |
| Intergenic region bssE-bssF | bssE-bssF_F bssE-bssF_R | GGCAGGTCTGGATGGATGAG CGTTTGGGAGGGGTATCGTC | 504 | this study |
| Intergenic region bssF-bssG | bssF-bssG_F bssF-bssG_R | GCCATTCGCCGTTTCAACAA GCGAACCTGGGAAAACATCG | 482 | this study |
| Intergenic region bssG-bssP | bssG-bssP_F bssG-bssP_R | CCTTGGATGAAGGCCGTAAC CCGAAGATCAGCCACTTTCC | 560 | this study |
| Intergenic region bssP-bssI | bssP-bssI_F bssP-bssI_R | CCCGTTTCAAGCGATGGTTC GAACTCCCTCCTTGCGGTAG | 493 | this study |
| Intergenic region bssI-bssJ | bssI-bssJ_F1 bssI-bssJ_R1 | CGAATTCCCCAGCGACTCA ATCTCGAACACGCCCACTC | 561 | this study |
| Intergenic region bssJ-bssK | bssJ-bssK_F bssJ-bssK_R | GCCCCCTACTTCAAATGGCT GTATCCCAATCCAGCGGAGG | 480 | this study |
| Intergenic region bssK-bssL | bssK-bssL_F bssK-bssL_R | AGTTGATTTCCTACGGGCGG GTGAGCAATCCACATGACGC | 515 | this study |
| Intergenic region tdiC-tdiR | tdiC-tdiR_F tdiC-tdiR_R | GAGATCATGACGACGGAGGT TAGAAAGATCAGCGGCAGCG | 500 | this study |
| Intergenic region tdiR-tdiS | tdiR-tdiS_F tdiR-tdiS_R | CGCCTCAGCAAGGAAGTGTT GGAAGCAAATGCCAACGGG | 435 | this study |
| xylR | xylR_F xylR_R | TGTCGAGCGTGGCTATTACTC CTTCCACCAAATTCTCGGGC | 545 | this study |
| Intergenic region bbsA-bbsB | bbsA-bbsB_F bbsA-bbsB_R | GGGCAGCTTGATTTTCCCAA GGCGATGAACACATCTCGTTG | 436 | this study |
| Intergenic region bbsB-bbsC | bbsB-bbsC_F bbsB-bbsC_R | GGCGGGATGTTGTCCTATGG ACCGATTCCGGAAGGAAAGG | 535 | this study |
| Intergenic region bbsC-bbsD | bbsC-bbsD_F bbsC-bbsD_R | CGGCAAGAGCGCCTATTTCT GGTGATTCCATTGCGTCCCA | 611 | this study |
| Intergenic region bbsD-bbsE | bbsD-bbsE_F bbsD-bbsE_R | CTGGGACGCAATGGAATCAC GCGCAAAAACACCTCCCTG | 542 | this study |
| Intergenic region bbsE-bbsF | bbsE-bbsF_F1 bbsE-bbsF_R1 | ATACGCCTATCGGACCTCGG GCTCTGCATGAACCACTTCC | 380 | this study |
| Intergenic region bbsF-bbsG | bbsF-bbsG_F bbsF-bbsG_R | CGCGGTGTTTTCCGATGAAG TTCAGCATTGCGTGCTCTTG | 550 | this study |
| Intergenic region bbsG-bbsH | bbsG-bbsH_F bbsG-bbsH_R | CGGGAAACCGGCATCGAATA TTCATCTGGGGGATGAGGGG | 428 | this study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer-Cifuentes, I.; Gruhl, S.; Haange, S.-B.; Lünsmann, V.; Jehmlich, N.; von Bergen, M.; Heipieper, H.J.; Müller, J.A. Benzylsuccinate Synthase is Post-Transcriptionally Regulated in the Toluene-Degrading Denitrifier Magnetospirillum sp. Strain 15-1. Microorganisms 2020, 8, 681. https://doi.org/10.3390/microorganisms8050681
Meyer-Cifuentes I, Gruhl S, Haange S-B, Lünsmann V, Jehmlich N, von Bergen M, Heipieper HJ, Müller JA. Benzylsuccinate Synthase is Post-Transcriptionally Regulated in the Toluene-Degrading Denitrifier Magnetospirillum sp. Strain 15-1. Microorganisms. 2020; 8(5):681. https://doi.org/10.3390/microorganisms8050681
Chicago/Turabian StyleMeyer-Cifuentes, Ingrid, Sylvie Gruhl, Sven-Bastiaan Haange, Vanessa Lünsmann, Nico Jehmlich, Martin von Bergen, Hermann J. Heipieper, and Jochen A. Müller. 2020. "Benzylsuccinate Synthase is Post-Transcriptionally Regulated in the Toluene-Degrading Denitrifier Magnetospirillum sp. Strain 15-1" Microorganisms 8, no. 5: 681. https://doi.org/10.3390/microorganisms8050681
APA StyleMeyer-Cifuentes, I., Gruhl, S., Haange, S.-B., Lünsmann, V., Jehmlich, N., von Bergen, M., Heipieper, H. J., & Müller, J. A. (2020). Benzylsuccinate Synthase is Post-Transcriptionally Regulated in the Toluene-Degrading Denitrifier Magnetospirillum sp. Strain 15-1. Microorganisms, 8(5), 681. https://doi.org/10.3390/microorganisms8050681