Metagenomic- and Cultivation-Based Exploration of Anaerobic Chloroform Biotransformation in Hypersaline Sediments as Natural Source of Chloromethanes
Abstract
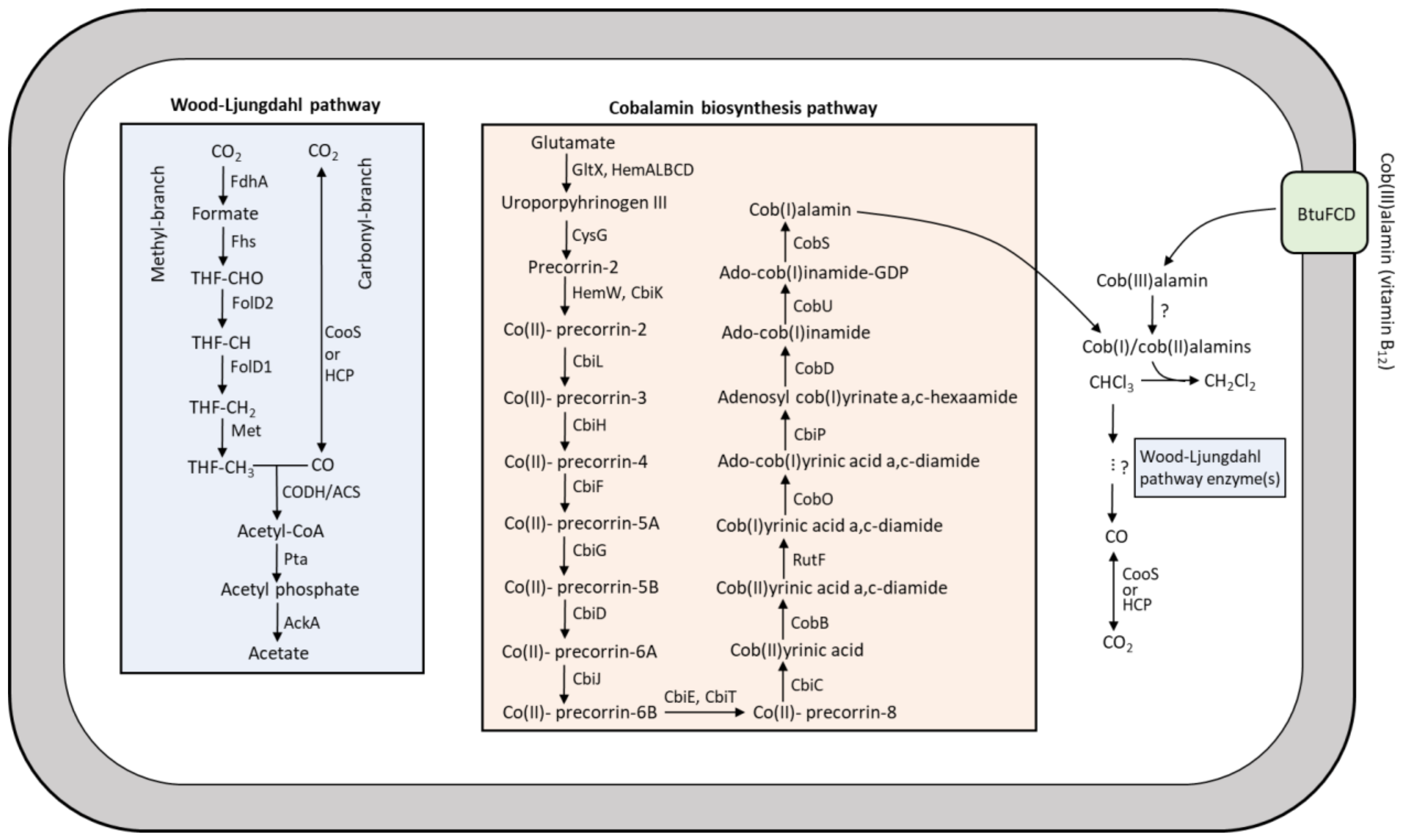
1. Introduction
2. Materials and Methods
2.1. Sediment Sampling
2.2. Physical Chemical Analysis
2.3. Microcosm Preparation
2.4. GC Analysis
2.5. Isotope Analysis
2.6. DNA Extraction
2.7. Quantitative PCR (qPCR)
2.8. Bacterial Community Analysis
2.9. Metagenomic Analysis
2.10. Sequence Deposition
3. Results
3.1. Physical Chemical Characteristics of Sediments
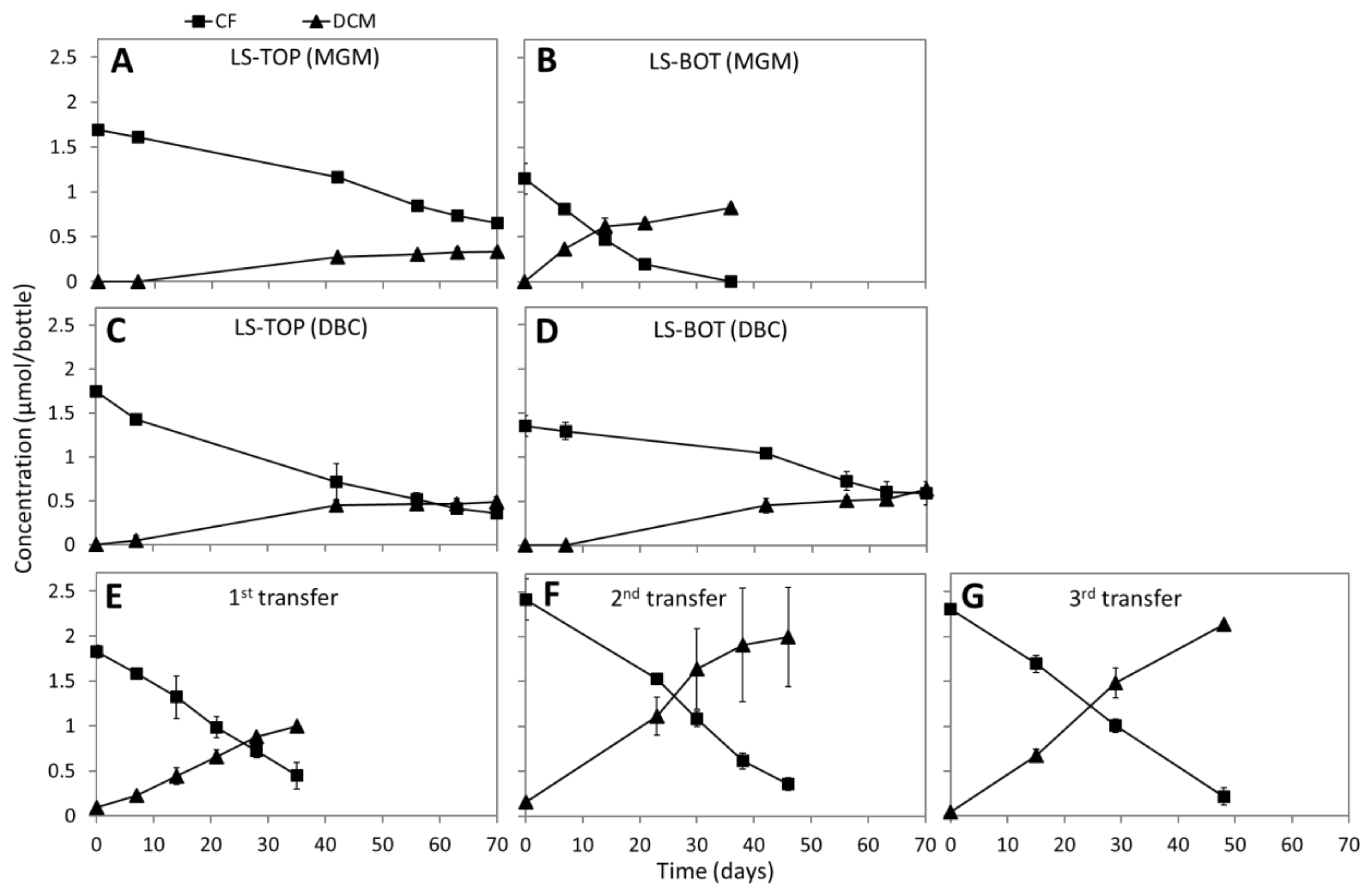
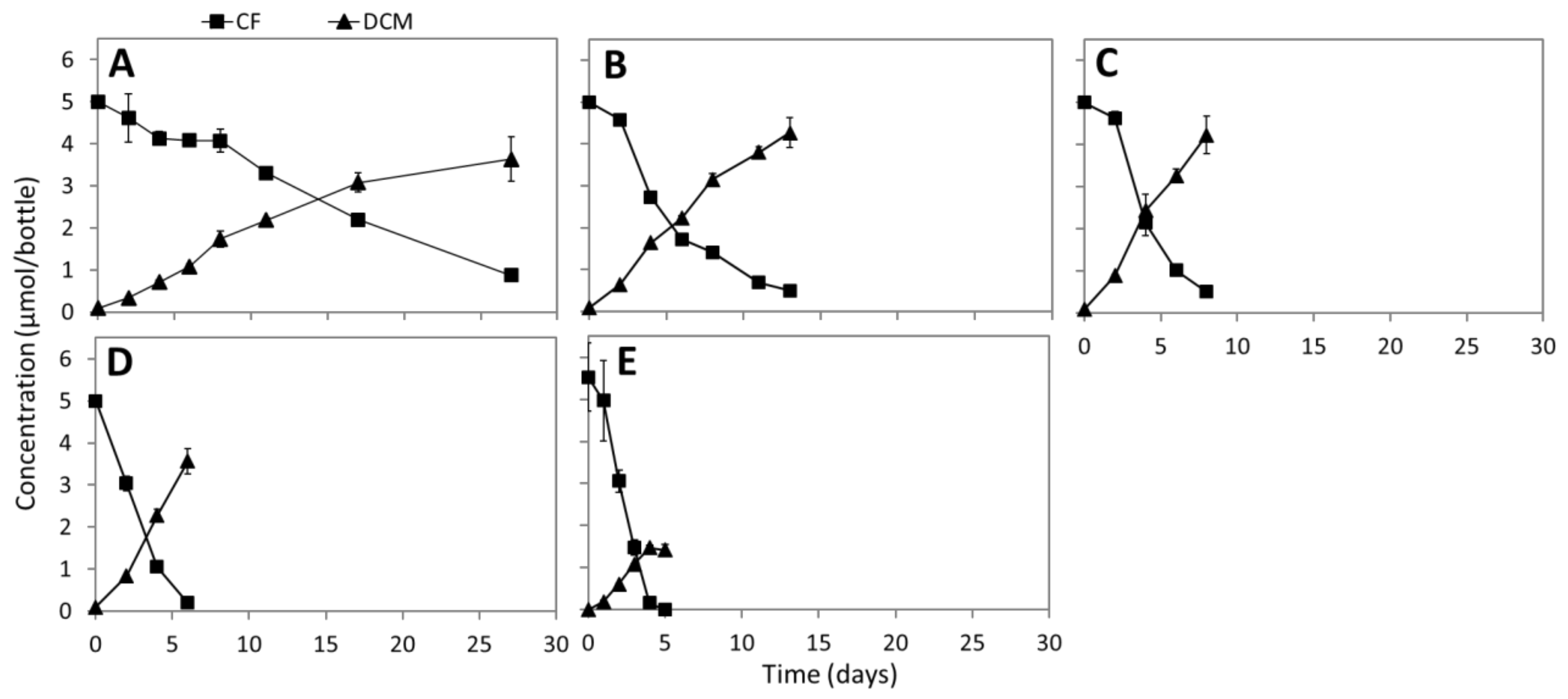
3.2. CF Dechlorination in Enrichment Cultures
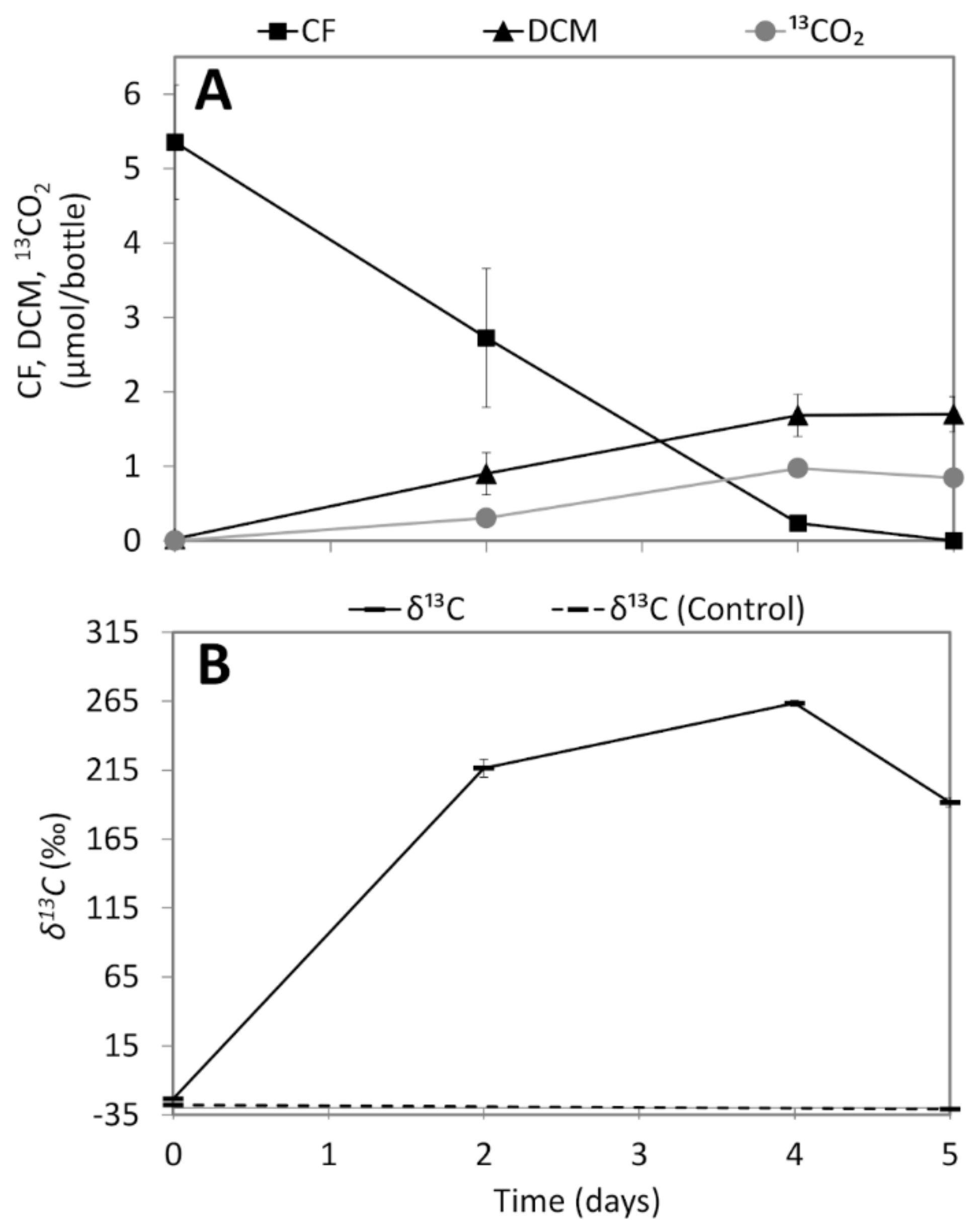
3.3. Analysis of 13CO2 Production from 13C-Labelled CF
3.4. qPCR and Bacterial Community Analysis
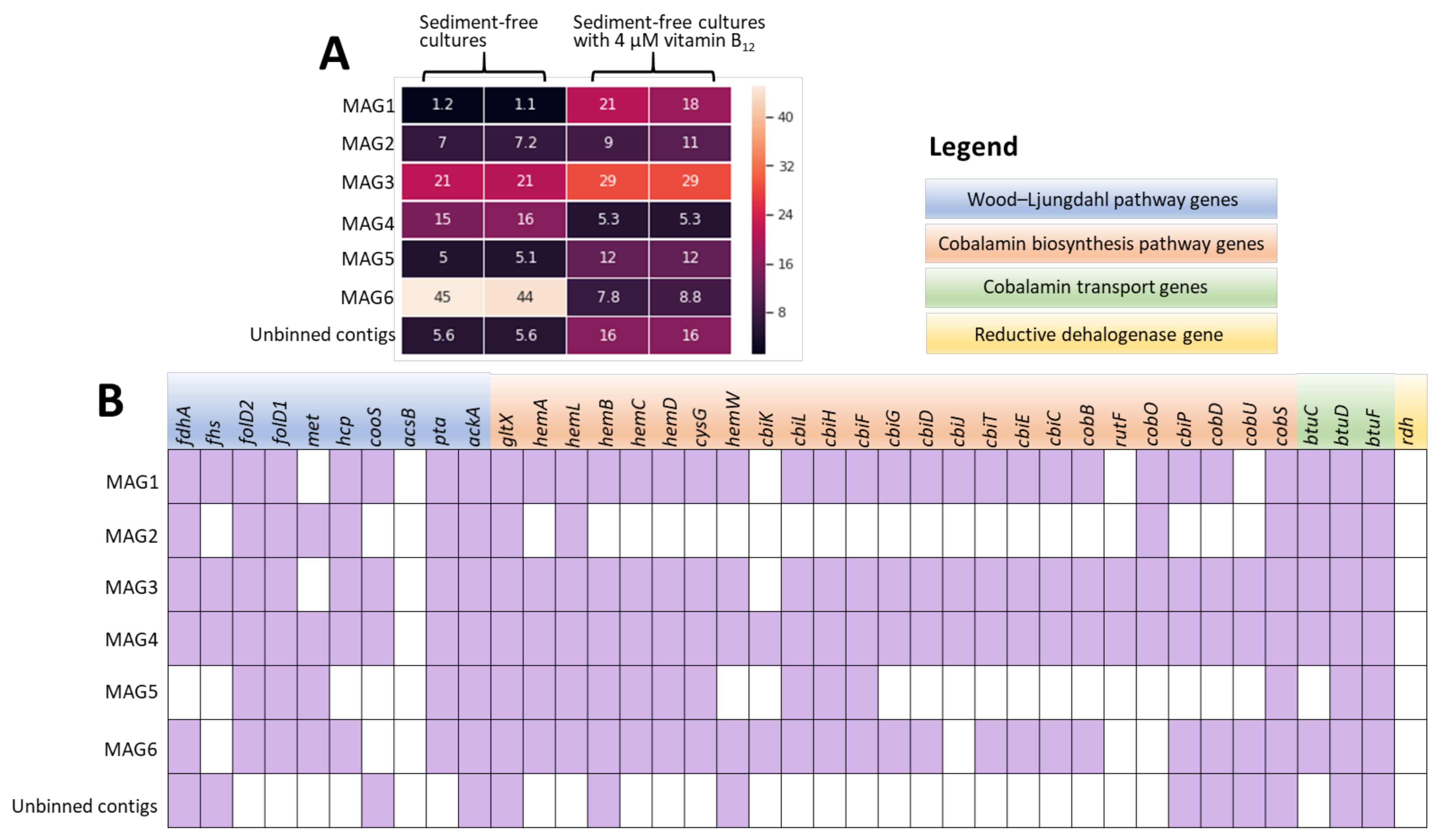
3.5. Metagenomic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CF | chloroform |
| CM | chloromethane |
| CODH/ACS | carbon monoxide dehydrogenase/acetyl-CoA synthase |
| DBC | DBCM2 medium |
| DCM | dichloromethane |
| DTT | dithiothreitol |
| GC/C-IRMS | gas chromatography combustion isotope ratio mass spectrometry |
| HCP | hybrid cluster protein |
| MAG | metagenome-assembled genome |
| MGM | modified growth medium |
| OHRB | organohalide-respiring bacteria |
| RDase | reductive dehalogenase |
| TOC | total organic carbon |
| VOX | volatile organohalogens |
| WLP | Wood-Ljungdahl pathway |
References
- Cicerone, R.J.; Stolarski, R.S.; Walters, S. Stratospheric ozone destruction by man-made chlorofluoromethanes. Science 1974, 185, 1165–1167. [Google Scholar] [CrossRef]
- Gribble, G.W. Naturally Occurring Organohalogen Compounds—A Comprehensive Update; Springer: Vienna, Austria, 2010; Volume 91. [Google Scholar]
- Rosenthal, S.L. A review of the mutagenicity of chloroform. Environ. Mol. Mutagen. 1987, 10, 211–226. [Google Scholar] [CrossRef] [PubMed]
- ATSDR. Toxicological Profile for Chloroform; U.S. Department of Health and Human Services: Atlanta, GA, USA, 1997.
- Field, J.A. Natural production of organohalide compounds in the environment. In Organohalide-Respiring Bacteria; Adrian, L., Löffler, F.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 7–29. [Google Scholar]
- Albers, C.N.; Hansen, P.E.; Jacobsen, O.S. Trichloromethyl compounds—Natural background concentrations and fates within and below coniferous forests. Sci. Total Environ. 2010, 408, 6223–6234. [Google Scholar] [CrossRef]
- Breider, F.; Albers, C.N.; Hunkeler, D. Assessing the role of trichloroacetyl-containing compounds in the natural formation of chloroform using stable carbon isotopes analysis. Chemosphere 2013, 90, 441–448. [Google Scholar] [CrossRef]
- Haselmann, K.F.; Laturnus, F.; Grøn, C. Formation of chloroform in soil. A year-round study at a Danish spruce forest site. Water Air Soil Pollut. 2002, 139, 35–41. [Google Scholar] [CrossRef]
- Osswald, A.; Poszwa, A.; Bueno, M.; Arnaudguilhem, C.; Billet, D.; Thiry, Y.; Leyval, C. Contribution of microbial activity to formation of organically bound chlorine during batch incubation of forest soil using 37Cl as a tracer. Soil Biol. Biochem. 2016, 100, 210–217. [Google Scholar] [CrossRef]
- Khalil, M.; Rasmussen, R.; Shearer, M.; Chen, Z.L.; Yao, H.; Yang, J. Emissions of methane, nitrous oxide, and other trace gases from rice fields in China. J. Geophys. Res. Atmos. 1998, 103, 25241–25250. [Google Scholar] [CrossRef]
- Hunkeler, D.; Laier, T.; Breider, F.; Jacobsen, O.S. Demonstrating a natural origin of chloroform in groundwater using stable carbon isotopes. Environ. Sci. Technol. 2012, 46, 6096–6101. [Google Scholar] [CrossRef][Green Version]
- Nightingale, P.D. Low Molecular Weight Halocarbons in Seawater. Ph.D. Thesis, University of East Anglia, Norfolk, UK, 1991. [Google Scholar]
- Ruecker, A.; Weigold, P.; Behrens, S.; Jochmann, M.; Laaks, J.; Kappler, A. Predominance of biotic over abiotic formation of halogenated hydrocarbons in hypersaline sediments in Western Australia. Environ. Sci. Technol. 2014, 48, 9170–9178. [Google Scholar] [CrossRef]
- Weissflog, L.; Lange, C.A.; Pfennigsdorff, A.; Kotte, K.; Elansky, N.; Lisitzyna, L.; Putz, E.; Krueger, G. Sediments of salt lakes as a new source of volatile highly chlorinated C1/C2 hydrocarbons. Geophys. Res. Lett. 2005, 32, 1–4. [Google Scholar] [CrossRef]
- Laturnus, F.; Haselmann, K.F.; Borch, T.; Grøn, C. Terrestrial natural sources of trichloromethane (chloroform, CHCl3)–an overview. Biogeochemistry 2002, 60, 121–139. [Google Scholar] [CrossRef]
- Fang, X.; Park, S.; Saito, T.; Tunnicliffe, R.; Ganesan, A.L.; Rigby, M.; Li, S.; Yokouchi, Y.; Fraser, P.J.; Harth, C.M. Rapid increase in ozone-depleting chloroform emissions from China. Nat. Geosci. 2019, 12, 89–93. [Google Scholar] [CrossRef]
- Cappelletti, M.; Frascari, D.; Zannoni, D.; Fedi, S. Microbial degradation of chloroform. Appl. Microbiol. Biotechnol. 2012, 96, 1395–1409. [Google Scholar] [CrossRef]
- Janssen, D.B.; Dinkla, I.J.; Poelarends, G.J.; Terpstra, P. Bacterial degradation of xenobiotic compounds: Evolution and distribution of novel enzyme activities. Environ. Microbiol. 2005, 7, 1868–1882. [Google Scholar] [CrossRef] [PubMed]
- Grostern, A.; Duhamel, M.; Dworatzek, S.; Edwards, E.A. Chloroform respiration to dichloromethane by a Dehalobacter population. Environ. Microbiol. 2010, 12, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Barajas, C.; Field, J.A. Riboflavin- and cobalamin-mediated biodegradation of chloroform in a methanogenic consortium. Biotechnol. Bioeng. 2005, 89, 539–550. [Google Scholar] [CrossRef]
- Justicia-Leon, S.D.; Higgins, S.; Mack, E.E.; Griffiths, D.R.; Tang, S.; Edwards, E.A.; Löffler, F.E. Bioaugmentation with distinct Dehalobacter strains achieves chloroform detoxification in microcosms. Environ. Sci. Technol. 2014, 48, 1851–1858. [Google Scholar] [CrossRef]
- Rodríguez-Fernández, D.; Torrentó, C.; Guivernau, M.; Viñas, M.; Hunkeler, D.; Soler, A.; Domènech, C.; Rosell, M. Vitamin B12 effects on chlorinated methanes-degrading microcosms: Dual isotope and metabolically active microbial populations assessment. Sci. Total Environ. 2018, 621, 1615–1625. [Google Scholar] [CrossRef]
- Shan, H.; Kurtz, H.D.; Mykytczuk, N.; Trevors, J.T.; Freedman, D.L. Anaerobic biotransformation of high concentrations of chloroform by an enrichment culture and two bacterial isolates. Appl. Environ. Microbiol. 2010, 76, 6463–6469. [Google Scholar] [CrossRef][Green Version]
- Egli, C.; Tschan, T.; Scholtz, R.; Cook, A.M.; Leisinger, T. Transformation of tetrachloromethane to dichloromethane and carbon dioxide by Acetobacterium woodii. Appl. Environ. Microbiol. 1988, 54, 2819–2824. [Google Scholar] [CrossRef]
- Gälli, R.; McCarty, P.L. Biotransformation of 1,1,1-trichloroethane, trichloromethane, and tetrachloromethane by a Clostridium sp. Appl. Environ. Microbiol. 1989, 55, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Baeseman, J.L.; Novak, P.J. Effects of various environmental conditions on the transformation of chlorinated solvents by Methanosarcina thermophila cell exudates. Biotechnol. Bioeng. 2001, 75, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Bagley, D.M.; Gossett, J.M. Chloroform degradation in methanogenic methanol enrichment cultures and by Methanosarcina barkeri 227. Appl. Environ. Microbiol. 1995, 61, 3195–3201. [Google Scholar] [CrossRef] [PubMed]
- Mikesell, M.D.; Boyd, S.A. Dechlorination of chloroform by Methanosarcina strains. Appl. Environ. Microbiol. 1990, 56, 1198–1201. [Google Scholar] [CrossRef]
- Holliger, C.; Schraa, G.; Stupperich, E.; Stams, A.; Zehnder, A. Evidence for the involvement of corrinoids and factor F430 in the reductive dechlorination of 1,2-dichloroethane by Methanosarcina barkeri. J. Bacteriol. 1992, 174, 4427–4434. [Google Scholar] [CrossRef]
- Gantzer, C.J.; Wackett, L.P. Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ. Sci. Technol. 1991, 25, 715–722. [Google Scholar] [CrossRef]
- Krone, U.E.; Laufer, K.; Thauer, R.K.; Hogenkamp, H.P. Coenzyme F430 as a possible catalyst for the reductive dehalogenation of chlorinated C1 hydrocarbons in methanogenic bacteria. Biochemistry 1989, 28, 10061–10065. [Google Scholar] [CrossRef]
- Krone, U.E.; Thauer, R.K.; Hogenkamp, H.P. Reductive dehalogenation of chlorinated C1-hydrocarbons mediated by corrinoids. Biochemistry 1989, 28, 4908–4914. [Google Scholar] [CrossRef]
- Fincker, M.; Spormann, A.M. Biochemistry of catabolic reductive dehalogenation. Annu. Rev. Biochem. 2017, 86, 357–386. [Google Scholar] [CrossRef]
- Schubert, T.; Adrian, L.; Sawers, R.G.; Diekert, G. Organohalide respiratory chains: Composition, topology and key enzymes. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Ding, C.; Zhao, S.; He, J. A Desulfitobacterium sp. strain PR reductively dechlorinates both 1,1,1-trichloroethane and chloroform. Environ. Microbiol. 2014, 16, 3387–3397. [Google Scholar] [CrossRef] [PubMed]
- Gerritse, J.; Drzyzga, O.; Kloetstra, G.; Keijmel, M.; Wiersum, L.P.; Hutson, R.; Collins, M.D.; Gottschal, J.C. Influence of different electron donors and acceptors on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl. Environ. Microbiol. 1999, 65, 5212–5221. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Low, A.; Zemb, O.; Koenig, J.; Michaelsen, A.; Manefield, M. Complete chloroform dechlorination by organochlorine respiration and fermentation. Environ. Microbiol. 2012, 14, 883–894. [Google Scholar] [CrossRef]
- Wong, Y.K.; Holland, S.I.; Ertan, H.; Manefield, M.; Lee, M. Isolation and characterization of Dehalobacter sp. strain UNSWDHB capable of chloroform and chlorinated ethane respiration. Environ. Microbiol. 2016, 18, 3092–3105. [Google Scholar] [CrossRef]
- Tang, S.; Edwards, E.A. Identification of Dehalobacter reductive dehalogenases that catalyse dechlorination of chloroform, 1,1,1-trichloroethane and 1,1-dichloroethane. Phil. Trans. R. Soc. B 2013, 368, 20120318. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wilson, J.; Su, C.; Wilkin, R. Review of abiotic degradation of chlorinated solvents by reactive iron minerals in aquifers. Groundw. Monit. Rem. 2015, 35, 57–75. [Google Scholar] [CrossRef]
- Rodríguez-Fernández, D.; Heckel, B.; Torrentó, C.; Meyer, A.; Elsner, M.; Hunkeler, D.; Soler, A.; Rosell, M.; Domènech, C. Dual element (CCl) isotope approach to distinguish abiotic reactions of chlorinated methanes by Fe(0) and by Fe (II) on iron minerals at neutral and alkaline pH. Chemosphere 2018, 206, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Torrentó, C.; Palau, J.; Rodríguez-Fernández, D.; Heckel, B.; Meyer, A.; Domènech, C.; Rosell, M.n.; Soler, A.; Elsner, M.; Hunkeler, D. Carbon and chlorine isotope fractionation patterns associated with different engineered chloroform transformation reactions. Environ. Sci. Technol. 2017, 51, 6174–6184. [Google Scholar] [CrossRef]
- Atashgahi, S.; Häggblom, M.M.; Smidt, H. Organohalide respiration in pristine environments: Implications for the natural halogen cycle. Environ. Microbiol. 2018, 20, 934–948. [Google Scholar] [CrossRef]
- Krzmarzick, M.J.; Crary, B.B.; Harding, J.J.; Oyerinde, O.O.; Leri, A.C.; Myneni, S.C.; Novak, P.J. Natural niche for organohalide-respiring Chloroflexi. Appl. Environ. Microbiol. 2012, 78, 393–401. [Google Scholar] [CrossRef]
- Atashgahi, S.; Liebensteiner, M.G.; Janssen, D.B.; Smidt, H.; Stams, A.; Sipkema, D. Microbial synthesis and transformation of inorganic and organic chlorine compounds. Front. Microbiol. 2018, 9, 3079. [Google Scholar] [CrossRef] [PubMed]
- Rhew, R.C.; Miller, B.R.; Weiss, R.F. Natural methyl bromide and methyl chloride emissions from coastal salt marshes. Nature 2000, 403, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: Implications for the functioning of salt lake ecosystems. In Saline Lakes; Melack, J.M., Jellison, R., Herbst, D.B., Eds.; Springer: Dordrecht, The Netherlands, 2001; pp. 61–72. [Google Scholar]
- Becker, J.G.; Freedman, D.L. Use of cyanocobalamin to enhance anaerobic biodegradation of chloroform. Environ. Sci. Technol. 1994, 28, 1942–1949. [Google Scholar] [CrossRef] [PubMed]
- Mebius, L.J. A rapid method for the determination of organic carbon in soil. Anal. Chim. Acta 1960, 22, 120–124. [Google Scholar] [CrossRef]
- Amstaetter, K.; Borch, T.; Kappler, A. Influence of humic acid imposed changes of ferrihydrite aggregation on microbial Fe(III) reduction. Geochim. Cosmochim. Acta 2012, 85, 326–341. [Google Scholar] [CrossRef]
- Stookey, L.L. Ferrozine—A new spectrophotometric reagent for iron. Anal. Chem. 1970, 42, 779–781. [Google Scholar] [CrossRef]
- Weigold, P.; Ruecker, A.; Loesekann-Behrens, T.; Kappler, A.; Behrens, S. Ribosomal tag pyrosequencing of DNA and RNA reveals “rare” taxa with high protein synthesis potential in the sediment of a hypersaline lake in Western Australia. Geomicrobiol. J. 2016, 33, 426–440. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Siroosi, M.; Atashgahi, S.; Smidt, H.; Ventosa, A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 2017, 163, 623–645. [Google Scholar] [CrossRef]
- Dyall-Smith, M. The Halohandbook: Protocols for Haloarchaeal Genetics; Haloarchaeal Genetics Laboratory: Melbourne, Australia, 2008; Volume 14. [Google Scholar]
- Assaf-Anid, N.; Hayes, K.F.; Vogel, T.M. Reduction dechlorination of carbon tetrachloride by cobalamin(II) in the presence of dithiothreitol: Mechanistic study, effect of redox potential and pH. Environ. Sci. Technol. 1994, 28, 246–252. [Google Scholar] [CrossRef]
- Chiu, P.-C.; Reinhard, M. Metallocoenzyme-mediated reductive transformation of carbon tetrachloride in titanium(III) citrate aqueous solution. Environ. Sci. Technol. 1995, 29, 595–603. [Google Scholar] [CrossRef]
- Atashgahi, S.; Maphosa, F.; Doğan, E.; Smidt, H.; Springael, D.; Dejonghe, W. Small-scale oxygen distribution determines the vinyl chloride biodegradation pathway in surficial sediments of riverbed hyporheic zones. FEMS Microbiol. Ecol. 2013, 84, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Atashgahi, S.; Lu, Y.; Zheng, Y.; Saccenti, E.; Suarez-Diez, M.; Ramiro-Garcia, J.; Eisenmann, H.; Elsner, M.; Stams, A.J.M.; Springael, D.; et al. Geochemical and microbial community determinants of reductive dechlorination at a site biostimulated with glycerol. Environ. Microbiol. 2017, 19, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Ramiro-Garcia, J.; Hermes, G.D.; Giatsis, C.; Sipkema, D.; Zoetendal, E.G.; Schaap, P.J.; Smidt, H. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000 Res. 2016, 5. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i88–i890. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2015, 32, 605–607. [Google Scholar] [CrossRef]
- Kang, D.; Li, F.; Kirton, E.S.; Thomas, A.; Egan, R.S.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peer J. 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Loman, N.J.; Andersson, A.F.; Quince, C. CONCOCT: Clustering Contigs on Coverage and Composition; Cornell University: Ithaca, NY, USA, 2013. [Google Scholar]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Matsen, F.A.; Kodner, R.B.; Armbrust, E.V. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 2010, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.L.M.; Gunturu, S.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. Nonpareil 3: Fast estimation of metagenomic coverage and sequence diversity. mSystems 2018, 3, e00039-e00018. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, W475–W478. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Valk, L.C.; Diender, M.; Stouten, G.R.; Petersen, J.F.; Nielsen, P.H.; Dueholm, M.S.; Pronk, J.T.; van Loosdrecht, M. “Candidatus Galacturonibacter soehngenii” Shows Acetogenic Catabolism of Galacturonic Acid but Lacks a Canonical Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase Complex. Front. Microbiol. 2020, 11, 63. [Google Scholar] [CrossRef]
- Read, K.A.; Mahajan, A.S.; Carpenter, L.J.; Evans, M.J.; Faria, B.V.; Heard, D.E.; Hopkins, J.R.; Lee, J.D.; Moller, S.J.; Lewis, A.C. Extensive halogen-mediated ozone destruction over the tropical Atlantic Ocean. Nature 2008, 453, 1232–1235. [Google Scholar] [CrossRef]
- Atashgahi, S.; Lu, Y.; Smidt, H. Overview of known organohalide-respiring bacteria—Phylogenetic diversity and environmental distribution. In Organohalide-Respiring Bacteria; Adrian, L., Löffler, F.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 63–105. [Google Scholar]
- Oren, A. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 1999, 63, 334–348. [Google Scholar] [CrossRef]
- Walker, G.; Murphy, S.; Huennekens, F. Enzymatic conversion of vitamin B12a to adenosyl-B12: Evidence for the existence of two separate reducing systems. Arch. Biochem. Biophys. 1969, 134, 95–102. [Google Scholar] [CrossRef]
- Weissbach, H.; Redfield, B.; Peterkofsky, A. Conversion of vitamin B12 to coenzyme B12 in cell-free extracts of Clostridium tetanomorphum. J. Biol. Chem. 1961, 236, PC40–PC42. [Google Scholar] [PubMed]
- Zhuang, W.-Q.; Yi, S.; Bill, M.; Brisson, V.L.; Feng, X.; Men, Y.; Conrad, M.E.; Tang, Y.J.; Alvarez-Cohen, L. Incomplete Wood–Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc. Natl. Acad. Sci. USA 2014, 111, 6419–6424. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, I.A.; Barnum, T.P.; Somasekhar, P.Y.; Carlström, C.I.; Engelbrektson, A.L.; Coates, J.D. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E92–E101. [Google Scholar] [CrossRef] [PubMed]
- Durán-Viseras, A.; Andrei, S.; Ghai, R.; Sánchez-Porro, C.; Ventosa, A. New Halonotius species provide genomics-based insights into cobalamin synthesis in haloarchaea. Front. Microbiol. 2019, 10, 1928. [Google Scholar] [CrossRef]
- Lu, X.; Heal, K.R.; Ingalls, A.E.; Doxey, A.C.; Neufeld, J.D. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 2019, 14, 53–66. [Google Scholar] [CrossRef]
- Roth, J.R.; Lawrence, J.; Bobik, T. Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol. 1996, 50, 137–181. [Google Scholar] [CrossRef]
- Romine, M.F.; Rodionov, D.A.; Maezato, Y.; Anderson, L.N.; Nandhikonda, P.; Rodionova, I.A.; Carre, A.; Li, X.; Xu, C.; Clauss, T.R. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc. Natl. Acad. Sci. USA 2017, 114, E1205–E1214. [Google Scholar] [CrossRef]
| Lake Strawbridge (LS) | Lake Whurr (LW) | |||||||
|---|---|---|---|---|---|---|---|---|
| LS1-TOP | LS2-TOP | LS1-BOT | LS2-BOT | LW1-TOP | LW2-TOP | LW1-BOT | LW2-BOT | |
| pH 1 | 8.2 | 8.3 | 8.5 | 8.5 | 5.4 | 5.4 | 4.5 | 4.6 |
| Water content (%) | 37.3 | 27.3 | 16.7 | 15.4 | 26.0 | 25.7 | 24.2 | 23.0 |
| Salinity (%) | 17 | 14 | 5 | 5 | 15 | 20 | 11 | 11 |
| TOC (g/kg dry sediment) | 21 | 15 | 5 | 5 | 12 | 14 | 6 | 6 |
| Na (mg/g dry sediment) | 57.0 | 48.5 | 17.5 | 18.1 | 55.0 | 71.1 | 35.0 | 35.8 |
| Ca (mg/g dry sediment) | 0.7 | 0.8 | 0.1 | 0.2 | 6.8 | 4.2 | 0.3 | 0.3 |
| K (mg/g dry sediment) | 2.0 | 2.0 | 1.0 | 0.9 | 1.7 | 1.8 | 1.1 | 1.2 |
| Mg (mg/g dry sediment) | 2.8 | 2.9 | 1.1 | 1.1 | 4.5 | 4.6 | 3.5 | 3.4 |
| Total Fe (mg/g dry sediment) | 6.5 | 6.3 | 2.2 | 1.9 | 1.5 | 3.2 | 0.3 | 0.6 |
| Cl- (mg/g dry sediment) | 101.3 | 84.7 | 31.9 | 33.1 | 93.1 | 123.5 | 64.8 | 64.0 |
| SO42- (mg/g dry sediment) | 3.9 | 3.6 | 1.5 | 1.8 | 19.6 | 14.8 | 4.3 | 4.4 |
| NO3- (mg/g dry sediment) | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| ClO3- (mg/g dry sediment) | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, P.; Lu, Y.; Bosma, T.N.P.; Nijenhuis, I.; Nijsse, B.; Shetty, S.A.; Ruecker, A.; Umanets, A.; Ramiro-Garcia, J.; Kappler, A.; et al. Metagenomic- and Cultivation-Based Exploration of Anaerobic Chloroform Biotransformation in Hypersaline Sediments as Natural Source of Chloromethanes. Microorganisms 2020, 8, 665. https://doi.org/10.3390/microorganisms8050665
Peng P, Lu Y, Bosma TNP, Nijenhuis I, Nijsse B, Shetty SA, Ruecker A, Umanets A, Ramiro-Garcia J, Kappler A, et al. Metagenomic- and Cultivation-Based Exploration of Anaerobic Chloroform Biotransformation in Hypersaline Sediments as Natural Source of Chloromethanes. Microorganisms. 2020; 8(5):665. https://doi.org/10.3390/microorganisms8050665
Chicago/Turabian StylePeng, Peng, Yue Lu, Tom N.P. Bosma, Ivonne Nijenhuis, Bart Nijsse, Sudarshan A. Shetty, Alexander Ruecker, Alexander Umanets, Javier Ramiro-Garcia, Andreas Kappler, and et al. 2020. "Metagenomic- and Cultivation-Based Exploration of Anaerobic Chloroform Biotransformation in Hypersaline Sediments as Natural Source of Chloromethanes" Microorganisms 8, no. 5: 665. https://doi.org/10.3390/microorganisms8050665
APA StylePeng, P., Lu, Y., Bosma, T. N. P., Nijenhuis, I., Nijsse, B., Shetty, S. A., Ruecker, A., Umanets, A., Ramiro-Garcia, J., Kappler, A., Sipkema, D., Smidt, H., & Atashgahi, S. (2020). Metagenomic- and Cultivation-Based Exploration of Anaerobic Chloroform Biotransformation in Hypersaline Sediments as Natural Source of Chloromethanes. Microorganisms, 8(5), 665. https://doi.org/10.3390/microorganisms8050665







