Siccibacter turicensis from Kangaroo Scats: Possible Implication in Cellulose Digestion
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Cellulose Degraders
2.2. MALDI-TOF Mass Spectroscopy Analysis
2.3. Partial 16S rRNA Gene Sequencing
2.4. Whole Genome Sequencing and Gene Annotation
2.5. Identification of the Cellulose Degrading Microorganism Using Genome Information
2.6. Analysis of Annotated Protein Sequences for Presence of Cellulose Operon Components
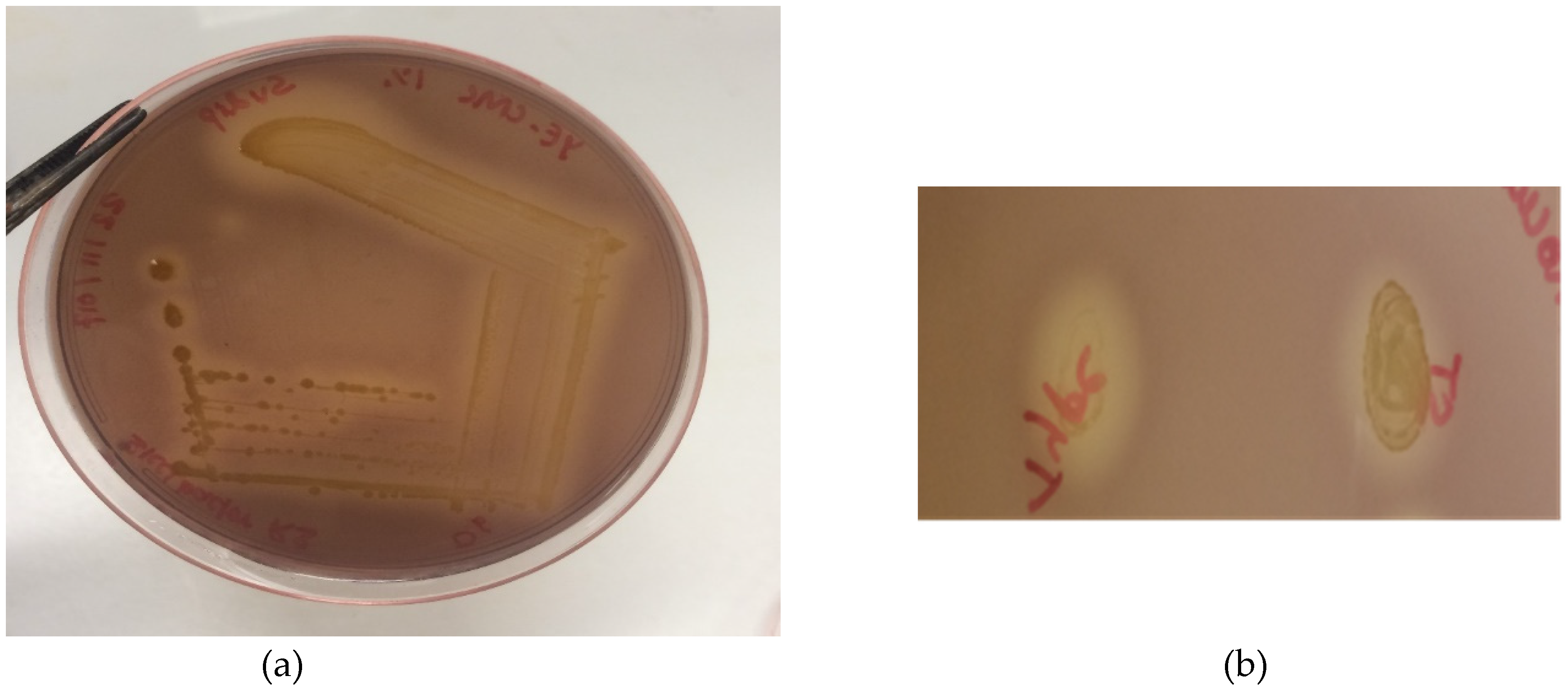
2.7. Cellulase Activity Assay
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barker, C.J.; Gillett, A.; Polkinghorne, A.; Timms, P. Investigation of the koala (Phascolarctos cinereus) hindgut microbiome via 16S pyrosequencing. Vet. Microbiol. 2013, 167, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Arman, S.D.; Prideaux, G.J. Dietary classification of extant kangaroos and their relatives (Marsupialia: Macropodoidea). Austral Ecol. 2015, 40, 909–922. [Google Scholar] [CrossRef]
- Meng, F.; Ma, L.; Ji, S.; Yang, W.; Cao, B. Isolation and characterization of Bacillus subtilis strain BY-3, a thermophilic and efficient cellulase-producing bacterium on untreated plant biomass. Lett. Appl. Microbiol. 2014, 59, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Poomai, N.; Siripornadulsil, W.; Siripornadulsil, S. Cellulase enzyme production from agricultural waste by Acinetobacter sp. KKU44. Adv. Mater. Res. 2014, 931–932, 1106–1110. [Google Scholar] [CrossRef]
- Wang, D.; Wang, Z.; He, F.; Kinchla, A.J.; Nugen, S.R. Enzymatic digestion for improved bacteria separation from leafy green vegetables. J. Food Prot. 2016, 79, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Xing, T.; Wu, N.; Xu, X.; Zhou, G. Removal of Salmonella biofilm formed under meat processing environment by surfactant in combination with bio-enzyme. LWT-Food Sci. Technol. 2016, 66, 298–304. [Google Scholar] [CrossRef]
- Zulkarnain, D.; Zuprizal, Z.; Wihandoyo, W.; Supadmo, S. Effect of cellulase supplementation on in vitro digestibility and energy, crude fiber and cellulose content of sago palm (Metroxylon sp.) waste as broiler chicken feed. Pak. J. Nutr. 2016, 15, 997–1002. [Google Scholar] [CrossRef][Green Version]
- Ghodadara, S.; Shilpkar, P.; Dungrechiya, A. Optimized production of cellulase by Aspergillus niger using ricinus communis seed coat waste. J. Pure Appl. Microbiol. 2015, 9, 623–629. [Google Scholar]
- Khan, M.N.; Luna, I.Z.; Islam, M.M.; Sharmeen, S.; Salem, K.S.; Rashid, T.U.; Zaman, A.; Haque, P.; Rahman, M.M. Cellulase in Waste Management Applications. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Cellulase System Properties and Applications; Elsevier: Amsterdam, The Netherlands, 2016; pp. 237–256. [Google Scholar] [CrossRef]
- Pawar, K.D.; Dar, M.A.; Rajput, B.P.; Kulkarni, G.J. Enrichment and identification of cellulolytic bacteria from the gastrointestinal tract of giant african snail, Achatina fulica. Appl. Biochem. Biotechnol. 2014, 175, 1971–1980. [Google Scholar] [CrossRef]
- Thongekkaew, J.; Patangtasa, W.; Jansri, A. Cellulase and xylanase production from Candida easanensis using agricultural wastes as a substrate. Songklanakarin J. Sci. Technol. 2014, 36, 607–613. [Google Scholar]
- Wong, H.C.; Fear, A.L.; Calhoon, R.D.; Eichinger, G.H.; Mayer, R.; Amikam, D.; Benziman, M.; Gelfand, D.H.; Meade, J.H.; Emerick, A.W. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 1990, 87, 8130. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Boath, J.M.; Dakhal, S.; Van, T.T.H.; Moore, R.J.; Dekiwadia, C.; Macreadie, I.G. Polyphasic characterisation of Cedecea colo sp. Nov., a new enteric bacterium isolated from the koala hindgut. Microorganisms 2020, 8, 309. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics (Oxf. Engl.) 2015, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- de Jong, A.; Pietersma, H.; Cordes, M.; Kuipers, O.P.; Kok, J. PePPER: A webserver for prediction of prokaryote promoter elements and regulons. BMC Genom. 2012, 13, 299. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Nova Science Publisher’s: Hauppauge, NY, USA, 2011; pp. 62–78. [Google Scholar]
- Stephan, R.; Van Trappen, S.; Cleenwerck, I.; Vancanneyt, M.; De Vos, P.; Lehner, A. Enterobacter turicensis sp. nov. and Enterobacter helveticus sp. nov., isolated from fruit powder. Int. J. Syst. Evol. Microbiol. 2007, 57, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Lepuschitz, S.; Pekard-Amenitsch, S.; Haunold, R.; Schill, S.; Schriebl, A.; Mach, R.; Allerberger, F.; Ruppitsch, W.; Forsythe, S.J. Draft genome sequence of the first documented clinical Siccibacter turicensis isolate in Austria. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Jackson, E.E.; Sonbol, H.; Masood, N.; Forsythe, S.J. Genotypic and phenotypic characteristics of Cronobacter species, with particular attention to the newly reclassified species Cronobacter helveticus, Cronobacter pulveris, and Cronobacter zurichensis. Food Microbiol. 2014, 44, 226–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stephan, R.; Grim, C.J.; Gopinath, G.R.; Mammel, M.K.; Sathyamoorthy, V.; Trach, L.H.; Chase, H.R.; Fanning, S.; Tall, B.D. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2014, 64, 3402–3410. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Rouf, S.F.; Sun, L.; Cimdins, A.; Shafeeq, S.; Guyon, S.; Schottkowski, M.; Rhen, M.; Römling, U. BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium. Microb. Cell Factories 2016, 15. [Google Scholar] [CrossRef]
- Solano, C.; García, B.; Valle, J.; Berasain, C.; Ghigo, J.M.; Gamazo, C.; Lasa, I. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 2002, 43, 793–808. [Google Scholar] [CrossRef]
- Morgan, J.L.W.; Strumillo, J.; Zimmer, J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 2013, 493, 181–186. [Google Scholar] [CrossRef]
- Sun, L.; Vella, P.; Schnell, R.; Polyakova, A.; Bourenkov, G.; Li, F.; Cimdins, A.; Schneider, T.R.; Lindqvist, Y.; Galperin, M.Y.; et al. Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella Typhimurium. J. Mol. Biol. 2018, 430, 3170–3189. [Google Scholar] [CrossRef] [PubMed]
- Yeoman, C.J.; Han, Y.; Dodd, D.; Schroeder, C.M.; Mackie, R.I.; Cann, I.K.O. Chapter 1-Thermostable Enzymes as Biocatalysts in the Biofuel Industry. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2010; Volume 70, pp. 1–55. [Google Scholar]
- Ryjenkov, D.A.; Tarutina, M.; Moskvin, O.V.; Gomelsky, M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005, 187, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Sarenko, O.; Klauck, G.; Wilke, F.M.; Pfiffer, V.; Richter, A.M.; Herbst, S.; Kaever, V.; Hengge, R. More than Enzymes That Make or Break Cyclic Di-GMP—Local Signaling in the Interactome of GGDEF/EAL Domain Proteins of Escherichia coli. mBio 2017, 8, e01639-17. [Google Scholar] [CrossRef] [PubMed]
- Bobrov, A.G.; Kirillina, O.; Perry, R.D. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia Pestis. FEMS Microbiol. Lett. 2005, 247, 123–130. [Google Scholar] [CrossRef] [PubMed][Green Version]

| Highly Similar Organism (16S rRNA Sequences) | Raw Score | Query Cover (%) | Identity (%) | E-Value | NCBI Accession Number |
|---|---|---|---|---|---|
| Cronobacter zurichensis strain LMG 23730 (Siccibacter turicensis) | 2627 | 98 | 99 | 0.0 | NR_104924.1 |
| Erwinia tasmaniensis strain Et1/99 | 2505 | 99 | 98 | 0.0 | NR_074869.1 |
| Yokenella regensburgei strain CIP 105435 | 2488 | 99 | 98 | 0.0 | NR_104934.1 |
| Escherichia hermannii strain CIP 103176 | 2475 | 99 | 98 | 0.0 | NR_104940.1 |
| Pantoea agglomerans strain NCTC9381 | 2471 | 99 | 97 | 0.0 | NR_114735.1 |
| Whole Genome | Average Nucleotide Identity (ANI) | |
|---|---|---|
| Two-Way ANI (%) | Standard Deviation | |
| Siccibacter turicensis LMG 23730 | 97.98 | 1.41 |
| Siccibacter colletis | 84.99 | 4.26 |
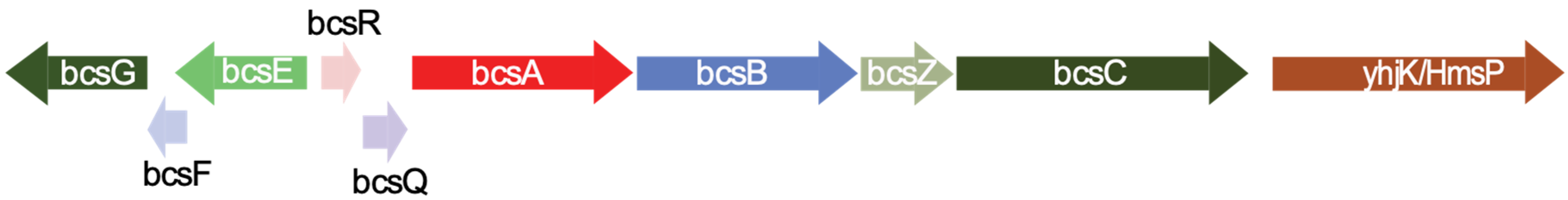
| Protein Names from RAST Annotation | Annotation from RAST Tool | Protein Family | Length (aa) | Most similar Protein in NCBI Database (BLASTP) | Accession Number |
|---|---|---|---|---|---|
| Prot_03025 scaffold_4_205556_203871 | FIG002337: predicted inner membrane protein | BcsG | 561 | Cellulose biosynthesis protein BcsG [S. turicensis] | WP_106877578.1 |
| Prot_03026 scaffold_4_205743_205549 | Not annotated | BcsF | 64 | BcsF [S. turicensis] | WP_031521981.1 |
| Prot_03027 scaffold_4_207095_205740 | FIG005274: hypothetical protein | BcsE | 415 | Cellulose biosynthesis protein BcsE [S. turicensis] | WP_106877577.1 |
| Prot_03028 scaffold_4_207471_207656 | FIG004405: putative cytoplasmic protein | BcsR * | 61 | Hypothetical protein from Enterobacteriaceae | WP_024549966.1 |
| Prot_03029 scaffold_4_207668_208402 | Cellulose synthase, putative | BcsQ | 244 | Cellulose biosynthesis protein BcsQ [S. turicensis] | WP_106877576.1 |
| Prot_03030 scaffold_4_208399_211017 | UDP-forming cellulose synthase catalytic subunit | BcsA | 872 | UDP-forming cellulose synthase catalytic subunit [S. turicensis] | WP_106877575.1 |
| Prot_03031 scaffold_4_211028_213472 | Cyclic di-GMP binding protein precursor | BcsB | 814 | Cellulose biosynthesis cyclic di-GMP-binding regulatory protein BcsB [S. turicensis] | WP_106877574.1 |
| Prot_03032 scaffold_4_213483_214586 | Endo-1,4-β-D-glucanase | BcsZ | 367 | Endoglucanase [S. turicensis] | WP_106877573.1 |
| Prot_03033 scaffold_4_214568_218068 | Cellulose synthase operon protein C | BcsC | 1166 | Cellulose biosynthesis protein BcsC [S. turicensis] | WP_106877572.1 |
| Prot_03034 scaffold_4_218174_220189 | Protein yhjK | GAPES3/GGDEF/EAL | 671 | Biofilm formation regulator HmsP [S. turicensis] | WP_106877571.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhakal, S.; Boath, J.M.; Van, T.T.H.; Moore, R.J.; Macreadie, I.G. Siccibacter turicensis from Kangaroo Scats: Possible Implication in Cellulose Digestion. Microorganisms 2020, 8, 635. https://doi.org/10.3390/microorganisms8050635
Dhakal S, Boath JM, Van TTH, Moore RJ, Macreadie IG. Siccibacter turicensis from Kangaroo Scats: Possible Implication in Cellulose Digestion. Microorganisms. 2020; 8(5):635. https://doi.org/10.3390/microorganisms8050635
Chicago/Turabian StyleDhakal, Sudip, Jarryd M. Boath, Thi Thu Hao Van, Robert J. Moore, and Ian G. Macreadie. 2020. "Siccibacter turicensis from Kangaroo Scats: Possible Implication in Cellulose Digestion" Microorganisms 8, no. 5: 635. https://doi.org/10.3390/microorganisms8050635
APA StyleDhakal, S., Boath, J. M., Van, T. T. H., Moore, R. J., & Macreadie, I. G. (2020). Siccibacter turicensis from Kangaroo Scats: Possible Implication in Cellulose Digestion. Microorganisms, 8(5), 635. https://doi.org/10.3390/microorganisms8050635







