Taxogenomics of the Genus Cyclobacterium: Cyclobacterium xiamenense and Cyclobacterium halophilum as Synonyms and Description of Cyclobacterium plantarum sp. nov.
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Taxophylogenomic Characterization
2.2.1. DNA Extraction, Purification, and Sequencing
2.2.2. Phylogenetic Analysis Based on 16S rRNA Gene Sequence Comparison
2.2.3. Genome Assembly and Annotation
2.2.4. Phylogenomic Comparative Analysis
2.2.5. In Silico DNA–DNA Hybridization (GGDC), Average Nucleotide Identity (ANI), and Average Amino Acid Identity (AAI)
2.3. Phenotypic Characterization
2.4. Antimicrobial Susceptibility
2.5. Chemotaxonomic Characterization
3. Results and Discussion
3.1. Phylogenetic Analysis Based on 16S rRNA Gene Sequence Comparison
3.2. Phylogenomic Comparative Analysis
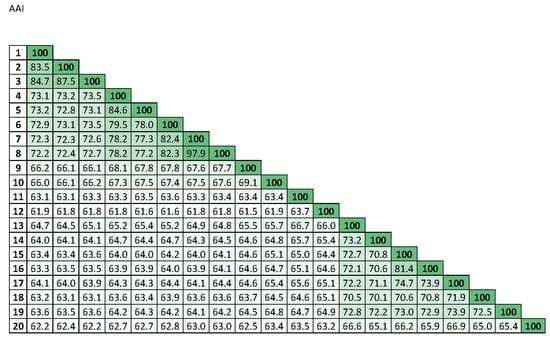
3.3. in silico DNA–DNA Hybridization (GGDC), ANI, and AAI Values
3.4. Phenotypic Characterization
3.5. Chemotaxonomic Characterization
4. Conclusions
4.1. Description of Cyclobacterium plantarum sp. nov.
4.2. Emended description of Cyclobacterium xiamenense Chen et al. 2014
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Parte, A.C. LPSN—List of Prokaryotic Names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Raj, H.D.; Maloy, S.R. Proposal of Cyclobacterium marinus gen. nov., comb. nov. for a marine bacterium previously assigned to the genus Flectobacillus. Int. J. Syst. Bacteriol. 1990, 40, 337–347. [Google Scholar] [CrossRef]
- Ying, J.Y.; Wang, B.J.; Yang, S.S.; Liu, S.J. Cyclobacterium lianum sp. nov., a marine bacterium isolated from sediment of an oilfield in the South China Sea, and emended description of the genus Cyclobacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 2927–2930. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.T.; Lee, J.S.; Yoon, J.H. Cyclobacterium caenipelagi sp. nov. isolated from a tidal flat sediment, and emended description of the genus Cyclobacterium. Int. J. Syst. Evol. Microbiol. 2013, 63, 3158–3163. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yang, L.; Li, Y.; Lai, Q.; Zhang, H.; Wei, J.; Zhou, Y.; Lei, X.; Zheng, W.; Tian, Y.; et al. Cyclobacterium xiamenense sp. nov., isolated from aggregates of Chlorella autotrophica, and emended description of the genus Cyclobacterium. Int. J. Syst. Evol. Microbiol. 2014, 64, 887–893. [Google Scholar] [CrossRef]
- Raj, H.D. A new species: Microcyclus marinus. Int. J. Syst. Bacteriol. 1976, 26, 528–544. [Google Scholar] [CrossRef]
- Borrall, R.; Larkin, J.M. Flectobacillus marinus (Raj) comb. nov., a marine bacterium previously assigned to Microcyclus. Int. J. Syst. Bacteriol. 1978, 28, 341–343. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.I.; Kim, S.B.; Lee, M.S.; Park, M.S.; Lee, K.H.; Lysenko, A.M.; Oh, H.W.; Mikhailov, V.V.; Bae, K.S. Cyclobacterium amurskyense sp. nov., a novel marine bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol. 2005, 55, 2391–2394. [Google Scholar] [CrossRef]
- Shivaji, S.; Reddy, P.V.; Rao, S.S.; Begum, Z.; Manasa, P.; Srinivas, T.N. Cyclobacterium qasimii sp. nov., a psychrotolerant bacterium isolated from Arctic marine sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 2133–2139. [Google Scholar] [CrossRef]
- Joung, Y.; Kim, H.; Kim, S.B.; Joh, K. Cyclobacterium jeungdonense sp. nov., isolated from a solar saltern. Int. J. Syst. Evol. Microbiol. 2014, 64, 11–15. [Google Scholar] [CrossRef]
- Shahinpei, A.; Amoozegar, M.A.; Sepahy, A.A.; Schumann, P.; Ventosa, A. Cyclobacterium halophilum sp. nov., a marine bacterium isolated from a coastal-marine wetland. Int. J. Syst. Evol. Microbiol. 2014, 64, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Kahng, H.Y. Cyclobacterium sediminis sp. nov. isolated from a sea cucumber aquaculture farm and emended description of the genus Cyclobacterium. J. Microbiol. 2017, 55, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Garrido, J.F.; Ramírez-Saad, H.C.; Toro, N.; Martínez-Abarca, F. Bacterial diversity in the soda saline crater lake from Isabel Island, Mexico. Microb. Ecol. 2016, 71, 68–77. [Google Scholar] [CrossRef]
- Van Trappen, S.; Mergaert, J.; Van Eygen, S.; Dawyndt, P.; Cnockaert, M.C.; Swings, J. Diversity of 746 heterotrophic bacteria isolated from microbial mats from ten Antarctic lakes. Syst. Appl. Microbiol. 2002, 25, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Stougaard, P.; Jørgensen, F.; Johnsen, M.G.; Hansen, O.-C. Microbial diversity in ikaite tufa columns: An alkaline, cold ecological niche in Greenland. Environ. Microbiol. 2002, 4, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Saba, F.; Noroozi, M.; Ghahremaninejad, F.; Sedghi, M.; Papizadeh, M. Isolation, purification and identification of three diatom species (Bacillariophyceae) from Gomishan wetland (N. Iran) using phylogeny and silica cell wall ultra-structure analysis. Rostaniha 2016, 17, 28–39. [Google Scholar]
- Marmur, J.A. Procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A laboratory Manual; Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.J. The clustral X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Rzhetsky, A.; Nei, M. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 1992, 9, 945–967. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Rodríguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2002, 32, 1792–1797. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Shimodaira, H.; Hasegawa, M. Multiple comparisons of loglikelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.G.E.; Doetsch, R.N.; Robinow, C.F. Determinative and cytological light microscopy. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 21–41. [Google Scholar]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Gutiérrez, C.; González, C. Method for simultaneous detection of proteinase and esterase in extremely halophilic bacteria. Appl. Microbiol. 1972, 24, 516–517. [Google Scholar] [CrossRef] [PubMed]
- Mata, J.A.; Martínez-Cánovas, J.; Quesada, E.; Béjar, V. A detailed phenotypic characterization of the type strains of Halomonas species. Syst. Appl. Microbiol. 2002, 25, 360–375. [Google Scholar] [CrossRef]
- Montes, M.J.; Bozal, N.; Mercade, E. Marinobacter guineae sp. nov., a novel moderately halophilic bacterium from an Antarctic environment. Int. J. Syst. Evol. Microbiol. 2008, 58, 1346–1349. [Google Scholar] [CrossRef]
- Ventosa, A.; Quesada, E.; Rodríguez-Valera, F.; Ruiz-Berraquero, F.; Ramos-Cormenzana, A. Numerical taxonomy of moderately halophilic Gram-negative rods. J. Gen. Microbiol. 1982, 128, 1959–1968. [Google Scholar]
- Kämpfer, P.; Kroppenstedt, R.M. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 1996, 42, 989–1005. [Google Scholar] [CrossRef]
- Growth, I.; Schumann, P.; Weiss, N.; Martin, K.; Rainey, F.A. Agrococcus jenensis gen. nov., sp. nov., a new genus of actinomycetes with diaminobutyric acid in the cell wall. Int. J. Syst. Bacteriol. 1996, 46, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Bacteriol. 2014, 64, 346. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 3160–3165. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef]
- Rodríguez-R, L.M.; Konstantinidis, K.T. Bypassing cultivation to identify bacterial species. Microbe 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Parker, C.T.; Tindall, B.J.; Garrity, G.M. International Code of Nomenclature of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2019, 69, S1–S111. [Google Scholar]


| Feature | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Size (bp) | 6,169,285 | 6,158,829 | 5,662,104 | 5,792,371 | 5,675,162 | 6,221,270 | 6,291,928 | 5,784,474 |
| Contigs | 37 | 1 | 30 | 41 | 31 | 1 | 202 | 98 |
| Genome coverage | 193X | 101X | 175X | 100X | 247X | 30X | 240X | 100X |
| G+C (mol%) | 43.0 | 38.3 | 48.4 | 44.0 | 45.5 | 38.1 | 38.8 | 48.5 |
| N50 (bp) | 547,880 | 6,158,829 | 350,204 | 381,560 | 266,214 | 6,221,273 | 107,474 | 137,064 |
| Total genes | 4943 | 4833 | 4689 | 4646 | 4736 | 4981 | 5997 | 4595 |
| Protein coding genes | 4818 | 4715 | 4635 | 4534 | 4687 | 4868 | 5958 | 4474 |
| rRNA | 6 | 12 | 5 | 7 | 5 | 9 | 4 | 7 |
| tRNA | 41 | 39 | 38 | 40 | 38 | 39 | 35 | 39 |
| Accession number | JAANYN000000000 | CP012040 | FNZH00000000 | WMCD00000000 | FRCY00000000 | NC_015914 | ATNM00000000 | WIOK00000000 |

| OrthoANI | |||||||||||||||||||||
| GGDC | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| 1 | 100 | 78.1 | 80.0 | 71.8 | 71.4 | 71.2 | 70.6 | 70.5 | 69.3 | 68.5 | 68.5 | 68.2 | 69.7 | 68.8 | 68.7 | 69.0 | 68.4 | 67.8 | 69.1 | 67.8 | |
| 2 | 26.9 | 100 | 82.4 | 72.0 | 71.1 | 71.4 | 70.6 | 70.5 | 68.8 | 68.1 | 68.3 | 68.4 | 69.6 | 68.6 | 68.7 | 69.0 | 68.6 | 67.7 | 68.9 | 67.6 | |
| 3 | 31.3 | 44.4 | 100 | 72.1 | 71.1 | 71.5 | 70.7 | 70.6 | 69.0 | 68.2 | 68.2 | 68.1 | 69.3 | 68.8 | 68.7 | 69.0 | 68.6 | 67.6 | 69.1 | 67.6 | |
| 4 | 14.3 | 14.6 | 14.5 | 100 | 79.2 | 76.0 | 74.3 | 74.1 | 69.9 | 69.1 | 68.4 | 67.7 | 69.0 | 69.0 | 68.7 | 68.7 | 68.9 | 68.0 | 69.0 | 67.5 | |
| 5 | 14.1 | 13.8 | 14.1 | 35.0 | 100 | 74.1 | 73.5 | 73.4 | 69.6 | 69.2 | 68.0 | 67.4 | 69.3 | 68.6 | 68.2 | 68.4 | 68.5 | 67.9 | 68.9 | 67.2 | |
| 6 | 14.2 | 14.3 | 14.4 | 20.8 | 17.8 | 100 | 76.6 | 76.4 | 69.3 | 69.2 | 68.0 | 67.5 | 68.6 | 68.8 | 68.3 | 68.2 | 68.7 | 67.9 | 68.7 | 67.3 | |
| 7 | 13.9 | 13.9 | 14 | 17.4 | 16.9 | 24.2 | 100 | 97.8 | 69.4 | 69.3 | 68.1 | 67.1 | 68.1 | 68.3 | 67.9 | 67.9 | 68.2 | 68.0 | 68.5 | 67.6 | |
| 8 | 14.0 | 13.9 | 14.1 | 17.4 | 16.7 | 23.9 | 81.6 | 100 | 69.2 | 69.2 | 68.0 | 67.1 | 68.3 | 68.2 | 68.1 | 68.2 | 68.1 | 67.7 | 68.7 | 67.6 | |
| 9 | 12.9 | 12.8 | 12.8 | 13 | 13 | 12.9 | 12.9 | 12.9 | 100 | 69.6 | 67.3 | 67.2 | 68.5 | 68.5 | 68.6 | 68.4 | 68.7 | 67.7 | 69.0 | 67.1 | |
| 10 | 12.9 | 12.8 | 12.9 | 12.9 | 12.8 | 12.9 | 13.1 | 13.0 | 13.1 | 100 | 68.0 | 67.4 | 68.8 | 68.3 | 68.9 | 68.3 | 69.2 | 68.1 | 68.9 | 67.8 | |
| 11 | 12.7 | 12.8 | 12.8 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 100 | 68.7 | 69.5 | 68.6 | 68.9 | 69.0 | 69.0 | 68.3 | 69.0 | 68.1 | |
| 12 | 12.7 | 12.7 | 12.7 | 12.6 | 12.5 | 12.6 | 12.6 | 12.6 | 12.6 | 12.6 | 12.7 | 100 | 69.7 | 69.0 | 68.7 | 68.9 | 68.6 | 68.0 | 69.0 | 67.5 | |
| 13 | 12.8 | 12.8 | 12.9 | 12.8 | 12.9 | 12.8 | 12.7 | 12.7 | 12.8 | 12.8 | 12.8 | 13.0 | 100 | 71.7 | 72.5 | 71.7 | 71.5 | 69.7 | 72.4 | 69.3 | |
| 14 | 12.8 | 12.8 | 12.8 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 12.8 | 12.8 | 13.6 | 100 | 70.9 | 70.7 | 70.8 | 70.0 | 71.7 | 68.1 | |
| 15 | 12.8 | 12.8 | 12.8 | 12.8 | 12.8 | 12.7 | 12.8 | 12.8 | 12.7 | 12.9 | 12.7 | 12.8 | 14.1 | 13.4 | 100 | 75.7 | 72.7 | 70.1 | 72.5 | 68.4 | |
| 16 | 12.8 | 12.7 | 12.8 | 12.7 | 12.7 | 12.7 | 12.8 | 12.8 | 12.7 | 12.7 | 12.9 | 12.8 | 13.6 | 13.2 | 21.2 | 100 | 72.2 | 69.9 | 72.4 | 68.2 | |
| 17 | 12.7 | 12.7 | 12.7 | 12.7 | 12.7 | 12.8 | 12.7 | 12.7 | 12.8 | 12.8 | 12.9 | 12.7 | 13.4 | 13.2 | 14.5 | 14.1 | 100 | 70.6 | 72.5 | 69.1 | |
| 18 | 12.7 | 12.7 | 12.6 | 12.7 | 12.6 | 12.7 | 12.7 | 12.6 | 12.7 | 12.7 | 12.8 | 12.7 | 12.9 | 12.9 | 13.1 | 13.2 | 12.8 | 100 | 70.9 | 68.1 | |
| 19 | 12.8 | 12.7 | 12.7 | 12.8 | 12.9 | 12.7 | 12.7 | 12.7 | 12.8 | 12.8 | 12.8 | 12.7 | 13.6 | 13.2 | 13.8 | 13.8 | 13.9 | 13.3 | 100 | 68.4 | |
| 20 | 12.7 | 12.7 | 12.7 | 12.7 | 12.6 | 12.7 | 12.7 | 12.7 | 12.6 | 12.7 | 12.8 | 12.7 | 13.1 | 12.8 | 12.8 | 12.8 | 13.1 | 12.7 | 12.8 | 100 | |
| 1 | 100 | Percentages of similarity | ||||||||||||||||||
| 2 | 83.5 | 100 |  | |||||||||||||||||
| 3 | 84.7 | 87.5 | 100 | |||||||||||||||||
| 4 | 73.1 | 73.2 | 73.5 | 100 | ||||||||||||||||
| 5 | 73.2 | 72.8 | 73.1 | 84.6 | 100 | |||||||||||||||
| 6 | 72.9 | 73.1 | 73.5 | 79.5 | 78.0 | 100 | ||||||||||||||
| 7 | 72.3 | 72.3 | 72.6 | 78.2 | 77.3 | 82.4 | 100 | |||||||||||||
| 8 | 72.2 | 72.4 | 72.7 | 78.2 | 77.2 | 82.3 | 97.9 | 100 | ||||||||||||
| 9 | 66.2 | 66.1 | 66.1 | 68.1 | 67.8 | 67.8 | 67.6 | 67.7 | 100 | |||||||||||
| 10 | 66.0 | 66.1 | 66.2 | 67.3 | 67.5 | 67.4 | 67.5 | 67.6 | 69.1 | 100 | ||||||||||
| 11 | 63.1 | 63.1 | 63.3 | 63.3 | 63.5 | 63.6 | 63.3 | 63.4 | 63.4 | 63.4 | 100 | |||||||||
| 12 | 61.9 | 61.8 | 61.8 | 61.8 | 61.6 | 61.6 | 61.8 | 61.8 | 61.5 | 61.9 | 63.7 | 100 | ||||||||
| 13 | 64.7 | 64.5 | 65.1 | 65.2 | 65.4 | 65.2 | 64.9 | 64.8 | 65.5 | 65.7 | 66.7 | 66.0 | 100 | |||||||
| 14 | 64.0 | 64.1 | 64.1 | 64.7 | 64.4 | 64.7 | 64.3 | 64.5 | 64.6 | 64.8 | 65.7 | 65.4 | 73.2 | 100 | ||||||
| 15 | 63.4 | 63.4 | 63.6 | 64.0 | 64.0 | 64.2 | 64.0 | 64.1 | 64.6 | 65.1 | 65.0 | 64.4 | 72.7 | 70.8 | 100 | |||||
| 16 | 63.3 | 63.5 | 63.5 | 63.9 | 63.9 | 64.0 | 63.9 | 64.1 | 64.6 | 64.7 | 65.1 | 64.6 | 72.1 | 70.6 | 81.4 | 100 | ||||
| 17 | 64.1 | 64.0 | 63.9 | 64.3 | 64.3 | 64.4 | 64.1 | 64.4 | 64.6 | 65.4 | 65.6 | 65.1 | 72.2 | 71.1 | 74.7 | 73.9 | 100 | |||
| 18 | 63.2 | 63.1 | 63.1 | 63.6 | 63.4 | 63.9 | 63.6 | 63.6 | 63.7 | 64.5 | 64.6 | 65.1 | 70.5 | 70.1 | 70.6 | 70.8 | 71.9 | 100 | ||
| 19 | 63.6 | 63.5 | 63.6 | 64.2 | 64.3 | 64.2 | 64.1 | 64.2 | 64.5 | 64.8 | 64.7 | 64.9 | 72.8 | 72.2 | 73.0 | 72.9 | 73.9 | 72.5 | 100 | |
| 20 | 62.2 | 62.4 | 62.2 | 62.7 | 62.7 | 62.8 | 63.0 | 63.0 | 62.5 | 63.4 | 63.5 | 63.2 | 66.6 | 65.1 | 66.2 | 65.9 | 66.9 | 65.0 | 65.4 | 100 |
| Characteristic | 1 | 2 | 3 | 4 | 5 * |
|---|---|---|---|---|---|
| Cell size (µm) | |||||
| Outer diameter-length | 0.8–1.9 | 1.5–1.8 | 1.5–1.8 | 0.8–1.7 | 1.5–2.0 |
| Width | 0.3–0.5 | 0.4–0.5 | 0.3–0.5 | 0.4–0.6 | 0.4–0.6 |
| Salinity range (% [w/v] NaCl) | 3–10 | 0.1–12 | 0–7 | 1–10 | 3–9 |
| Growth temperature (°C): | |||||
| Range | 4–40 | 15–40 | 15–35 | 4–35 | 4–40 |
| Optimum | 25 | 30 | 25 | 25 | 28 |
| pH growth range | 6.5–9.0 | 6.5–9.0 | 7.0–8.0 | 6.0–9.0 | 6.0–10.0 |
| Nitrate reduction | + | - | + | - | - |
| Hydrolysis of: | |||||
| Aesculin | + | + | + | - | + |
| Tween 20 | - | + | - | - | + |
| Acid production from: | |||||
| D-Arabinose | - | + | - | - | ND |
| D-Glucose | - | + | + | + | ND |
| Starch | - | + | + | - | ND |
| D-Xylose | - | + | - | - | ND |
| Utilization of: | |||||
| Cellobiose | + | + | - | - | ND |
| D-Mannose | + | + | - | - | w |
| myo-Inositol | + | - | - | - | - |
| L-Glutamic acid | - | + | - | - | ND |
| L-alanine | + | - | - | + | ND |
| DNA G+C content (mol%)† | 43.0 | 45.4 | 45.6 | 48.4 | 48.5 |
| Fatty Acid | 1 | 2 | 3 |
|---|---|---|---|
| iso-C15:1 G | 1.0 | - | 2.7 |
| iso-C15:0 | 26.3 | 29.1 | 34.6 |
| anteiso-C15:0 | 12.1 | 9.9 | 8.8 |
| C16:1ω5c | - | 5.3 | 2.3 |
| iso-C15:0 3-OH | 2.7 | 4.2 | 1.6 |
| iso-C17:1ω9c | 9.6 | 8.1 | 12.3 |
| C17:1ω6c | 1.2 | 2.1 | 3.8 |
| C16:0 3-OH | 1.0 | 1.2 | - |
| iso-C17:0 3-OH | 12.5 | 10.3 | 7.4 |
| C17:0 2-OH | 3.6 | 1.0 | 4.2 |
| Summed feature 3 | 23.9 | 24.5 | 17.7 |
| Summed feature 4 | 3.4 | 2.1 | 3.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahinpei, A.; Amoozegar, M.A.; Mirfeizi, L.; Nikou, M.M.; Ventosa, A.; Sánchez-Porro, C. Taxogenomics of the Genus Cyclobacterium: Cyclobacterium xiamenense and Cyclobacterium halophilum as Synonyms and Description of Cyclobacterium plantarum sp. nov. Microorganisms 2020, 8, 610. https://doi.org/10.3390/microorganisms8040610
Shahinpei A, Amoozegar MA, Mirfeizi L, Nikou MM, Ventosa A, Sánchez-Porro C. Taxogenomics of the Genus Cyclobacterium: Cyclobacterium xiamenense and Cyclobacterium halophilum as Synonyms and Description of Cyclobacterium plantarum sp. nov. Microorganisms. 2020; 8(4):610. https://doi.org/10.3390/microorganisms8040610
Chicago/Turabian StyleShahinpei, Azadeh, Mohammad Ali Amoozegar, Leila Mirfeizi, Mahdi Moshtaghi Nikou, Antonio Ventosa, and Cristina Sánchez-Porro. 2020. "Taxogenomics of the Genus Cyclobacterium: Cyclobacterium xiamenense and Cyclobacterium halophilum as Synonyms and Description of Cyclobacterium plantarum sp. nov." Microorganisms 8, no. 4: 610. https://doi.org/10.3390/microorganisms8040610
APA StyleShahinpei, A., Amoozegar, M. A., Mirfeizi, L., Nikou, M. M., Ventosa, A., & Sánchez-Porro, C. (2020). Taxogenomics of the Genus Cyclobacterium: Cyclobacterium xiamenense and Cyclobacterium halophilum as Synonyms and Description of Cyclobacterium plantarum sp. nov. Microorganisms, 8(4), 610. https://doi.org/10.3390/microorganisms8040610