Comparative Transcriptomic Analysis Uncovers Genes Responsible for the DHA Enhancement in the Mutant Aurantiochytrium sp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Cultivation
2.2. UV-Mediated Mutagenesis
2.3. Biomass Determination
2.4. Fatty Acid Extraction
2.5. Fatty Acid Structure and Composition Analysis
2.5.1. Fourier Transform Infrared (FTIR) Spectrometer Analysis
2.5.2. Gas Chromatography and Mass Spectrometry (GC/MS) Analysis
2.6. Comparative Transcriptomic Analysis
2.6.1. RNA Extraction and cDNA Library Construction
2.6.2. Illumina Sequencing, Assembly, and Annotation
2.6.3. qRT-PCR Analysis
2.6.4. Statistical Analysis
3. Results
3.1. Cell Mutagenesis
3.2. Screening of the Mutant Aurantiochytrium sp.
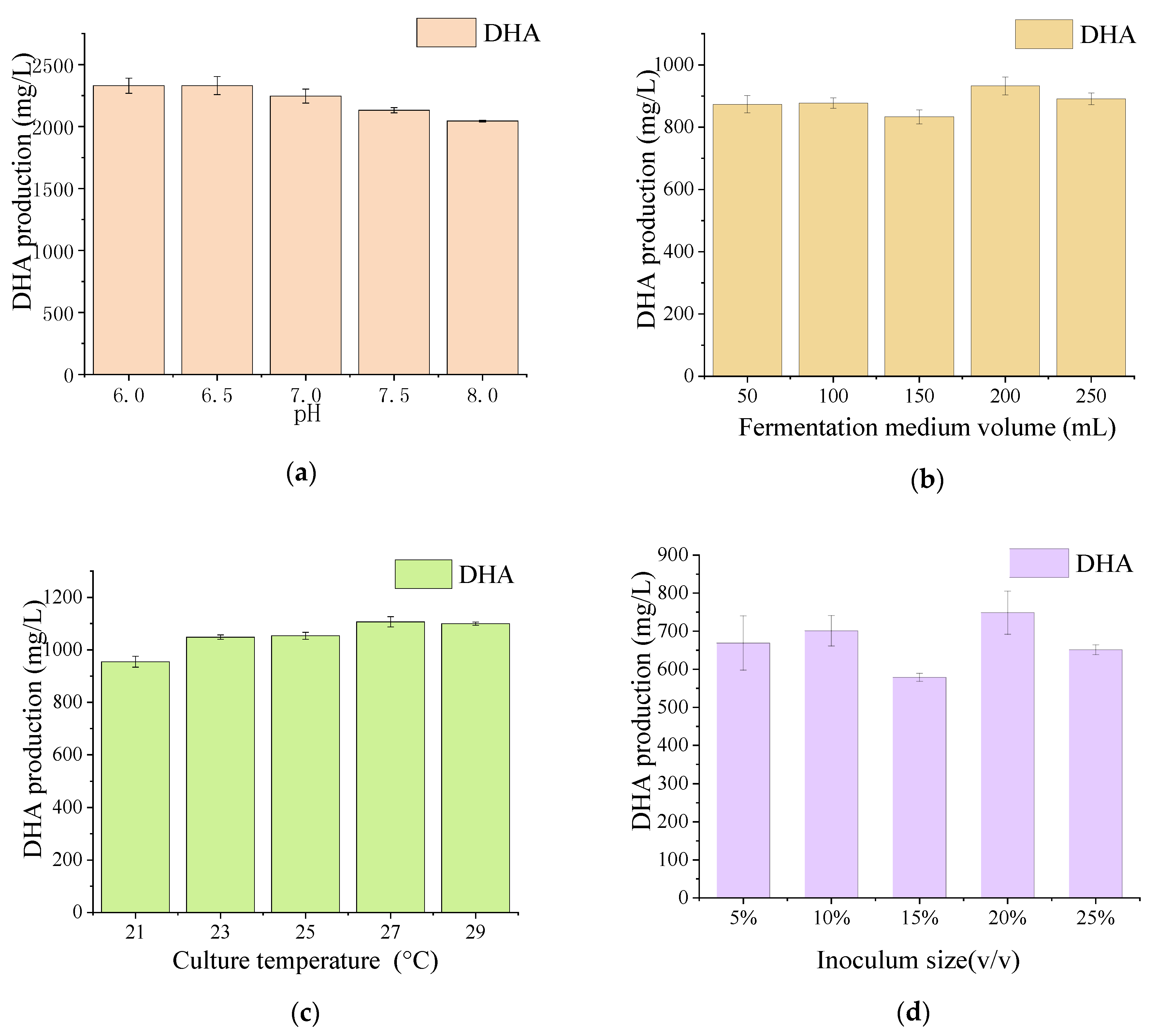
3.3. PUFAs Production by the Mutant Aurantiochytrium sp.
3.4. Sequence Analysis and Assembly
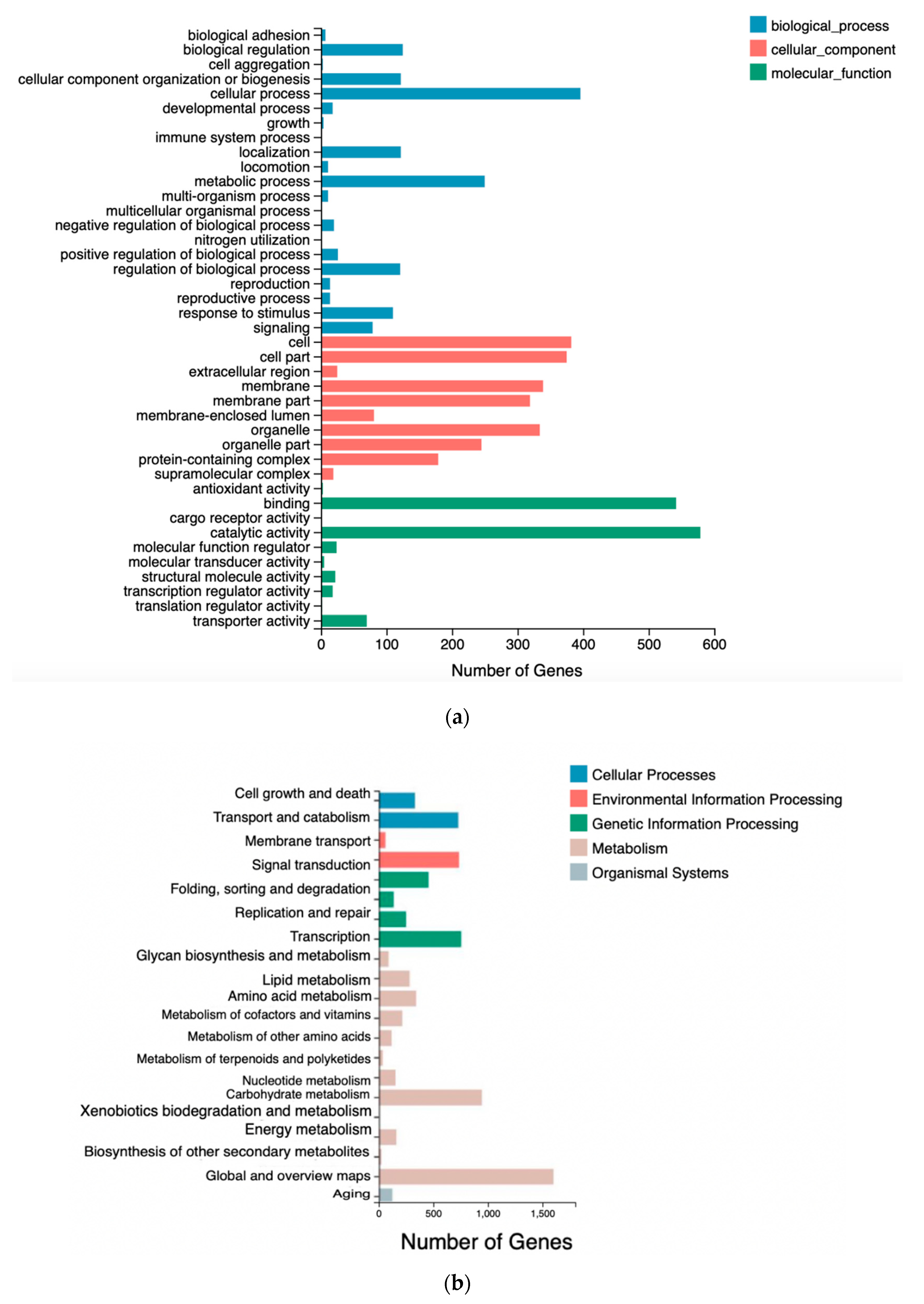
3.5. Differentially Expressed Gene Analysis by RNA-Seq
3.6. Identification and Characterization of Genes Involved in DHA Biosynthesis
3.7. mRNA Expression Level of the Mutant X2 and WT
4. Discussion
4.1. DHA Synthesis Enhancement
4.2. PKS Pathway
4.3. Transcriptional Responses of the PKS Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Simopoulos, A.P. Evolutionary aspects of diet, the omega-6/omega-3 ratio and genetic variation: Nutritional implications for chronic diseases. Biomed. Pharmacotherapy 2006, 60, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Bormann, J.L.; Meyer, E.S.; Voigt, E.M.; Whiteheart, S.W.; Larson, M.K. Attenuation of EPA and DHA-Mediated Platelet Inhibition Effects Following Heterogeneous Agonist Treatment. FASEB J. 2016, 30, 722. [Google Scholar]
- Malan, L.; Baumgartner, J.; Zandberg, L.; Calder, P.C.; Smuts, C.M. Iron and a mixture of DHA and EPA supplementation, alone and in combination, affect bioactive lipid signalling and morbidity of iron deficient south African school children in a two-by-two randomised controlled trial. Prostaglandins Leukot. Essent. Fat. Acids 2016, 105, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Cottin, S.C.; Alsaleh, A.; Sanders, T.A.B.; Hall, W.L. Lack of effect of supplementation with EPA or DHA on platelet-monocyte aggregates and vascular function in healthy men. Nutr. Metab. Cardiovasc Dis. 2016, 26, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Q.; Yang, M.; Wang, T.; Yao, J.; Cheng, J.; Yuan, J.; Lin, X.; Zhao, J.; Tickner, J.; et al. Dihydroartemisinin, an anti-malaria drug, suppresses estrogen deficiency-induced osteoporosis, osteoclast formation, and RANKL-induced signaling pathways. J. Bone Miner Res. 2016, 31, 964–974. [Google Scholar] [CrossRef]
- Létisse, M.; Rozières, M.; Hiol, A.; Sergent, M.; Comeau, L. Enrichment of EPA and DHA from sardine by supercritical fluid extraction without organic modifier: I. Optimization of extraction conditions. J. Supercritical Fluids 2006, 38, 27–36. [Google Scholar] [CrossRef]
- Codabaccus, M.B.; Ng, W.K.; Nichols, P.D.; Carter, C.G. Restoration of EPA and DHA in rainbow trout (Oncorhynchus mykiss) using a finishing fish oil diet at two different water temperatures. Food Chem. 2013, 141, 236–244. [Google Scholar] [CrossRef]
- Scholefield, A.M.; Lipids, K.A. Cell Proliferation and Long Chain Polyunsaturated Fatty Acid Metabolism in a Cell Line from Southern Bluefin Tuna (Thunnus maccoyii). Lipids 2014, 49, 703–714. [Google Scholar] [CrossRef]
- Regulska-Ilow, B.; Ilow, R.; Konikowska, K.; Kawicka, A.; Bochińska, A. FATTY ACIDS PROFILE OF THE FAT IN SELECTED SMOKED MARINE FISH. Rocz Panstw Zakl Hig. 2013, 64, 299–307. [Google Scholar]
- Kitessa, S.M.; Abeywardena, M.; Wijesundera, C.; Nichols, P.D. DHA-containing oilseed: A timely solution for the sustainability issues surrounding fish oil sources of the health-benefitting long-chain omega-3 oils. Nutrients 2014, 6, 2035–2058. [Google Scholar] [CrossRef]
- Shapira, N.; Weill, P.; Loewenbach, R. Egg fortification with n-3 polyunsaturated fatty acids (PUFA): Nutritional benefits versus high n-6 PUFA western diets, and consumer acceptance. Isr. Med Assoc J. 2008, 10, 262–265. [Google Scholar] [PubMed]
- Feng, D.; Chen, Z.; Xue, S.; Zhang, W. Increased lipid production of the marine oleaginous microalgae Isochrysis zhangjiangensis (Chrysophyta) by nitrogen supplement. Bioresour. Technol. 2011, 102, 6710–6716. [Google Scholar] [CrossRef] [PubMed]
- Tsirigoti, A.; Tzovenis, I.; Koutsaviti, A.; Economou-Amilli, A.; Ioannou, E.; Melkonian, M. Biofilm cultivation of marine dinoflagellates under different temperatures and nitrogen regimes enhances DHA productivity. J. Appl. Phycol. 2020. [Google Scholar] [CrossRef]
- Nanjappa, D.; d’Ippolito, G.; Gallo, C.; Zingone, A.; Fontana, A.J. Oxylipin diversity in the diatom family Leptocylindraceae reveals DHA derivatives in marine diatoms. Mar. Drugs 2014, 12, 368–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Singh, P.; Sun, Y.; Luan, S.; Wang, G. Culturable diversity and biochemical features of thraustochytrids from coastal waters of Southern China. Appl. Microbiol. Biotechnol. 2013, 98, 3241–3255. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-M.; Geng, L.-J.; Ren, L.-J.; Ji, X.-J.; Hao, N.; Chen, K.-Q.; Huang, H. Influence of oxygen on the biosynthesis of polyunsaturated fatty acids in microalgae. Bioresour. Technol. 2018, 250, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.B.; DiMasi, D.; Hansen, J.M.; Mirrasoul, P.J.; Ruecker, C.M.; Veeder III, G.T.; Kaneko, T.; Barclay, W.R. Enhanced production of lipids containing polyenoic fatty acid by very high density cultures of eukaryotic microbes in fermentors. U.S. Patent 6,607,900, 19 August 2012. [Google Scholar]
- Patel, A.; Liefeldt, S.; Rova, U.; Christakopoulos, P.; Matsakas, L. Co-production of DHA and squalene by thraustochytrid from forest biomass. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Wang, Q.; Ye, H.; Sen, B.; Xie, Y.; He, Y.; Park, S.; Wang, G.J.S.; Biotechnology, S. Improved production of docosahexaenoic acid in batch fermentation by newly-isolated strains of Schizochytrium sp. and Thraustochytriidae sp. through bioprocess optimization. Synth Syst Biotechnol. 2018, 3, 121–129. [Google Scholar] [CrossRef]
- Li, D.; Zhang, K.; Chen, L.; Ding, M.; Zhao, M.; Chen, S. Selection of Schizochytrium limacinum mutants based on butanol tolerance. Electron. J. Biotechnol. 2017, 30, 58–63. [Google Scholar] [CrossRef]
- Marques, T.S.; Pires, F.; Magalhães-Mota, G.; Ribeiro, P.A.; Raposo, M.; Mason, N. Development of a DNA Biodosimeter for UV Radiation. In Proceedings of the 6th International Conference on Photonics, Optics and Laser Technology, Funchal, Madeira, Portugal, 25–27 January 2018; pp. 328–333. [Google Scholar]
- Shi-Xiong, X.U.; Jiang, Y.; Zhan, X.B.; Zheng, Z.Y.; Jian-Rong, W.U.J.I.M. Breeding of Schizochytrium sp.by diethyl sulfate-UV mutagenesis for high docosahexaenoic acids production. Ind. Microbiol. 2013, 43, 64–70. [Google Scholar]
- Hoang, M.H.; Nguyen, C.; Pham, H.Q.; Van Nguyen, L.; Hoang Duc, L.; Van Son, L.; Hai, T.N.; Ha, C.H.; Nhan, L.D.; Anh, H.T.L.; et al. Transcriptome sequencing and comparative analysis of Schizochytrium mangrovei PQ6 at different cultivation times. Biotechnol. Lett. 2016, 38, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhou, P.-p.; Zhang, M.; Zhu, Y.-m.; Wang, X.-p.; Luo, X.-a.; Bao, Z.-d.; Yu, L.-j. Transcriptome analysis reveals that up-regulation of the fatty acid synthase gene promotes the accumulation of docosahexaenoic acid in Schizochytrium sp. S056 when glycerol is used. Algal Res. 2016, 15, 83–92. [Google Scholar] [CrossRef]
- Ma, Z.; Tan, Y.; Cui, G.; Feng, Y.; Cui, Q.; Song, X. Transcriptome and gene expression analysis of DHA producer Aurantiochytrium under low temperature conditions. Sci. Rep. 2015, 5, 14446. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, S.; Liu, L.; Li, S.; Luo, Y.; Lv, C.; Wang, B.; Cheng, C.H.; Chen, H.; Yang, X. Genome Sequencing and Analysis of Thraustochytriidae sp. SZU445 Provides Novel Insights into the Polyunsaturated Fatty Acid Biosynthesis Pathway. Mar. Drugs 2020, 18, 118. [Google Scholar]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, Y.; Liu, Z.; Zhu, X.; Zhai, L.; Xu, L.; Yu, R.; Gong, Y.; Liu, L. De novo transcriptome sequencing of radish (Raphanus sativus L.) and analysis of major genes involved in glucosinolate metabolism. BMC Genom. 2013, 14, 836. [Google Scholar]
- Zheng, M.; Tian, J.; Yang, G.; Zheng, L.; Chen, G.; Chen, J.; Wang, B. Transcriptome sequencing, annotation and expression analysis of Nannochloropsis sp. at different growth phases. Gene 2013, 523, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.H.; Ha, N.C.; Thom, L.T.; Tam, L.T.; Anh, H.T.L.; Thu, N.T.H.; Hong, D.D. Extraction of squalene as value-added product from the residual biomass of Schizochytrium mangrovei PQ6 during biodiesel producing process. J. Biosci. Bioeng. 2014, 118, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Rismani-Yazdi, H.; Haznedaroglu, B.Z.; Hsin, C.; Peccia, J. Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnol. Biofuels 2012, 5, 74. [Google Scholar] [CrossRef]
- O’neill, E.C.; Trick, M.; Hill, L.; Rejzek, M.; Dusi, R.G.; Hamilton, C.J.; Zimba, P.V.; Henrissat, B.; Field, R.A. The transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry. Mol. Biosyst. 2015, 11, 2808–2820. [Google Scholar] [CrossRef]
- Li, H.; Wang, W.; Wang, Z.; Lin, X.; Zhang, F.; Yang, L. De novo transcriptome analysis of carotenoid and polyunsaturated fatty acid metabolism in Rhodomonas sp. J. Appl. Phycol. 2016, 28, 1649–1656. [Google Scholar] [CrossRef]
- Aasen, I.M.; Ertesvåg, H.; Heggeset, T.M.B.; Liu, B.; Brautaset, T.; Vadstein, O.; Ellingsen, T.E. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl Microbiol. Biotechnol. 2016, 100, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A.; Sherman, D.H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 1990, 24, 37–62. [Google Scholar] [CrossRef] [PubMed]
- Meesapyodsuk, D.; Qiu, X.J. Biosynthetic mechanism of very long chain polyunsaturated fatty acids in Thraustochytrium sp. 26185. J. Lipid Res. 2016, 57, 1854–1864. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, S.; Li, S.; Liu, L.; Hu, Z. De Novo Transcriptome Analysis of Polyunsaturated Fatty Acid Metabolism in Marine Protist Thraustochytriidae sp. PKU# Mn16. Trends Food Sci. Technol. 2020, 97, 35–48. [Google Scholar]
- Patell, V.M. Delta 6 Desaturase From Thraustochytrid and its Uses Thereof. U.S. Patent 20,090,118,371, 18 January 2009. [Google Scholar]
- Song, Z.; Stajich, J.E.; Xie, Y.; Liu, X.; He, Y.; Chen, J.; Hicks, G.R.; Wang, G. Comparative analysis reveals unexpected genome features of newly isolated Thraustochytrids strains: On ecological function and PUFAs biosynthesis. BMC Genomics 2018, 19, 541. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, T.; Tanaka, S.; Yokochi, T.; Nakahara, T.; Higashihara, T. Production of high yields of docosahexaenoic acid by Schizochytrium sp. strain SR 21. J. Am. Oil Chem. Soc. 1997, 74, 1431–1434. [Google Scholar] [CrossRef]
- Chesler, S.N.; Emery, A.P.; Duewer, D.L. Recovery of diesel fuel from soil by supercritical fluid extraction–gas chromatography. J. Chromatogr. A 1997, 790, 125–130. [Google Scholar] [CrossRef]
- Kumari, P.; Reddy, C.R.K.; Jha, B. Comparative evaluation and selection of a method for lipid and fatty acid extraction from macroalgae. Anal. Biochem. 2011, 415, 134–144. [Google Scholar] [CrossRef]
- Lun, L.W.; Gunny, A.A.N.; Kasim, F.H.; Arbain, D. Fourier transform infrared spectroscopy (FTIR) analysis of paddy straw pulp treated using deep eutectic solvent. Proceedings of ADVANCED MATERIALS ENGINEERING AND TECHNOLOGY V: International Conference on Advanced Material Engineering and Technology, Jeju Island, South Korea, 18–20 November 2016. [Google Scholar]
- Wang, H.Q.; Liang, F.; Qiao, N.; Dong, J.X.; Zhang, L.Y.; Guo, Y.F.; Wang, H.Q.; Liang, F.; Qiao, N.; Dong, J.X. Chemical Composition of Volatile Oil from Two Emergent Plants and Their Algae Inhibition Activity. Pol. J. Environ. Stud. 2014, 23, 2371–2374. [Google Scholar]
- Gasic, K.; Hernandez, A.; Reporter, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 2012, 22, 437–438. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402. [Google Scholar] [CrossRef]
- Moeller, R.; Douki, T.; Rettberg, P.; Reitz, G.; Cadet, J.; Nicholson, W.L.; Horneck, G. Genomic bipyrimidine nucleotide frequency and microbial reactions to germicidal UV radiation. Arch. Microbiol. 2010, 192, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.A.; Jung, J.Y.; Kim, K.; Lee, J.S.; Kwon, J.H.; Kim, S.W.; Yang, J.W.; Park, J.Y. Acid-catalyzed hot-water extraction of docosahexaenoic acid (DHA)-rich lipids from Aurantiochytrium sp. KRS101. Bioresour. Technol. 2014, 161, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Sun, Z.; Cui, G.; Song, X.; Qiu, C.; Cheng, Y.; Sun, Z.; Cui, G.; Song, X.; Qiu, C.J.E.; et al. A new strategy for strain improvement of Aurantiochytrium sp. based on heavy-ions mutagenesis and synergistic effects of cold stress and inhibitors of enoyl-ACP reductase. Enzyme Microb. Technol. 2016, 93–94, 182–190. [Google Scholar] [CrossRef]
- Rahman, S.N.S.A.; Kalil, M.S.; Hamid, A.A. The significance of fructose and MSG in affecting lipid and docosahexaenoic acid (DHA) production of Aurantiochytrium sp. SW1. In The 2017 UKM FST Postgraduate Colloquium: Proceedings of the Universiti Kebangsaan Malaysia, Faculty of Science and Technology 2017 Postgraduate Colloquium; AIP Publishing: Melville, NY, USA.
- Lian, M.; Huang, H.; Ren, L.; Ji, X.; Zhu, J.; Jin, L. Increase of Docosahexaenoic Acid Production by Schizochytrium sp. Through Mutagenesis and Enzyme Assay. Appl. Biochem. Biotechnol. 2010, 162, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Christophe, G.; Fontanille, P.; Larroche, C. Research and Production of Microbial Polyunsaturated Fatty Acids. Bioprocess. Biomol. Prod. 2019. [Google Scholar] [CrossRef]
- Gago, G.; Diacovich, L.; Arabolaza, A.; Tsai, S.-C.; Gramajo, H. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol. Rev. 2011, 35, 475–497. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.G.; Roessler, P.; Facciotti, D.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domergue, F.; Yamada, A.; et al. Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, D.; Isabel, M. Lipidomic methodologies for biomarkers of chronic inflammation in nutritional research: ω-3 and ω-6 lipid mediators. Free. Radic. Biol. Med. 2019, 144, 90–109. [Google Scholar]
- Rippe, J.K. Functional Food in the Marketplace: New Products, Availability, and Implications for the Consumer. Nutritional Health 2012, 451–475. [Google Scholar]
- Gupta, A.; Barrow, C.J.; Puri, M. Omega-3 biotechnology: Thraustochytrids as a novel source of omega-3 oils. Biotechnol Adv. 2012, 30, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Engström, M.K.; Saldeen, A.-S.; Yang, B.; Mehta, J.L.; Saldeen, T. Effect of fish oils containing different amounts of EPA, DHA, and antioxidants on plasma and brain fatty acids and brain nitric oxide synthase activity in rats. Ups J. Med. Sci. 2009, 114, 206–213. [Google Scholar] [CrossRef]
- Wang, Q.; Sen, B.; Liu, X.; He, Y.; Xie, Y.; Wang, G. Enhanced saturated fatty acids accumulation in cultures of newly-isolated strains of Schizochytrium sp. and Thraustochytriidae sp. for large-scale biodiesel production. Sci. Total Environ. 2018, 631, 994–1004. [Google Scholar] [CrossRef]
- Meireles, L.A.; Guedes, A.C.; Malcata, F.X. Lipid class composition of the microalga Pavlova lutheri: Eicosapentaenoic and docosahexaenoic acids. J. Agric. Food Chem. 2003, 51, 2237–2241. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Liu, Y.; Frear, C.; Chen, S. Study of a two-stage growth of DHA-producing marine algae Schizochytrium limacinum SR21 with shifting dissolved oxygen level. Appl. Microbiol. Biotechnol. 2009, 81, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Gravius, B.; Bezmalinović, T.; Hranueli, D.; Cullum, J. Genetic instability and strain degeneration in Streptomyces rimosus. Appl. Environ. Microbiol. 1993, 59, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Meireles, L.A.; Guedes, A.C.; Malcata, F.X. Increase of the yields of eicosapentaenoic and docosahexaenoic acids by the microalga Pavlova lutheri following random mutagenesis. Biotechnol. Bioeng. 2003, 81, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Alonso, D.L.; Belarbi, E.-H.; Fernández-Sevilla, J.M.; Rodríguez-Ruiz, J.; Grima, E.M. Acyl lipid composition variation related to culture age and nitrogen concentration in continuous culture of the microalga Phaeodactylum tricornutum. Phytochemistry 2000, 54, 461–471. [Google Scholar] [CrossRef]
- Beacham, T.A.; Macia, V.M.; Rooks, P.; White, D.A.; Ali, S.T. Altered lipid accumulation in Nannochloropsis salina CCAP849/3 following EMS and UV induced mutagenesis. Biotechnol. Rep. 2015, 16, 87–94. [Google Scholar] [CrossRef]
- Forján, E.; Garbayo, I.; Henriques, M.; Rocha, J.; Vega, J.M.; Vílchez, C. UV-A Mediated Modulation of Photosynthetic Efficiency, Xanthophyll Cycle and Fatty Acid Production of Nannochloropsis. Mar. Biotechnol. 2010, 13, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Podevels, A.M.; Kevany, B.M.; Thomas, M.G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009, 26, 90–114. [Google Scholar] [CrossRef]
- Ye, H.; He, Y.; Xie, Y.; Sen, B.; Wang, G. Fed-batch fermentation of mixed carbon source significantly enhances the production of docosahexaenoic acid in Thraustochytriidae sp. PKU# Mn16 by differentially regulating fatty acids biosynthetic pathways. Bioresour. Technol. 2020, 297, 122402. [Google Scholar]
- Henderson, R.J.; Phytochemistry, E.E. Polyunsaturated fatty acid metabolism in the marine dinoflagellate Crypthecodinium cohnii. Phytochemistry 1991, 30, 1781–1787. [Google Scholar] [CrossRef]
- Jeon, E.; Lee, S.; Won, J.I.; Han, S.O.; Kim, J.; Lee, J. Development of Escherichia coli MG1655 strains to produce long chain fatty acids by engineering fatty acid synthesis (FAS) metabolism. Enzym. Microb. Technol. 2011, 49, 44–51. [Google Scholar] [CrossRef]
- Hayashi, S.; Satoh, Y.; Ujihara, T.; Takata, Y.; Dairi, T. Enhanced production of polyunsaturated fatty acids by enzyme engineering of tandem acyl carrier proteins. Sci. Rep. 2016, 6, 35441. [Google Scholar] [CrossRef]
- Napier, J.A. Plumbing the depths of PUFA biosynthesis: A novel polyketide synthase-like pathway from marine organisms. Trends Plant Sci. 2002, 7, 0–54. [Google Scholar] [CrossRef]
- Santín, O.; Moncalián, G. Loading of malonyl-CoA onto tandem acyl carrier protein domains of polyunsaturated fatty acid synthases. J. Biol. Chem. 2018, 293, 12491–12501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Adams, I.P.; Ratledge, C. Malic enzyme: The controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 2007, 153, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.P.; Volkman, J.K.; Jackson, A.E.; Blackburn, S.I. The fatty acid and sterol composition of five marine dinoflagellates. J. Phycol. 1999, 35, 710–720. [Google Scholar] [CrossRef]
- Chang, K.J.L.; Nichols, C.M.; Blackburn, S.I.; Dunstan, G.A.; Koutoulis, A.; Nichols, P.D. Comparison of thraustochytrids Aurantiochytrium sp., Schizochytrium sp., Thraustochytrium sp., and Ulkenia sp. for production of biodiesel, long-chain omega-3 oils, and exopolysaccharide. Mar. Biotechnol. 2014, 16, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, G. Mechanisms of fatty acid synthesis in marine fungus-like protists. Appl. Microbiol. Biotechnol. 2015, 99, 8363–8375. [Google Scholar] [CrossRef] [PubMed]
- Robertsen, H.L.; Musiol-Kroll, E.M.; Ding, L.; Laiple, K.J.; Hofeditz, T.; Wohlleben, W.; Lee, S.Y.; Grond, S.; Weber, T. Filling the gaps in the kirromycin biosynthesis: Deciphering the role of genes involved in ethylmalonyl-CoA supply and tailoring reactions. Sci. Rep. 2018, 8, 3230. [Google Scholar] [CrossRef]
- Bi, Z.-Q.; Ren, L.-J.; Hu, X.-C.; Sun, X.-M.; Zhu, S.-Y.; Ji, X.-J.; Huang, H. Transcriptome and gene expression analysis of docosahexaenoic acid producer Schizochytrium sp. under different oxygen supply conditions. Biotechnol. Biofuels 2018, 11, 249. [Google Scholar] [CrossRef]





| Generation | TFAs (%DCW) | DHA (% TFAs) | DHA Content (mg/L) |
|---|---|---|---|
| WT | 41.07 ± 2.05 | 40.55 ± 2.05 | 321.37 ± 14.98 |
| X2-2nd | 50.00 ± 3.87 | 44.39 ± 3.87 | 624.34 ± 2.95 |
| X2-6th | 48.10 ± 0.74 | 44.98 ± 0.74 | 623.40 ± 7.14 |
| X2-10th | 48.53 ± 1.40 | 44.09 ± 1.40 | 624.93 ± 13.32 |
| Sample | Total Raw | Total Clean | Clean Reads | |||
|---|---|---|---|---|---|---|
| Reads (Mb) 1 | Reads (Mb) 2 | Bases (Gb) 3 | Q20 (%) 4 | Q30 (%) 5 | Ratio (%) 6 | |
| Mn16 | 50.94 | 44.69 | 6.70 | 96.82 | 88.53 | 87.72 |
| X2 | 47.43 | 43.14 | 6.47 | 96.67 | 87.97 | 90.94 |
| Quality Metrics | Uni-genes | |
|---|---|---|
| Mn16 | X2 | |
| Total Number | 20,874 | 18,952 |
| Total Length | 23,991,758 | 22,844,185 |
| Mean Length | 1149 | 1205 |
| N50 1 | 1880 | 2032 |
| N70 2 | 1264 | 1328 |
| N90 3 | 480 | 504 |
| GC (%) 4 | 48.36 | 48.75 |
| Gene ID | Protein Name | Gene Name | Control FPKM | Treat FPKM | log2 |
|---|---|---|---|---|---|
| CL555.Contig2_All | Enoyl-(Acyl carrier protein) reductase | ER | 3.85 | 10.1 | 1.400242079 |
| CL663.Contig2_All | CoA-transferase | CoAT | 1.14 | 3.04 | 1.394908036 |
| Unigene10491_All | Acyltransferase | AT | 74.10 | 149.53 | 1.006525578 |
| Unigene2419_All | Methyltransferase domain | MT | 8.46 | 18.11 | 1.083865306 |
| Unigene4591_All | Dehydratase family | DH | 0.01 | 1.66 | 7.211944308 |
| CL94.Contig2_All | Fatty acid desaturase | FAD | 2.02 | 4.34 | 1.101442064 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Hu, Z.; Li, S.; Yang, H.; Li, S.; Lv, C.; Zaynab, M.; Cheng, C.H.K.; Chen, H.; Yang, X. Comparative Transcriptomic Analysis Uncovers Genes Responsible for the DHA Enhancement in the Mutant Aurantiochytrium sp. Microorganisms 2020, 8, 529. https://doi.org/10.3390/microorganisms8040529
Liu L, Hu Z, Li S, Yang H, Li S, Lv C, Zaynab M, Cheng CHK, Chen H, Yang X. Comparative Transcriptomic Analysis Uncovers Genes Responsible for the DHA Enhancement in the Mutant Aurantiochytrium sp. Microorganisms. 2020; 8(4):529. https://doi.org/10.3390/microorganisms8040529
Chicago/Turabian StyleLiu, Liangxu, Zhangli Hu, Shuangfei Li, Hao Yang, Siting Li, Chuhan Lv, Madiha Zaynab, Christopher H. K. Cheng, Huapu Chen, and Xuewei Yang. 2020. "Comparative Transcriptomic Analysis Uncovers Genes Responsible for the DHA Enhancement in the Mutant Aurantiochytrium sp." Microorganisms 8, no. 4: 529. https://doi.org/10.3390/microorganisms8040529
APA StyleLiu, L., Hu, Z., Li, S., Yang, H., Li, S., Lv, C., Zaynab, M., Cheng, C. H. K., Chen, H., & Yang, X. (2020). Comparative Transcriptomic Analysis Uncovers Genes Responsible for the DHA Enhancement in the Mutant Aurantiochytrium sp. Microorganisms, 8(4), 529. https://doi.org/10.3390/microorganisms8040529