Engineering Improves Enzymatic Synthesis of L-Tryptophan by Tryptophan Synthase from Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Strains and Culture Conditions
2.2. Mutation Site
2.3. Site-Saturated Mutation Library
2.4. Error-Prone PCR Mutant Library
2.5. Screening of Mutated Enzymes
2.6. Protein Purification
2.7. Synthesis and Identification of L-tryptophan
3. Results
3.1. Selection of Mutation Sites
3.2. Mutation Library Screening
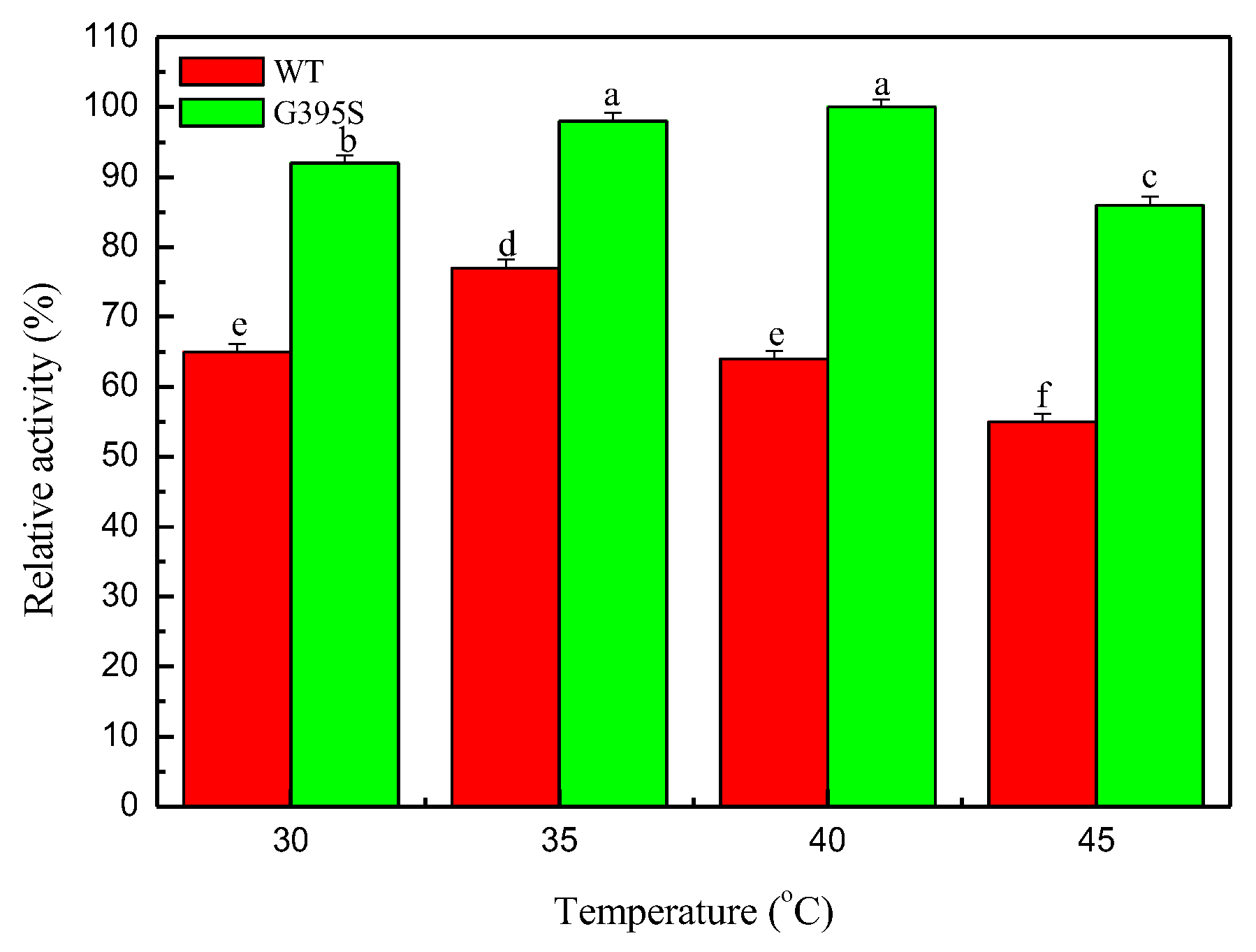
3.3. Optimum Temperature of the Mutant Enzyme
3.4. Directed Evolution
3.5. Synthesis of L-tryptophan
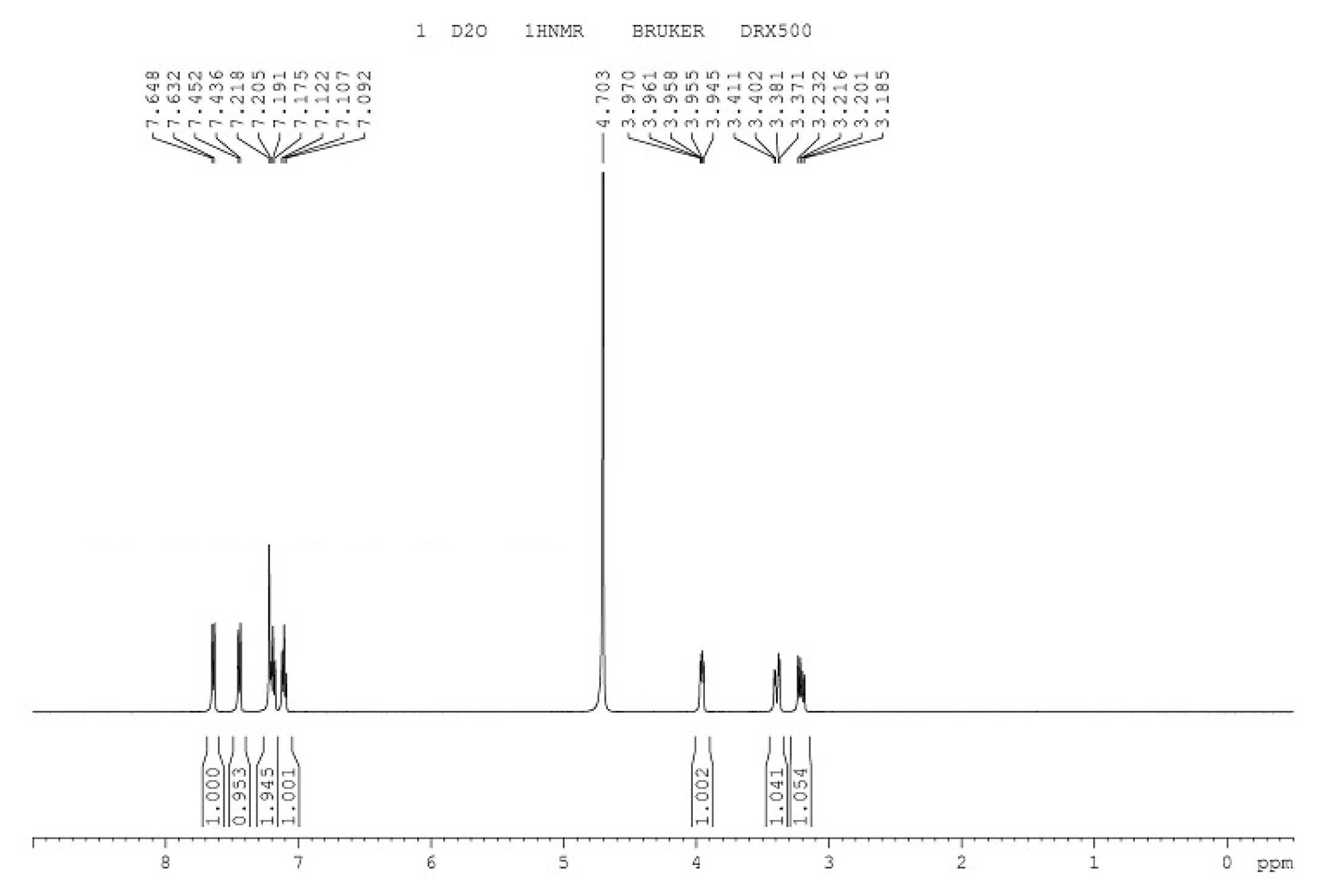
3.6. Identification of L-tryptophan
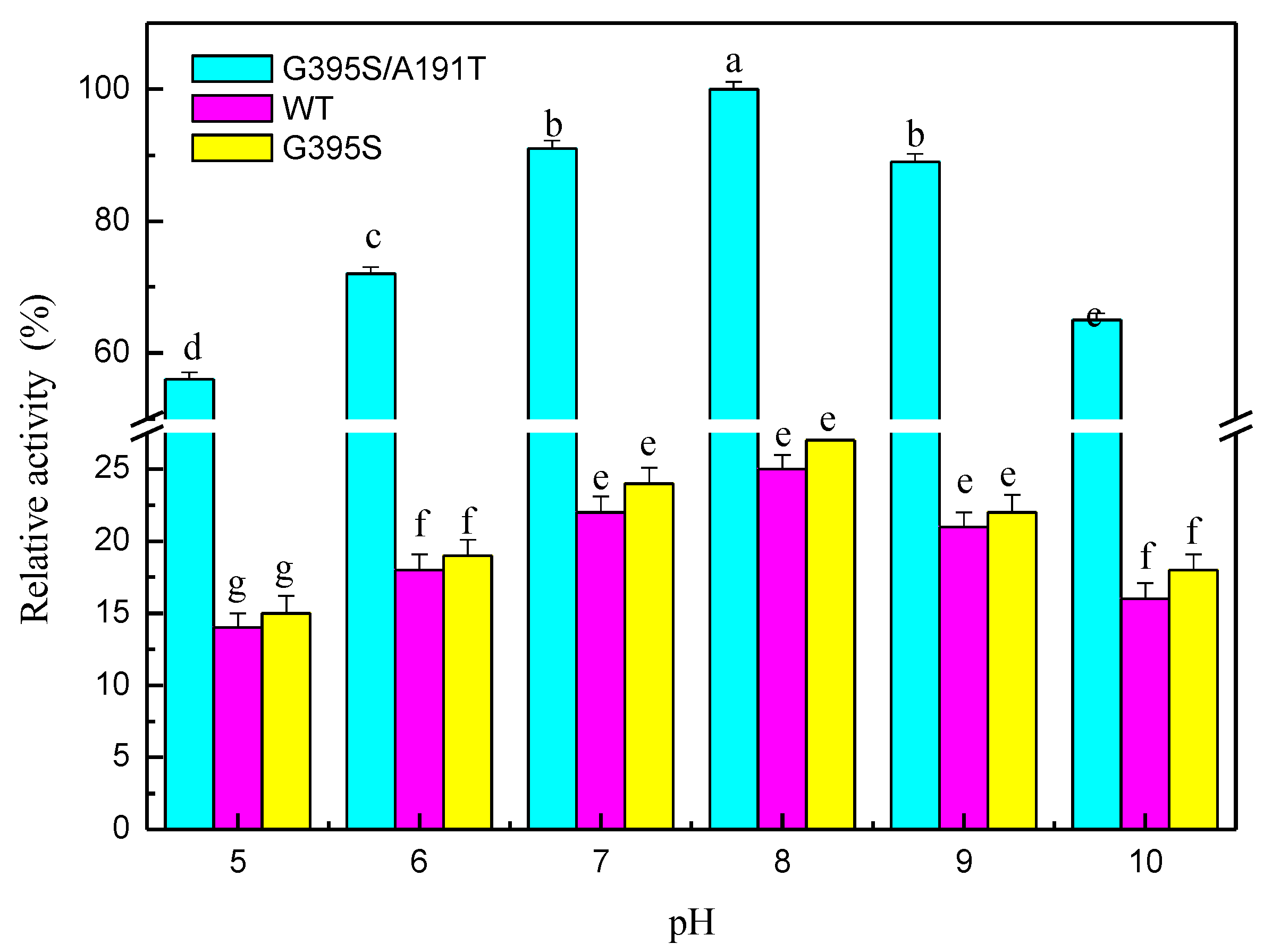
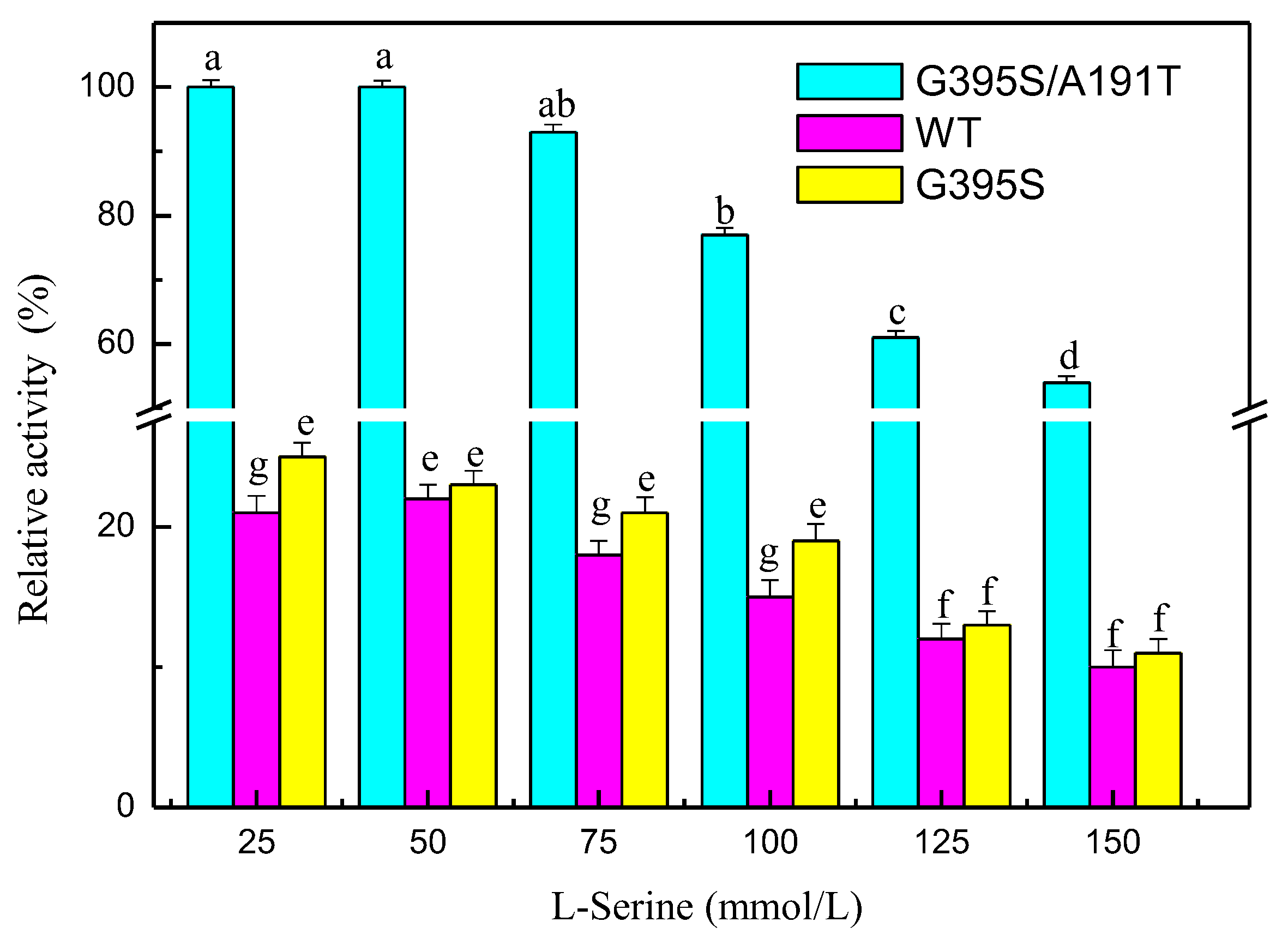
3.7. Mutation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Murciano-Calles, J.; Romney, D.K.; Brinkmann-Chen, S.; Buller, A.R.; Arnold, F.H. A panel of TrpB biocatalysts derived from tryptophan synthase through the transfer of mutations that mimic allosteric activation. Angew. Chem. Int. Ed. 2016, 55, 11577–11581. [Google Scholar] [CrossRef] [PubMed]
- Young, R.P.; Caulkins, B.G.; Borchardt, D.; Bulloch, D.N.; Larive, C.K.; Dunn, M.F.; Mueller, L.J. Solution-state (17) O quadrupole central-transition NMR spectroscopy in the active site of tryptophan synthase. Angew. Chem. Int. Ed. 2016, 55, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Kadumuri, R.V.; Gullipalli, J.; Subramanian, S.; Jaipuria, G.; Atreya, H.S.; Vadrevu, R. Crowding interactions perturb structure and stability by destabilizing the stable core of the α-subunit of tryptophan synthase. FEBS Lett. 2016, 590, 2096–2105. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.M.; Willemse, T.; Gkotsi, D.S.; Schepens, W.; Maes, B.U.W.; Ballet, S.; Goss, R.J.M. The first one-pot synthesis of L-7-iodotryptophan from 7-iodoindole and serine, and an improved synthesis of other L-7-halotryptophans. Org. Lett. 2014, 16, 2622–2625. [Google Scholar] [CrossRef] [PubMed]
- Buller, A.R.; Brinkmann-Chen, S.; Romney, D.K.; Herger, M.; Murciano-Calles, J.; Arnold, F.H. Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation. Proc. Natl. Aacd. Sci. USA 2015, 112, 14599–14604. [Google Scholar] [CrossRef]
- Xu, L.S.; Gao, G.Z.; Cao, W.G.; Zhao, L.; Chen, J.; Bao, N.N. Enzymatic synthesis of L-2-methyltrytophan catalyzed by recombinant Escherichia coli with tryptophan synthase activity. Int. J. Simul. Syst. Sci. Technol. 2016, 45, 1–4. [Google Scholar]
- Xu, L.S.; Wang, Z.Y.; Mao, P.T.; Liu, J.Z.; Zhang, H.J.; Liu, Q.; Jiao, Q.C. Enzymatic synthesis of S-phenyl-L-cysteine from keratin hydrolysis industries wastewater with tryptophan synthase. Bioresour. Technol. 2013, 133, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.S.; Zhang, X.T.; Gao, G.Z.; Sun, Y. Highly efficient preparation of active S-phenyl-L-cysteine with tryptophan synthase using a chemoenzymatic method. BMC Biotechnol. 2019, 19, 49. [Google Scholar] [CrossRef]
- Lin, B.X.; Fan, K.Q.; Zhao, J.; Ji, J.J.; Wu, L.J.; Yang, K.Q.; Tao, Y. Reconstitution of TCA cycle with DAOCS to engineer Escherichia coli into an efficient whole cell catalyst of penicillin G. Proc. Natl. Aacd. Sci. USA 2015, 112, 9855–9859. [Google Scholar] [CrossRef]
- Wijma, H.J.; Floor, R.J.; Bjelic, S.; Marrink, S.J.; Baker, D.; Janssen, D.B. Enantioselective enzymes by computational design and in silico screening. Angew. Chem. Int. Ed. 2015, 54, 3726–3730. [Google Scholar] [CrossRef]
- Payne, J.T.; Catherine, B.P.; Lewis, J.C. Directed evolution of RebH for site-selective halogenation of large biologically active molecules. Angew. Chem. Int. Ed. 2015, 54, 4226–4230. [Google Scholar] [CrossRef] [PubMed]
- Belsare, K.D.; Andorfer, M.C.; Cardenas, F.S.; Chael, J.R.; Park, H.J.; Lewis, J.C. A Simple combinatorial codon mutagenesis method for targeted protein engineering. ACS Synth. Biol. 2017, 6, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.Y.; Jiang, Z.N.; Zhang, J.H.; Zheng, H.H.; Jiang, X.; Gong, K.; Liang, Q.F.; Wang, Q.; Qi, Q.S. Stable and efficient biosynthesis of 5-aminolevulinic acid using plasmid-free Escherichia coli. J. Agric. Food Chem. 2019, 67, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.C.; Gao, X.; Liu, X.B.; Wang, Y.; Yang, S.L.; Wang, F.Q.; Ren, Y.H. Enhancing biosynthesis of a ginsenoside precursor by self-assembly of two key enzymes in Pichia pastoris. J. Agric. Food Chem. 2016, 64, 3380–3385. [Google Scholar] [CrossRef]
- Mu, D.D.; Li, H.W.; Chen, Q.; Zhu, J.; Wu, X.F.; Luo, S.Z.; Zhao, Y.Y.; Wang, L.; Jiang, S.T.; Li, X.J.; et al. Secretion of Bacillus amyloliquefaciens γ-glutamyltranspeptidase from Bacillus subtilis and its application in enzymatic synthesis of L-theanine. J. Agric. Food Chem. 2019, 67, 14129–14136. [Google Scholar] [CrossRef]
- Chen, K.; Huang, X.Y.; Jennifer Kan, S.B.; Zhang, R.K.; Arnold, F.H. Enzymatic construction of highly strained carbocycles. Science 2018, 360, 71–75. [Google Scholar] [CrossRef]
- Li, G.; Zhang, H.; Sun, Z.; Liu, X.; Reetz, M.T. Multiparameter optimization in directed evolution: Engineering thermostability, enantioselectivity, and activity of an epoxide hydrolase. ACS Catal. 2016, 6, 3679–3687. [Google Scholar] [CrossRef]
- Xu, J.; Cen, Y.; Singh, W.; Fan, J.; Wu, L.; Lin, X.; Zhou, J.; Huang, M.; Reetz, M.T.; Wu, Q. Stereodivergent protein engineering of a Lipase to access all possible stereoisomers of chiral esters with two stereocenters. J. Am. Chem. Soc. 2019, 141, 7934–7945. [Google Scholar] [CrossRef]
- Chen, Q.J.; Chen, X.; Feng, J.H.; Wu, Q.Q.; Zhu, D.M.; Ma, Y.H. Improving and inverting C β-stereoselectivity of threonine aldolase via substrate-binding-guided mutagenesis and a stepwise visual screening. ACS Catal. 2019, 9, 4462–4469. [Google Scholar] [CrossRef]
- Wu, Q.; Soni, P.; Reetz, M.T. Laboratory Evolution of Enantiocomplementary candida antarctica Lipase B mutants with broad substrate scope. J. Am. Chem. Soc. 2013, 135, 1872–1881. [Google Scholar] [CrossRef]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tam, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J.; et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Martínez, J.A.; Flores, N.; Escalante, A.; Gosset, G.; Bolivar, F. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Factories 2014, 13, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Caulkins, B.G.; Young, R.P.; Kudla, R.A.; Yang, C.; Bittbauer, T.J.; Bastin, B.; Hilario, E.; Fan, L.; Marsella, M.J.; Dunn, M.F.; et al. NMR crystallography of a carbanionic intermediate in tryptophan synthase: Chemical structure, tautomerization, and reaction specificity. J. Am. Chem. Soc. 2016, 138, 15214–15226. [Google Scholar] [CrossRef]
- Huang, Y.M.; You, W.; Caulkins, B.G.; Dunn, M.F.; Mueller, L.J.; Chang, C.A. Protonation states and catalysis: Molecular dynamics studies of intermediates in tryptophan synthase. Protein sci. 2016, 25, 166–183. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Lu, J.; Shu, C.; Li, H.; Li, X.; Cai, J.; Luo, S.; Yang, P.; Jiang, S.; Zheng, Z. Improvement of the activity and thermostability of microbial transglutaminase by multiple-site mutagenesis. Biosci. Biotechnol. Biochem. 2018, 82, 106–109. [Google Scholar] [CrossRef]
- Gao, X.; Wu, J.; Wu, D. Rational design of the beta-galactosidase from Aspergillus oryzae to improve galactooligosaccharide production. Food Chem. 2019, 286, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.X.; Chen, H.; Liu, J. Rational design of a synthetic Entner-Doudoroff pathway for enhancing glucose transformation to isobutanol in Escherichia coli. J. Ind. Microbiol. Biot. 2018, 45, 187–199. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, X.H.; Wu, Y.K. Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications. Metab. Eng. 2018, 50, 109–121. [Google Scholar] [CrossRef]
- Nie, Y.; Wang, S.; Xu, Y.; Luo, S.; Zhao, Y.-L.; Xiao, R.; Montelione, G.T.; Hunt, J.F.; Szyperski, T. Enzyme engineering based on Xray structures and kinetic profiling of substrate libraries: Alcohol dehydrogenases for stereospecific synthesis of a broad range of chiral alcohols. ACS Catal. 2018, 8, 5145–5152. [Google Scholar] [CrossRef]
- Chen, F.-F.; Zheng, G.-W.; Liu, L.; Li, H.; Chen, Q.; Li, F.-L.; Li, C.-X.; Xu, J.-H. Reshaping the active pocket of amine dehydrogenases for asymmetric synthesis of bulky aliphatic amines. ACS Catal. 2018, 8, 2622–2628. [Google Scholar] [CrossRef]
- Yang, B.; Wang, H.; Song, W.; Chen, X.L.; Liu, J.; Luo, Q.L.; Liu, L.M. Engineering of the conformational dynamics of lipase to increase enantioselectivity. ACS Catal. 2017, 7, 7593–7599. [Google Scholar] [CrossRef]
- Chen, G.S.; Siao, S.W.; She, C.R. Saturated mutagenesis of ketoisovalerate decarboxylase V461 enabled specific synthesis of L-pentanol via the ketoacid elongation cycle. SCI REP-UK 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.M.; Zhao, G.L.; Ng, K.R.; Mark, R.; Chen, W.N. Metabolic engineering of Saccharomyces cerevisiae for de novo production of kaempferol. J. Agric. Food Chem. 2019, 67, 5596–5606. [Google Scholar] [CrossRef] [PubMed]





| Primer | Sequence (5′ - 3′) |
|---|---|
| G395S For | (5′-AACTTTCGTGCTNNNCTTTAGACT-3′) |
| G395S Rev | (5′-TTTTGCTGGTACANNNAAATCCTTC-3′) |
| Rank | Residue Sequence Number | Amino Acid | B-Factor Value |
|---|---|---|---|
| 1 | 395 | Gly | 95.24 |
| 2 | 394 | Arg | 93.55 |
| 3 | 382 | Lys | 93.34 |
| 4 | 393 | Ala | 92.39 |
| 5 | 392 | Lys | 89.52 |
| 6 | 383 | Asp | 88.7 |
| 7 | 313 | His | 88.7 |
| 8 | 257 | Pro | 87.61 |
| 9 | 329 | Asp | 86.18 |
| 10 | 233 | Gly | 86.17 |
| 11 | 191 | Ala | 86.06 |
| 12 | 234 | Gly | 85.96 |
| 13 | 18 | Pro | 85.83 |
| 14 | 258 | Gly | 85.79 |
| 15 | 388 | His | 85.68 |
| 16 | 17 | Val | 85.64 |
| 17 | 378 | Gly | 85.18 |
| 18 | 190 | Thr | 85.01 |
| 19 | 256 | Glu | 83.42 |
| 20 | 232 | Gly | 82.93 |
| Mutations | Km (mM) | kcat (s−1) | kcat/Km (mM−1∙s −1) | Fold Change over WT |
|---|---|---|---|---|
| WT | 0.31 ± 0.11 | 0.35 ± 0.09 | 1.12 ± 0.07 | 1 |
| G395S | 0.32 ± 0.14 | 0.42 ± 0.11 | 1.31 ± 0.10 | 1.16 |
| G395S/A191T | 0.21 ± 0.10 | 1.13 ± 0.12 | 5.38 ± 0.11 | 4.80 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Han, F.; Dong, Z.; Wei, Z. Engineering Improves Enzymatic Synthesis of L-Tryptophan by Tryptophan Synthase from Escherichia coli. Microorganisms 2020, 8, 519. https://doi.org/10.3390/microorganisms8040519
Xu L, Han F, Dong Z, Wei Z. Engineering Improves Enzymatic Synthesis of L-Tryptophan by Tryptophan Synthase from Escherichia coli. Microorganisms. 2020; 8(4):519. https://doi.org/10.3390/microorganisms8040519
Chicago/Turabian StyleXu, Lisheng, Fangkai Han, Zeng Dong, and Zhaojun Wei. 2020. "Engineering Improves Enzymatic Synthesis of L-Tryptophan by Tryptophan Synthase from Escherichia coli" Microorganisms 8, no. 4: 519. https://doi.org/10.3390/microorganisms8040519
APA StyleXu, L., Han, F., Dong, Z., & Wei, Z. (2020). Engineering Improves Enzymatic Synthesis of L-Tryptophan by Tryptophan Synthase from Escherichia coli. Microorganisms, 8(4), 519. https://doi.org/10.3390/microorganisms8040519






