Role of Sinorhizobium meliloti and Escherichia coli Long-Chain Acyl-CoA Synthetase FadD in Long-Term Survival
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. DNA Manipulations
2.3. In vivo Labeling of S. meliloti and E. coli with 14C-Acetate and Analysis of Lipid Extracts by Thin-Layer Chromatography (TLC)
2.4. Surface Motility Assays
2.5. Cell Viable Counts of S. meliloti and E. coli
3. Results
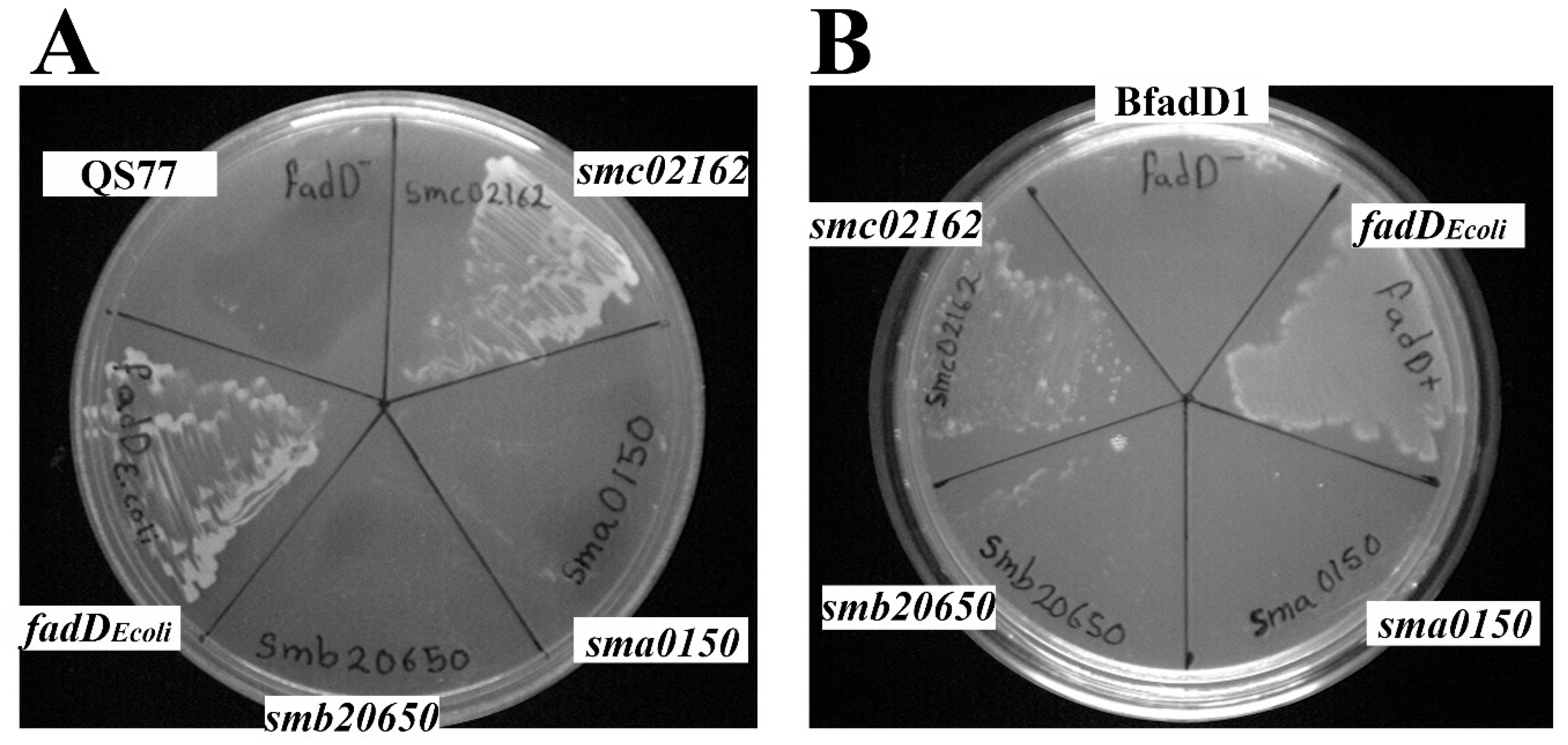
3.1. Growth of Sinorhizobium meliloti and Escherichia coli fadD Mutants on Oleate Cannot be Complemented by sma0150 or smb20650
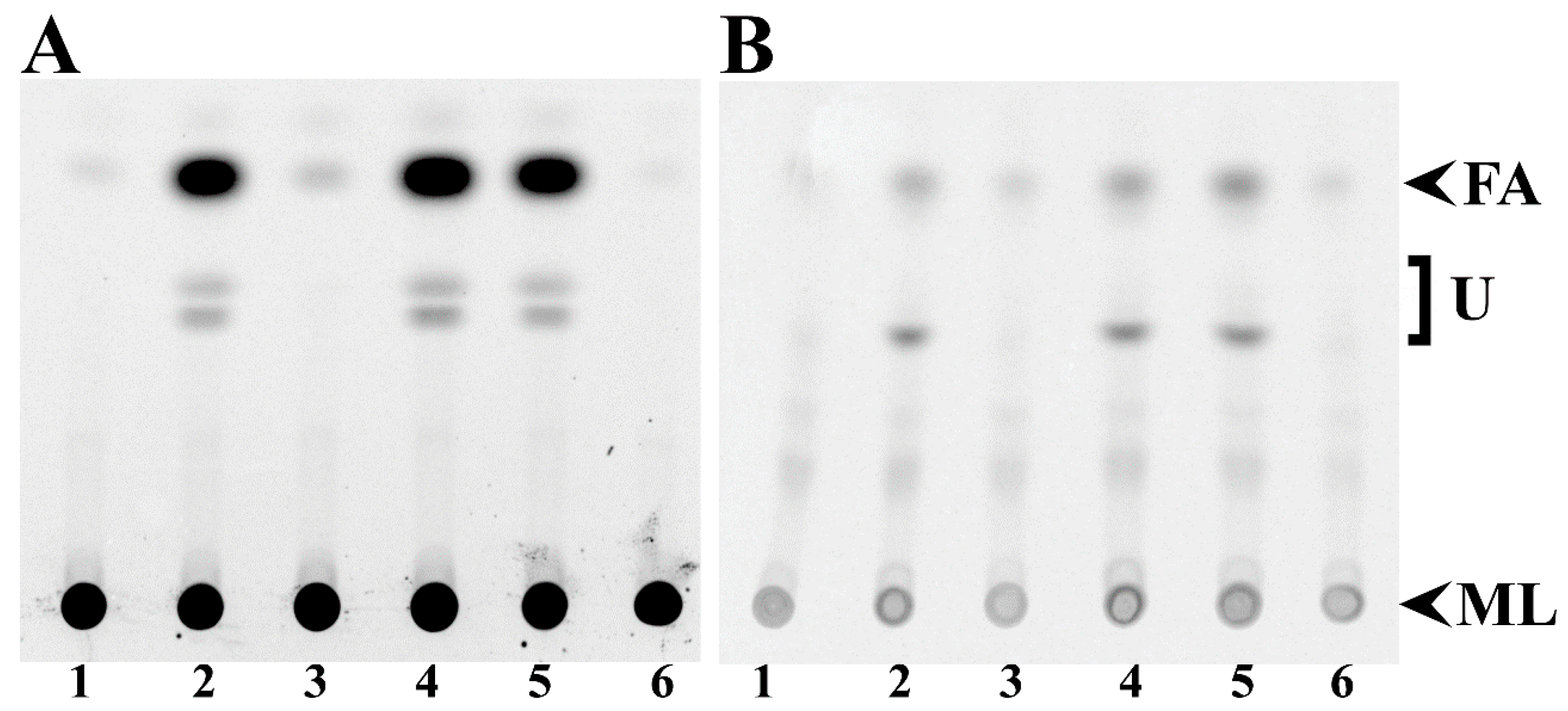
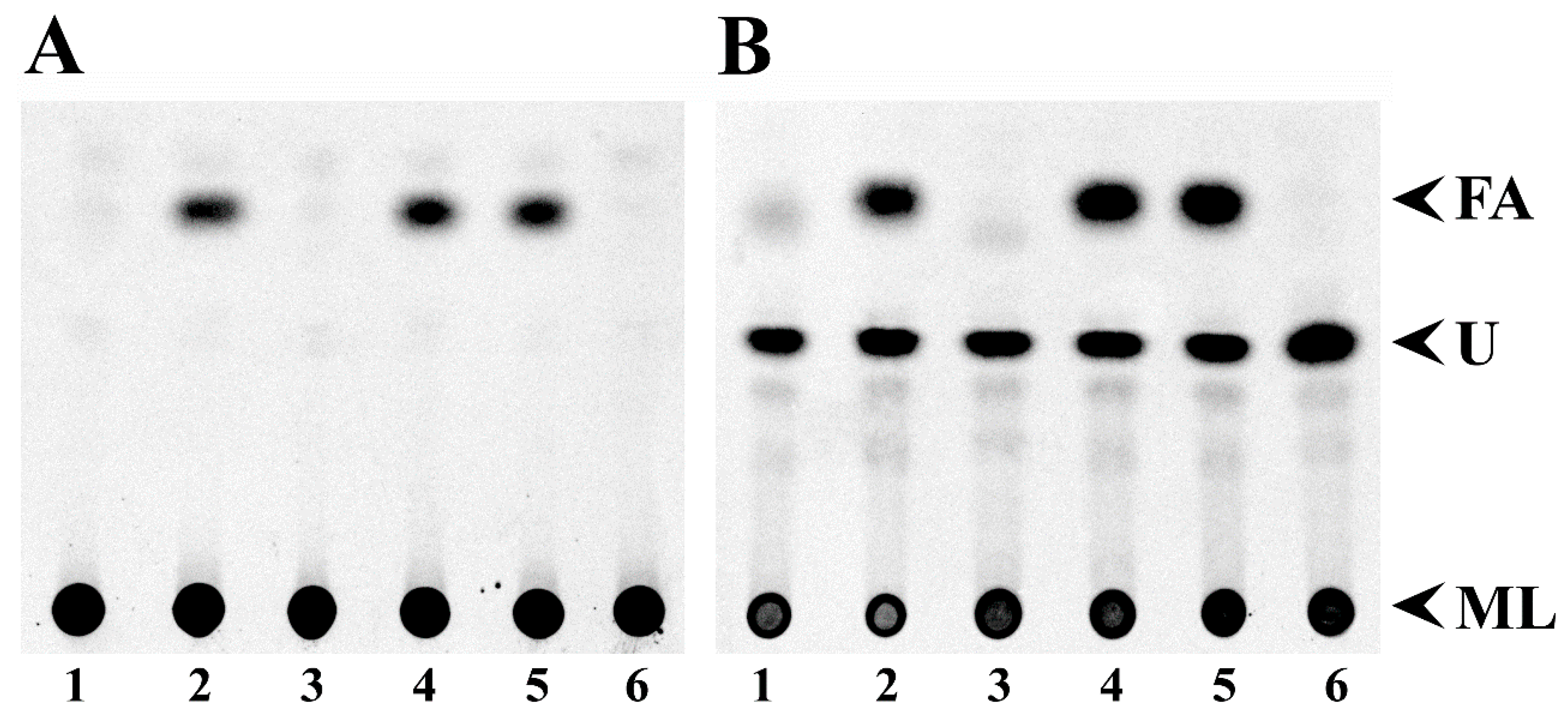
3.2. Free Fatty Acid Accumulation in Sinorhizobium meliloti and Escherichia coli fadD Mutants Cannot be Reverted by sma0150 or smb20650
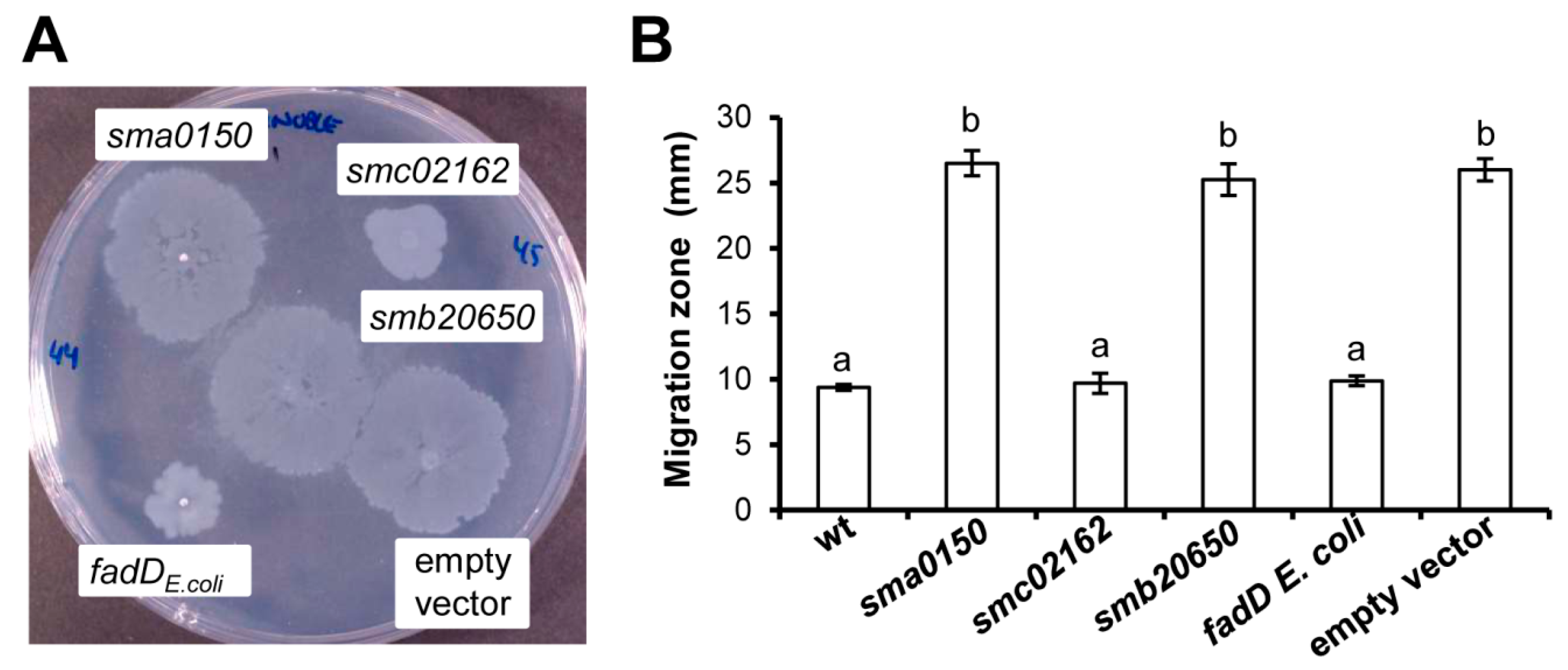
3.3. Effect of the E. coli fadD, S. meliloti fadD (smc02162), and S. meliloti Genes sma0150 and smb20650 on the Surface Motility of a fadD Mutant of S. meliloti (QS77)
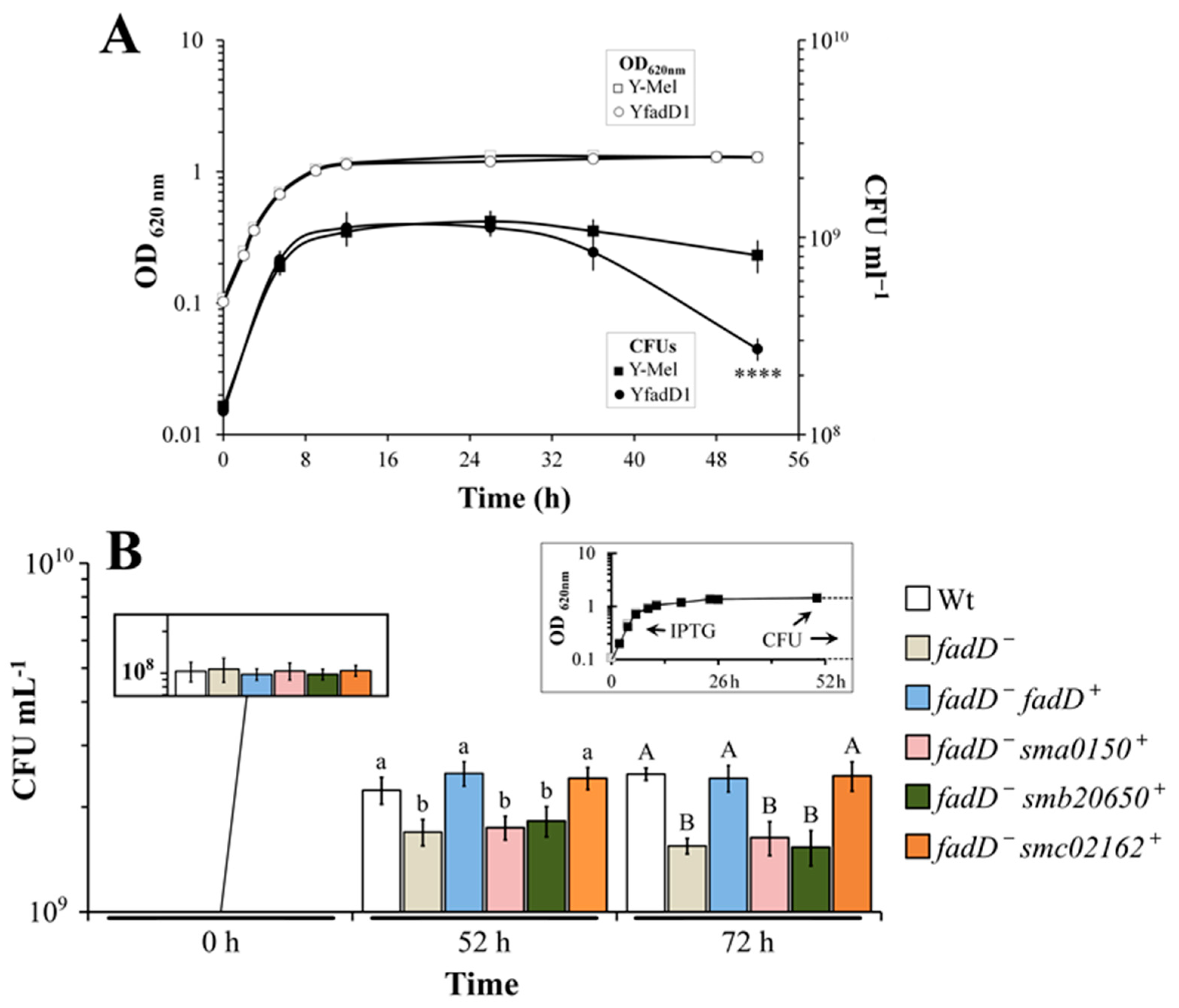
3.4. Absence of fadD Reduces Survival Rates in the Stationary Phase of Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rock, C.O. Fatty acid and phospholipid metabolism in prokaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes, 5th ed.; Vance, D.E., Vance, J.E., Eds.; Elsevier: San Diego, CA, USA, 2008; pp. 59–96. [Google Scholar] [CrossRef]
- Soto, M.J.; Fernández-Pascual, M.; Sanjuan, J.; Olivares, J. A fadD mutant of Sinorhizobium meliloti shows multicellular swarming migration and is impaired in nodulation efficiency on alfalfa roots. Mol. Microbiol. 2002, 43, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Pech-Canul, Á.; Nogales, J.; Miranda-Molina, A.; Álvarez, L.; Geiger, O.; Soto, M.J.; López-Lara, I.M. FadD is required for utilization of endogenous fatty acids released from membrane lipids. J. Bacteriol. 2011, 193, 6295–6304. [Google Scholar] [CrossRef] [PubMed]
- López-Lara, I.M.; Nogales, J.; Pech-Canul, Á.; Calatrava-Morales, N.; Bernabéu-Roda, L.M.; Durán, P.; Cuéllar, V.; Olivares, J.; Alvarez, L.; Palenzuela-Bretones, D.; et al. 2-Tridecanone impacts surface-associated bacterial behaviours and hinders plant–bacteria interactions. Environ. Microbiol. 2018, 20, 2049–2065. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Vega, A.L.; Dávila-Martínez, Y.; Sohlenkamp, C.; Contreras-Martínez, S.; Encarnación, S.; Geiger, O.; López-Lara, I.M. SMb20651 is another acyl carrier protein from Sinorhizobium meliloti. Microbiology 2009, 155, 257–267. [Google Scholar] [CrossRef] [PubMed]
- An, J.H.; Lee, G.Y.; Jung, J.-W.; Lee, W.; Kim, Y.S. Identification of residues essential for a two-step reaction by malonyl-CoA synthetase from Rhizobium trifolii. Biochem. J. 1999, 344, 159–166. [Google Scholar] [CrossRef]
- Koo, H.M.; Kim, Y.S. Identification of active-site residues in Bradyrhizobium japonicum malonyl-Coenzyme A synthetase. Arch. Biochem. Biophys. 2000, 378, 167–174. [Google Scholar] [CrossRef]
- Hughes, A.J.; Keatinge-Clay, A. Enzymatic extender unit generation for in vitro polyketide synthase reactions: Structural and functional showcasing of Streptomyces coelicolor MatB. Chem. Biol. 2011, 18, 165–176. [Google Scholar] [CrossRef]
- Witkowski, A.; Thweatt, J.; Smith, S. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J. Biol. Chem. 2011, 286, 33729–33736. [Google Scholar] [CrossRef]
- Chen, H.; Kim, H.U.; Weng, H.; Browse, J. Malonyl-CoA synthetase, encoded by Acyl Activating Enzyme13, is essential for growth and development of Arabidopsis. Plant Cell 2011, 23, 2247–2262. [Google Scholar] [CrossRef]
- Chen, A.-M.; Wang, Y.-B.; Jie, S.; Yu, A.-Y.; Luo, L.; Yu, G.-Q.; Zhu, J.-B.; Wang, Y.-Z. Identification of a TRAP transporter for malonate transport and its expression regulated by GtrA from Sinorhizobium meliloti. Res. Microbiol. 2010, 161, 556–564. [Google Scholar] [CrossRef]
- Pohl, N.L.; Hans, M.; Lee, H.Y.; Kim, Y.S.; Cane, D.E.; Khosla, C. Remarkably broad substrate tolerance of malonyl-CoA synthetase, an enzyme capable of intracellular synthesis of polyketide precursors. J. Am. Chem. Soc. 2001, 123, 5822–5823. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zarzycki-Siek, J.; Walton, C.B.; Norris, M.H.; Hoang, T.T. Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa. PLoS ONE 2010, 5, e13557. [Google Scholar] [CrossRef]
- Zarzycki-Siek, J.; Norris, M.H.; Kang, Y.; Sun, Z.; Bluhm, A.P.; McMillan, I.A.; Hoang, T.T. Elucidating the Pseudomonas aeruginosa fatty acid degradation pathway: Identification of additional fatty acyl-CoA synthetase homologues. PLoS ONE 2013, 8, e64554. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Russell, D. Molecular Cloning: A Laboratory Manual (3-Volume Set), 3rd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001; Volume 1, p. 2100. [Google Scholar]
- Beringer, J.E. R Factor Transfer in Rhizobium leguminosarum. Microbiology 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Robertsen, B.K.; Åman, P.; Darvill, A.G.; McNeil, M.; Albersheim, P. Host-symbiont interactions. V. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981, 67, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, M.T. Improved synthetic medium for the growth of Rhizobium. J. Appl. Bacteriol. 1970, 33, 708–713. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Thomason, L.C.; Costantino, N.; Court, D.L. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007, 79, 11–18. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Rickenberg, H.V.; Lester, G. The preferential synthesis of β-galactosidase in Escherichia coli. Microbiology 1955, 13, 279–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Studier, F.W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991, 219, 37–44. [Google Scholar] [CrossRef]
- Casadesús, J.; Olivares, J. Rough and fine linkage mapping of the Rhizobium meliloti chromosome. Mol. Gen. Genet. 1979, 174, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Ditta, G.; Schmidhauser, T.; Yakobson, E.; Lu, P.; Liang, X.-W.; Finlay, D.R.; Guiney, D.; Helinski, D.R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 1985, 13, 149–153. [Google Scholar] [CrossRef]
- González-Silva, N.; López-Lara, I.M.; Reyes-Lamothe, R.; Taylor, A.M.; Sumpton, D.; Thomas-Oates, J.; Geiger, O. The dioxygenase-encoding olsD gene from Burkholderia cenocepacia causes the hydroxylation of the amide-linked fatty acyl moiety of ornithine-containing membrane lipids. Biochemistry 2011, 50, 6396–6408. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Bernabéu-Roda, L.; Calatrava-Morales, N.; Cuéllar, V.; Soto, M.J. Characterization of surface motility in Sinorhizobium meliloti: Regulation and role in symbiosis. Symbiosis 2015, 67, 79–90. [Google Scholar] [CrossRef]
- Hoben, H.J.; Somasegaran, P. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. App. Environ. Microbiol. 1982, 44, 1246–1247. [Google Scholar] [CrossRef]
- Nogales, J.; Domínguez-Ferreras, A.; Amaya-Gómez, C.V.; van Dillewijn, P.; Cuéllar, V.; Sanjuán, J.; Olivares, J.; Soto, M.J. Transcriptome profiling of a Sinorhizobium meliloti fadD mutant reveals the role of rhizobactin 1021 biosynthesis and regulation genes in the control of swarming. BMC Genomics 2010, 11, 157. [Google Scholar] [CrossRef]
- Sahonero-Canavesi, D.X.; Sohlenkamp, C.; Sandoval-Calderón, M.; Lamsa, A.; Pogliano, K.; López-Lara, I.M.; Geiger, O. Fatty acid-releasing activities in Sinorhizobium meliloti include unusual diacylglycerol lipase. Environ. Microbiol. 2015, 17, 3391–3406. [Google Scholar] [CrossRef] [PubMed]
- Fulda, M.; Heinz, E.; Wolter, F.P. The fadD gene of Escherichia coli K12 is located close to rnd at 39.6 min of the chromosomal-map and is a new member of the AMP-binding protein family. MGG 1994, 242, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Caviglia, J.M.; Li, L.O.; Wang, S.; DiRusso, C.C.; Coleman, R.A.; Lewin, T.M. Rat long chain acyl-CoA synthetase 5, but not 1, 2, 3, or 4, complements Escherichia coli fadD. J. Biol. Chem. 2004, 279, 11163–11169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, P.; Qi, Q. Molecular effect of FadD on the regulation and metabolism of fatty acid in Escherichia coli. FEMS Microbiol. Lett. 2006, 259, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Nyström, T. Stationary-phase physiology. Annu. Rev. Microbiol. 2004, 58, 161–181. [Google Scholar] [CrossRef]
- Farewell, A.; Diez, A.A.; DiRusso, C.C.; Nyström, T. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J. Bacteriol. 1996, 178, 6443–6450. [Google Scholar] [CrossRef]
- Sauviac, L.; Philippe, H.; Phok, K.; Bruand, C. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 2007, 189, 4204–4216. [Google Scholar] [CrossRef]
- Available online: http://iant.toulouse.inra.fr/bacteria/annotation/site/prj/rhime/doc/Bruand2007/ (accessed on 4 February 2020).
- Campbell, J.W.; Morgan-Kiss, R.M.; E. Cronan, J., Jr. A new Escherichia coli metabolic competency: Growth on fatty acids by a novel anaerobic β-oxidation pathway. Mol. Microbiol. 2003, 47, 793–805. [Google Scholar] [CrossRef]
- Lucas, R.L.; Lostroh, C.P.; DiRusso, C.C.; Spector, M.P.; Wanner, B.L.; Lee, C.A. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2000, 182, 1872–1882. [Google Scholar] [CrossRef]
- Jamet, A.; Euphrasie, D.; Martin, P.; Nassif, X. Identification of genes involved in Neisseria meningitidis colonization. Infect. Immun. 2013, 81, 3375–3381. [Google Scholar] [CrossRef]
- Banchio, C.; Gramajo, H.C. Medium- and long-chain fatty acid uptake and utilization by Streptomyces coelicolor A3(2): First characterization of a Gram-positive bacterial system. Microbiology 1997, 143, 2439–2447. [Google Scholar] [CrossRef] [PubMed]
- Tejima, K.; Ishiai, M.; Murayama, S.O.; Iwatani, S.; Kajiwara, S. Candida albicans fatty acyl-CoA synthetase, CaFaa4p, is involved in the uptake of exogenous long-chain fatty acids and cell activity in the biofilm. Curr. Genet. 2018, 64, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Bullock, H.A.; Shen, H.; Boynton, T.O.; Shimkets, L.J. Fatty acid oxidation is required for Myxococcus xanthus development. J. Bacteriol. 2018, 200, e00572-17. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, H.; Hirooka, K.; Fujita, Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 2007, 282, 5180–5194. [Google Scholar] [CrossRef] [PubMed]
- Black, P.N.; DiRusso, C.C. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. BBA-Mol. Cell Biol. Lipids 2007, 1771, 286–298. [Google Scholar] [CrossRef]

| Strain or Plasmid | Relevant Characteristicsa | Reference or Source |
|---|---|---|
| Escherichia coli | ||
| DH5α | recA1, Φ80 lacZ∆M1; cloning strain | [21] |
| S17-1 | thi pro recA hsdR− hsdM+ RP4 integrated in the chromosome, 2-Tc::Mu, Km::Tn7 (TpR/SmR) | [22] |
| Y-Mel | Wild type strain | [23] |
| YfadD1 | Y-Mel fadD::kan | [3] |
| JW1794-1 | BW25113 fadD::kan | [19] |
| BL21(DE3) | F−ompT hsdSB (rB−, mB−) gal dcm (DE3) | [24] |
| BfadD1 | BL21(DE3) fadD::kan | This work |
| Sinorhizobium meliloti | ||
| GR4 | Wild type strain | [25] |
| QS77 | fadD::Tn5 insertion mutant derivative of GR4, NmR | [2] |
| Plasmids | ||
| pLysS | CmR; causes repression of T7 polymerase | [24] |
| pET16b | Expression vector, CbR | Novagen |
| pET17b | Expression vector, CbR | Novagen |
| pAL55 | smb20650 in pBBR1MCS-5, GmR | [5] |
| pECH1 | sma0150 in pET16b, CbR | This work |
| pECH6 | smc02162 in pET17b, CbR | This work |
| pECH7 | smb20650 in pET17b, CbR | This work |
| pECH8 | E. coli fadD pET16b, CbR | This work |
| pBBR1MCS-3 | Broad-host range vector, TcR | [26] |
| pBBRD4 | pBBR1MCS-3 derivative harbouring the fadD gene of S. meliloti GR4, TcR | [2] |
| pRK404 | Broad-host range vector, TcR | [27] |
| pRCanul1 | pECH1 cloned as a BamHI fragment into pRK404, sma0150 | This work |
| pRCanul2 | pECH6 cloned as a HindIII fragment into pRK404, smc02162 | This work |
| pRCanul3 | pECH7 cloned as a BglII fragment into pRK404, smb20650 | This work |
| pRCanul4 | pECH8 cloned as a BamHI fragment into pRK404, fadDEcoli | This work |
| pNG28 | pET17b cloned in pRK404 | [28] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pech-Canul, Á.d.l.C.; Rivera-Hernández, G.; Nogales, J.; Geiger, O.; Soto, M.J.; López-Lara, I.M. Role of Sinorhizobium meliloti and Escherichia coli Long-Chain Acyl-CoA Synthetase FadD in Long-Term Survival. Microorganisms 2020, 8, 470. https://doi.org/10.3390/microorganisms8040470
Pech-Canul ÁdlC, Rivera-Hernández G, Nogales J, Geiger O, Soto MJ, López-Lara IM. Role of Sinorhizobium meliloti and Escherichia coli Long-Chain Acyl-CoA Synthetase FadD in Long-Term Survival. Microorganisms. 2020; 8(4):470. https://doi.org/10.3390/microorganisms8040470
Chicago/Turabian StylePech-Canul, Ángel de la Cruz, Geovanny Rivera-Hernández, Joaquina Nogales, Otto Geiger, María J. Soto, and Isabel M. López-Lara. 2020. "Role of Sinorhizobium meliloti and Escherichia coli Long-Chain Acyl-CoA Synthetase FadD in Long-Term Survival" Microorganisms 8, no. 4: 470. https://doi.org/10.3390/microorganisms8040470
APA StylePech-Canul, Á. d. l. C., Rivera-Hernández, G., Nogales, J., Geiger, O., Soto, M. J., & López-Lara, I. M. (2020). Role of Sinorhizobium meliloti and Escherichia coli Long-Chain Acyl-CoA Synthetase FadD in Long-Term Survival. Microorganisms, 8(4), 470. https://doi.org/10.3390/microorganisms8040470






