Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Analysis of the BSPM4 Phage Lysis Cassette
2.2. Bacterial Strains, Media, and Growth Conditions
2.3. Cloning and Expression of the Lysis Proteins
2.4. Overproduction and Purification of the M4LysΔTMD Protein
2.5. Lytic Activity and Host Range Test
2.6. Target Site Identification of M4Lys with HPLC
2.7. Functional Analysis of the M4Lys TMD
2.8. Phylogenetic Analysis and Sequence Alignment of M4Lys
3. Results and Discussion
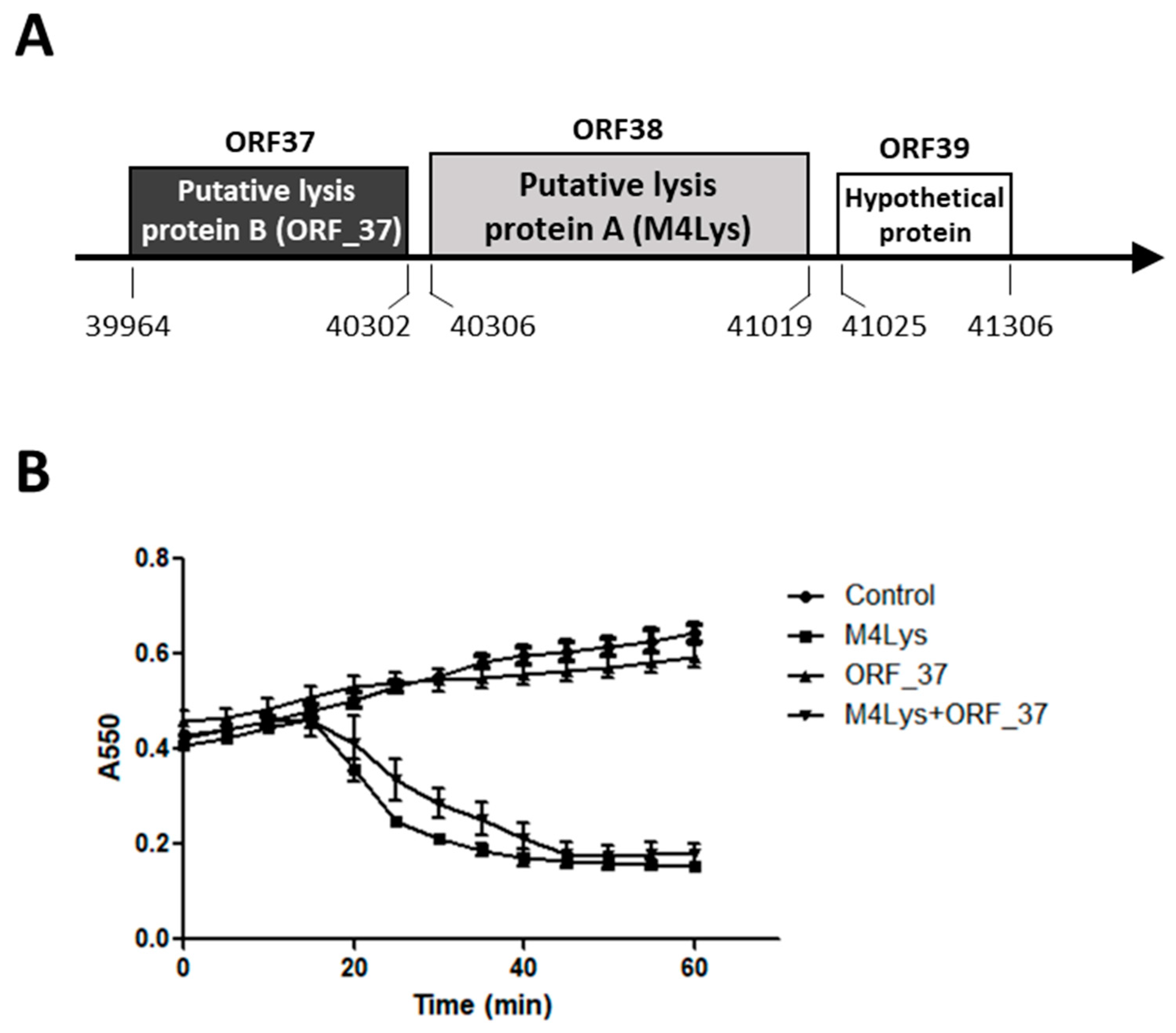
3.1. In Silico Analysis of the BSPM4 Lysis Cassette Showed Two Putative Lysis Genes
3.2. Only One Putative Lysis Gene Caused Cell Lysis when Overproduced in E. coli
3.3. Sec Machinery is Not Involved in M4Lys-Mediated Lysis In Vitro
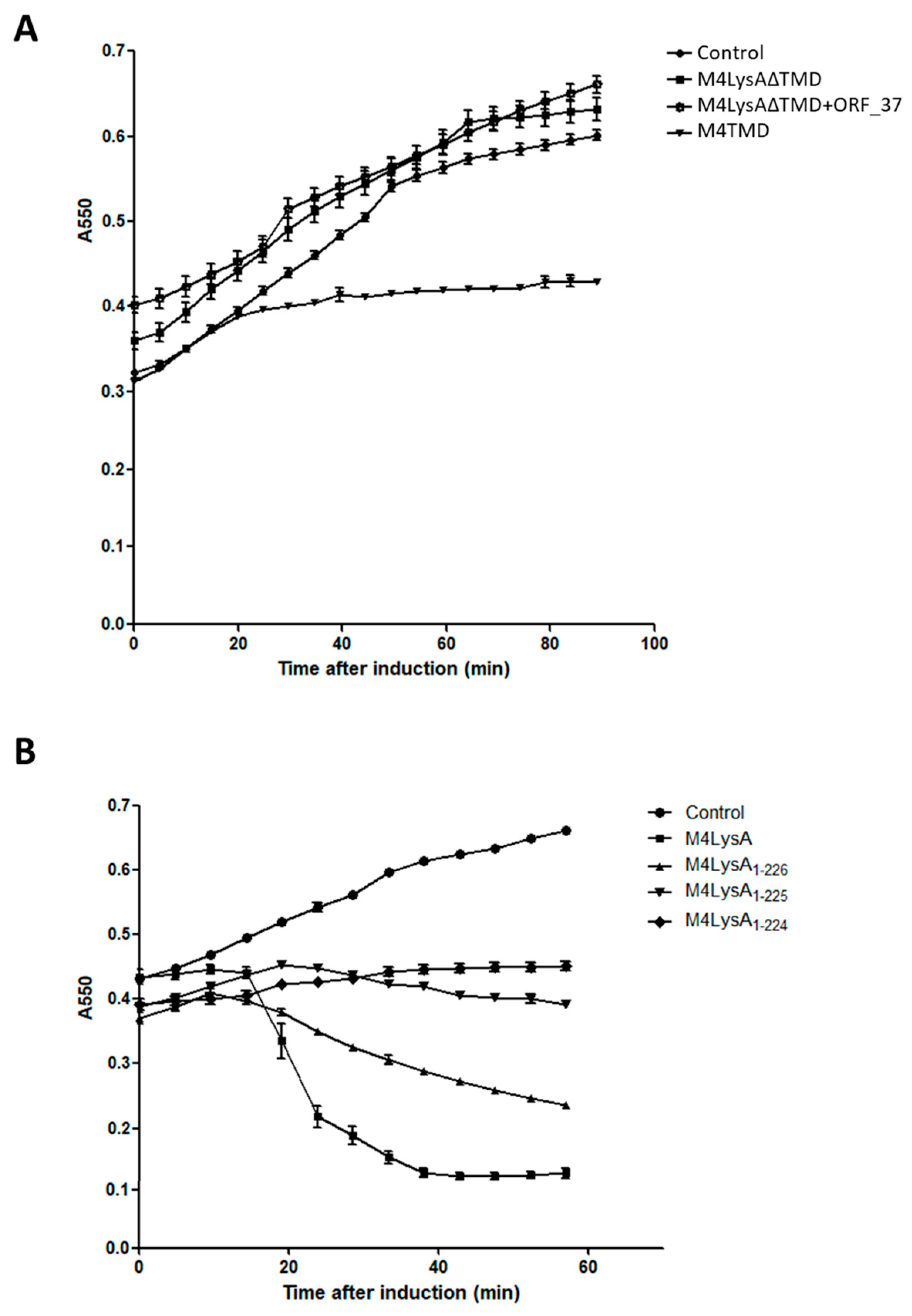
3.4. Domain Analysis of M4Lys
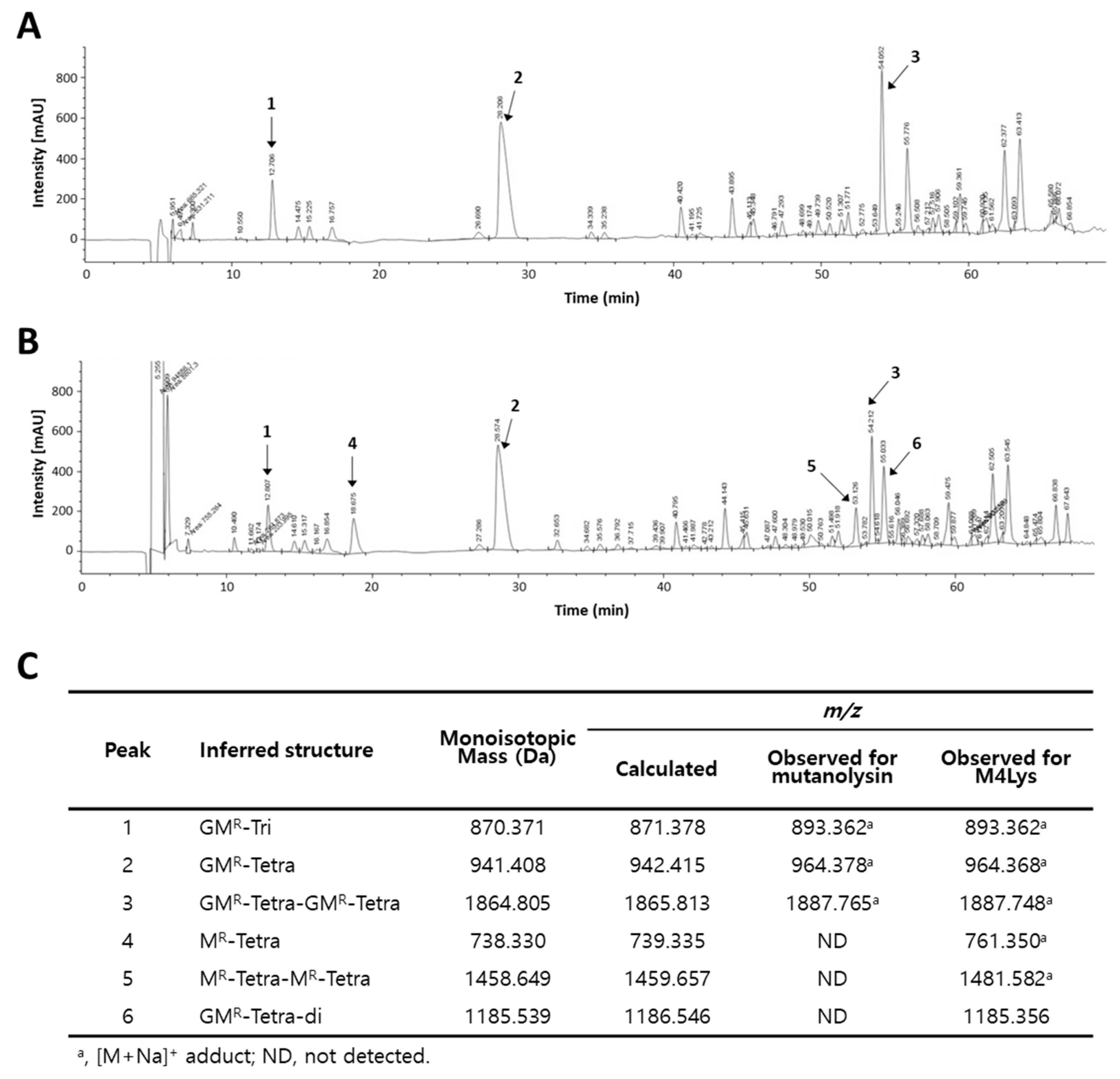
3.5. Catalytic Activity of M4Lys and Its Target Site Identification
3.6. The M4Lys TMD is Important for Cell Lysis by M4Lys In Vitro
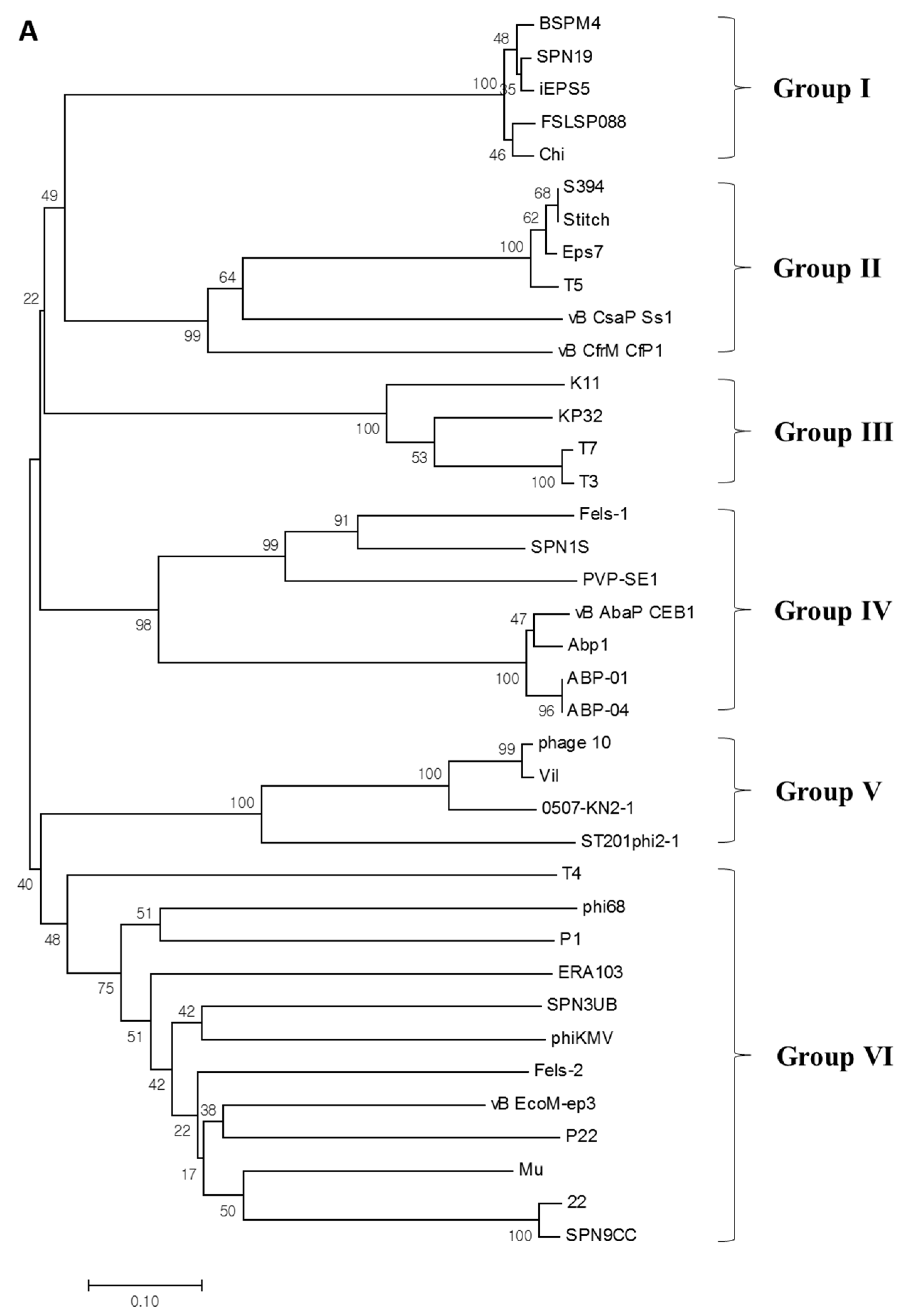
3.7. Phylogenetic Analysis of M4Lys
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; Roof, W.D.; Young, R. Genetic evidence that the bacteriophage phi X174 lysis protein inhibits cell wall synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 4297–4302. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; Struck, D.K.; Young, R. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J. Biol. Chem. 2001, 276, 6093–6097. [Google Scholar] [CrossRef] [PubMed]
- Young, I.; Wang, I.; Roof, W.D. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000, 8, 120–128. [Google Scholar] [CrossRef]
- Ackermann, H.W. Bacteriophage observations and evolution. Res. Microbiol. 2003, 154, 245–251. [Google Scholar] [CrossRef]
- Xu, M.; Arulandu, A.; Struck, D.K.; Swanson, S.; Sacchettini, J.C.; Young, R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 2005, 307, 113–117. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Xu, M.; Struck, D.K.; Deaton, J.; Wang, I.N.; Young, R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. USA 2004, 101, 6415–6420. [Google Scholar] [CrossRef]
- Sun, Q.; Kuty, G.F.; Arockiasamy, A.; Xu, M.; Young, R.; Sacchettini, J.C. Regulation of a muralytic enzyme by dynamic membrane topology. Nat. Struct. Mol. Biol. 2009, 16, 1192–1194. [Google Scholar] [CrossRef]
- Briers, Y.; Peeters, L.M.; Volckaert, G.; Lavigne, R. The lysis cassette of bacteriophage phiKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage 2011, 1, 25–30. [Google Scholar] [CrossRef]
- Sao-Jose, C.; Parreira, R.; Vieira, G.; Santos, M.A. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 2000, 182, 5823–5831. [Google Scholar] [CrossRef] [PubMed]
- Catalao, M.J.; Gil, F.; Moniz-Pereira, J.; Pimentel, M. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 2010, 77, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Jeon, B.; Ryu, S. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol. 2019, 77, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007, 35, D237–D240. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of Bacteriophage-T7 Rna-Polymerase to Direct Selective High-Level Expression of Cloned Genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Chang, Y.; Ryu, S. Characterization of a novel cell wall binding domain-containing Staphylococcus aureus endolysin LysSA97. Appl. Microbiol. Biotechnol. 2017, 101, 147–158. [Google Scholar] [CrossRef]
- Fein, J.E.; Rogers, H.J. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J. Bacteriol. 1976, 127, 1427–1442. [Google Scholar] [CrossRef]
- Lim, J.A.; Shin, H.; Kang, D.H.; Ryu, S. Characterization of endolysin from a Salmonella Typhimurium-infecting bacteriophage SPN1S. Res. Microbiol. 2012, 163, 233–241. [Google Scholar] [CrossRef]
- Lee, J.H.; Bai, J.; Shin, H.; Kim, Y.; Park, B.; Heu, S.; Ryu, S. A Novel Bacteriophage Targeting Cronobacter sakazakii Is a Potential Biocontrol Agent in Foods. Appl. Environ. Microbiol. 2016, 82, 192–201. [Google Scholar] [CrossRef]
- Desmarais, S.M.; Cava, F.; de Pedro, M.A.; Huang, K.C. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J. Vis. Exp. 2014, 83, e51183. [Google Scholar] [CrossRef]
- Singh, S.K.; SaiSree, L.; Amrutha, R.N.; Reddy, M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of E scherichia coli K 12. Mol. Microbiol. 2012, 86, 1036–1051. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.J. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments. Available online: http://www.psc.edu/biomed/genedoc (accessed on 17 February 2017).
- Choi, Y.; Shin, H.; Lee, J.H.; Ryu, S. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 2013, 79, 4829–4837. [Google Scholar] [CrossRef]
- Moreno Switt, A.I.; Orsi, R.H.; den Bakker, H.C.; Vongkamjan, K.; Altier, C.; Wiedmann, M. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genom. 2013, 14, 481. [Google Scholar] [CrossRef]
- Chang, Y.; Yoon, H.; Kang, D.H.; Chang, P.S.; Ryu, S. Endolysin LysSA97 is synergistic with carvacrol in controlling Staphylococcus aureus in foods. Int. J. Food Microbiol. 2017, 244, 19–26. [Google Scholar] [CrossRef]
- Fernandes, S.; Sao-Jose, C. More than a hole: The holin lethal function may be required to fully sensitize bacteria to the lytic action of canonical endolysins. Mol. Microbiol. 2016, 102, 92–106. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Valent, Q.A. Signal recognition particle mediated protein targeting in Escherichia coli. Antonie Van Leeuwenhoek 2001, 79, 17–31. [Google Scholar] [CrossRef]
- Briers, Y.; Lavigne, R. Breaking barriers: Expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol. 2015, 10, 377–390. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Server, T.V. 2.0. TMHMM. Available online: http://www.cbs.dtu.dk/services (accessed on 31 January 2019).
- Young, R.; Blasi, U. Holins: Form and function in bacteriophage lysis. FEMS Microbiol. Rev. 1995, 17, 191–205. [Google Scholar] [CrossRef]
- Borysowski, J.; Weber-Dabrowska, B.; Gorski, A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef]
- Zhang, W.H.; Mi, Z.Q.; Yin, X.Y.; Fan, H.; An, X.P.; Zhang, Z.Y.; Chen, J.K.; Tong, Y.G. Characterization of Enterococcus faecalis Phage IME-EF1 and Its Endolysin. PLoS ONE 2013, 8, e80435. [Google Scholar] [CrossRef]
- Yuan, Y.H.; Peng, Q.; Gao, M.Y. Characteristics of a broad lytic spectrum endolysin from phage BtCS33 of Bacillus thuringiensis. BMC Microbiol. 2012, 12, 297. [Google Scholar] [CrossRef]
- Kong, M.; Ryu, S. Bacteriophage PBC1 and Its Endolysin as an Antimicrobial Agent against Bacillus cereus. Appl. Environ. Microbiol. 2015, 81, 2274–2283. [Google Scholar] [CrossRef]
- Whitley, P.; Gafvelin, G.; Vonheijne, G. Seca-Independent Translocation of the Periplasmic N-Terminal Tail of an Escherichia-Coli Inner Membrane-Protein—Position-Specific Effects on Translocation of Positively Charged Residues and Construction of a Protein with a C-Terminal Translocation Signal. J. Biol. Chem. 1995, 270, 29831–29835. [Google Scholar]
- Lee, J.H.; Shin, H.; Choi, Y.; Ryu, S. Complete genome sequence analysis of bacterial-flagellum-targeting bacteriophage chi. Arch. Virol. 2013, 158, 2179–2183. [Google Scholar] [CrossRef]
- Shin, H.; Lee, J.H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS ONE 2012, 7, e43392. [Google Scholar] [CrossRef]
- Phothaworn, P.; Dunne, M.; Supokaivanich, R.; Ong, C.; Lim, J.; Taharnklaew, R.; Vesaratchavest, M.; Khumthong, R.; Pringsulaka, O.; Ajawatanawong, P.; et al. Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms. Viruses 2019, 11, 520. [Google Scholar] [CrossRef]



| Strains | Description | Reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | F- Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rk-, mk+) phoA supE44 thi-1 gyrA96 relA1 λ- | Invitrogen |
| BL21 (DE3) | F– ompT hsdSB (rB– mB–) gal dcm (DE3) | [18] |
| Plasmids | ||
| pET28a (+) | Expression vector with a hexahistidine tag, Kanr | Novagen |
| pETDuet-1 | Dual-expression vector with a hexahistidine tag, Ampr | Novagen |
| pM4LysΔTMD | pET28a-orf38ΔTMD1–206 | This study |
| M4Lys | pETDuet-1-orf38 | This study |
| ORF_37 | pETDuet-1-orf37 | This study |
| M4Lys-ORF_37 | pETDuet-1-orf37.orf38 | This study |
| M4LysΔTMD | pETDuet-1-orf38ΔTMD1–206 | This study |
| M4LysΔTMD-ORF_37 | pETDuet-1- orf38ΔTMD1–206.orf37 | This study |
| M4TMD | pETDuet-1-orf38TMD158–237 | This study |
| M4LysΔTMD-TMD | pETDuet-1- orf38ΔTMD1–206.TMD158–237 | This study |
| M4Lys1–226 | pET28a-orf381–226 | This study |
| M4Lys1–225 | pET28a-orf381–225 | This study |
| M4Lys1–224 | pET28a-orf381–224 | This study |
| Primer | Nucleotide Sequence [5′–3′] † | Restriction Site |
|---|---|---|
| BSPM4Lys-F-NdeI | ATG AGG AAA TAA CAT ATG GCT AAA CAG AAG | NdeI |
| BSPM4Lys-R-XhoI | GAA AAC GAG CGC CTC GAG GAC GCC CGT CTT | XhoI |
| BSPM4Lys2-R-XhoI | GGC CGC GCC GAA CTC GAG GCG TAC TCA ATC ACC | XhoI |
| BSPM4Lys5_3-R-XhoI | CTT CCG GTA CTC GAG TCA TCC CAT GTA CAG | XhoI |
| BSPM4Lys5_4-R-XhoI | CCG ATG CTT CTC GAG CGC TCA CCA TCC CAT | XhoI |
| BSPM4Lys6-R-XhoI | CCC GGC CCG ATG CTC GAG GTA CTA GAC CCA TCC | XhoI |
| BSPM4orf37-F-NcoI | CCC GCT AAT TTT TTG TGA GGA CCA TGG GC A TGA GCG AAA TGG AAC G | NcoI |
| BSPM4orf37-R-HindIII | GTT ATT GCG AAT CCC GCG AAG CTT CTG TTT AGC CAT CGG | HindIII |
| pETDuet-M4TMD-NcoI-F | CGA GGA AGG TCT GCC CAT GGG GGG CAT CGT TAA G | NcoI |
| pETDuet-M4TMD-HindIII-R | AAA ACG AGC GCC GCA AGC TTG CCC GTC TTG ATC C | HindIII |
| Duet-UP1-F | GAT GCG TCC GGC GTA GAG G | - |
| Duet-DOWN1-R | CGA TTA TGC GGC CGT GTA CAA T | - |
| Duet-UP2-F | ATT GTA CAC GGC CGC ATA ATC G | - |
| T7-promoter-F | TAATACGACTCACTATAGGG | - |
| T7-terminator-R | GCT AGT TAT TGC TCA GCG GTG | - |
| Bacterial Strain | BSPM4 (phage)* | M4LysΔTMD (endolysin)# |
|---|---|---|
| Salmonella enterica | ||
| serovar Typhimurium | ||
| SL1344 | CC | ++ |
| UK1 | CCC | ++ |
| LT2 | C | + |
| LT2C | C | + |
| ATCC14028† | C | ++ |
| ATCC19586 | C | + |
| ATCC43147 | CCC | + |
| ATCC13076 | C | ++ |
| DT104 | C | + |
| serovar Paratyphi | ||
| A IB 211 | - | + |
| B IB 231 | - | ++ |
| serovar Dublin IB 2973 | - | ++ |
| E. coli | ||
| BL21 | - | +++ |
| E. coliO157:H7 | ||
| ATCC35150 | - | +++ |
| ATCC43890 | - | + |
| ATCC43894 | - | +++ |
| ATCC43895 | - | ++ |
| O157:NM 3204-92 | - | ++ |
| O157:NM H-0482 | - | + |
| Gram-negative bacteria | ||
| Vibrio fischeri ES-114 ATCC 700601 | - | - |
| Pseudomonas aeruginosa ATCC 27853 | - | +++ |
| Cronobacter sakazakii ATCC29544 | - | + |
| Gram-positive bacteria | ||
| Staphylococcus aureus ATCC 29213 | - | - |
| Staphylococcus epidermis ATCC 35983 | - | - |
| Bacillus subtilis ATCC 23857 | - | - |
| Bacillus cereus ATCC 14579 | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, J.; Lee, S.; Ryu, S. Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria. Microorganisms 2020, 8, 447. https://doi.org/10.3390/microorganisms8030447
Bai J, Lee S, Ryu S. Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria. Microorganisms. 2020; 8(3):447. https://doi.org/10.3390/microorganisms8030447
Chicago/Turabian StyleBai, Jaewoo, Sangmi Lee, and Sangryeol Ryu. 2020. "Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria" Microorganisms 8, no. 3: 447. https://doi.org/10.3390/microorganisms8030447
APA StyleBai, J., Lee, S., & Ryu, S. (2020). Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria. Microorganisms, 8(3), 447. https://doi.org/10.3390/microorganisms8030447






