The Structure and Diversity of Nitrogen Functional Groups from Different Cropping Systems in Yellow River Delta
Abstract
1. Introduction
2. Materials and Methods
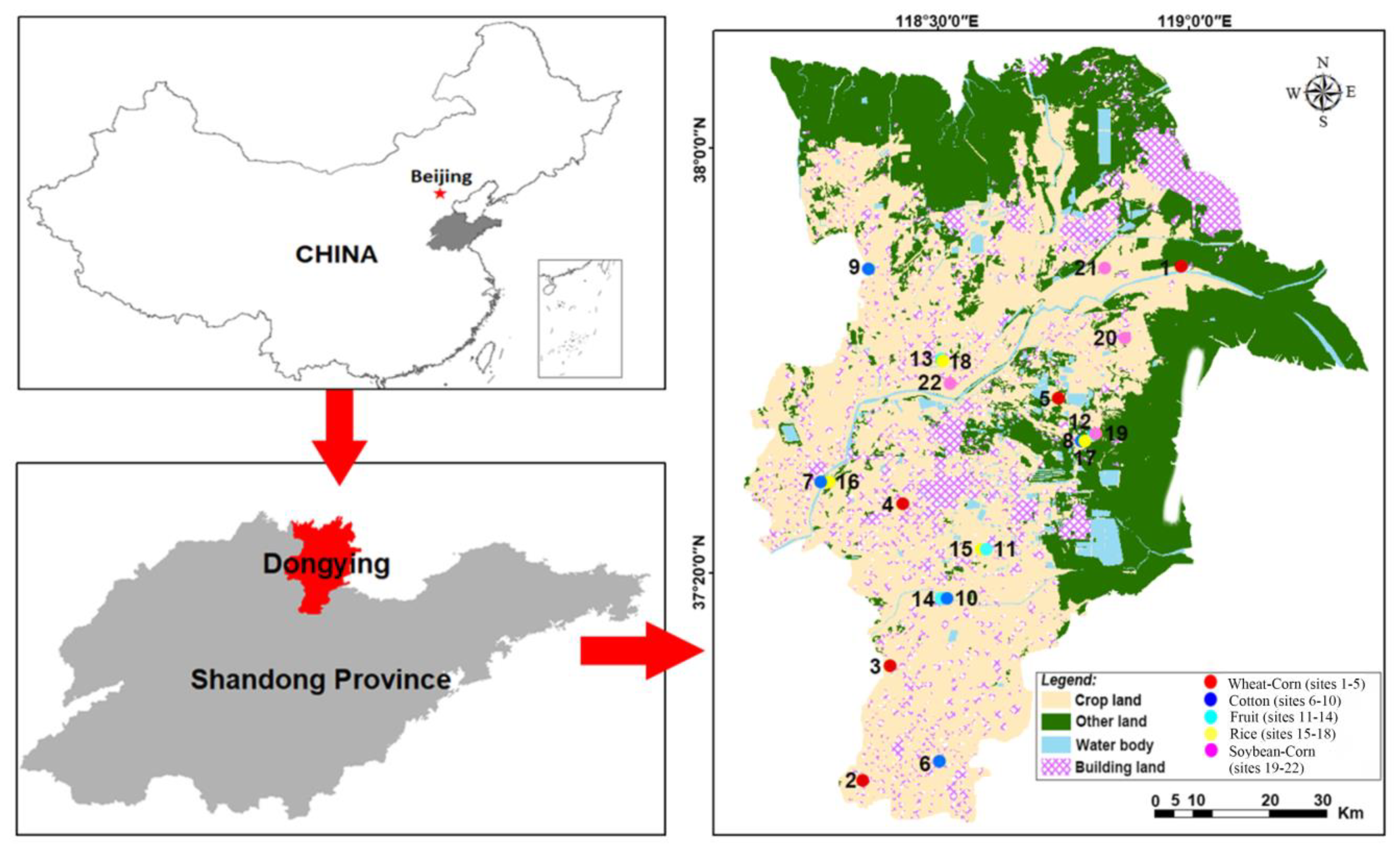
2.1. Study Area
2.2. Soil Sampling and Preparation
2.3. Analysis of Soil Properties
2.4. Potential Nitrification Rate (PNR)
2.5. Soil DNA Extraction and Real-Time Quantitative PCR
2.6. High–Throughput Sequencing and Data Analysis
2.7. Accession Numbers
3. Results
3.1. Soil Physicochemical Properties
3.2. Diversity of Bacteria and N–Cycling Functional Groups in the Five Cropping Systems
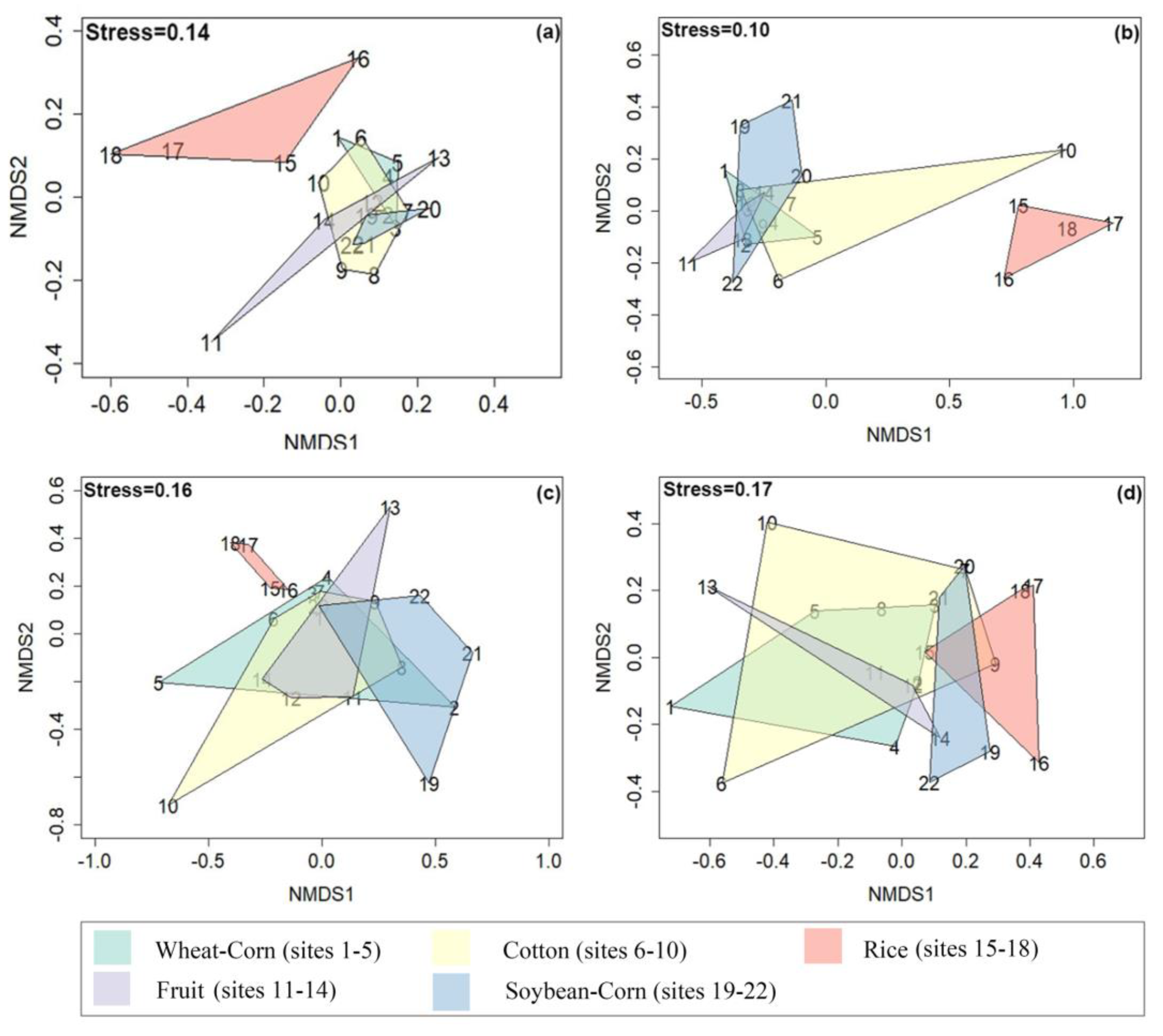
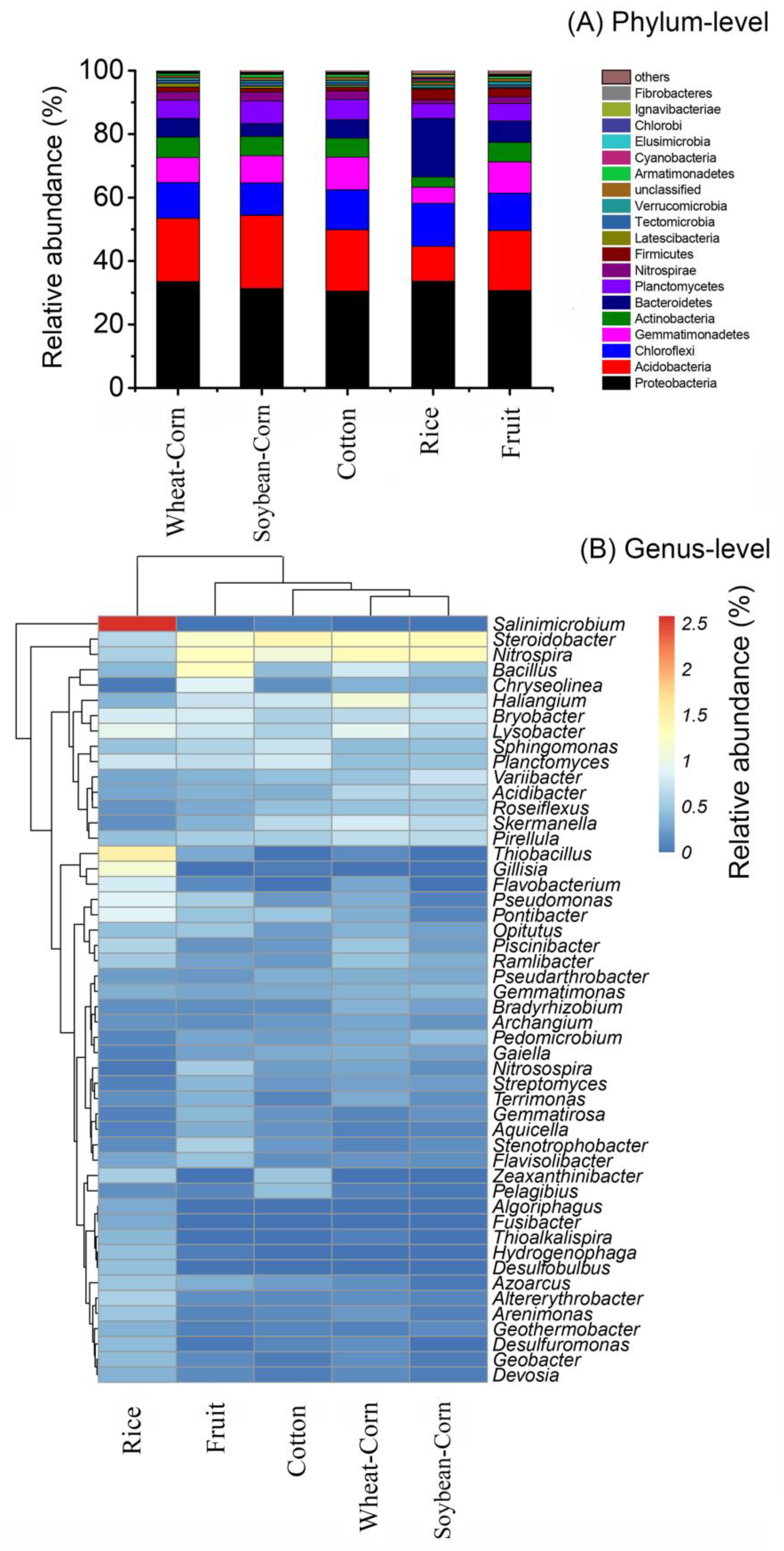
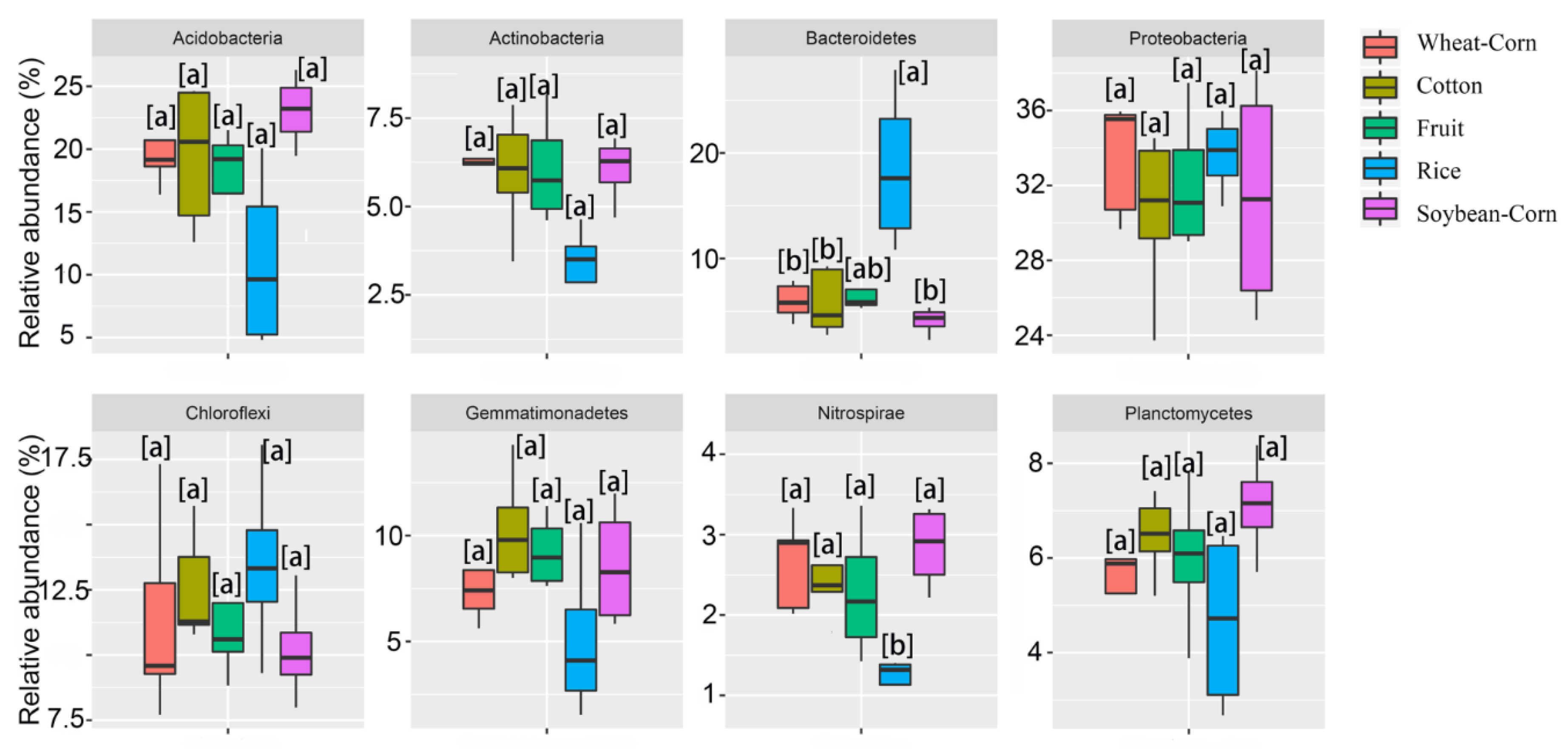
3.3. Composition and Difference in the Bacterial Community
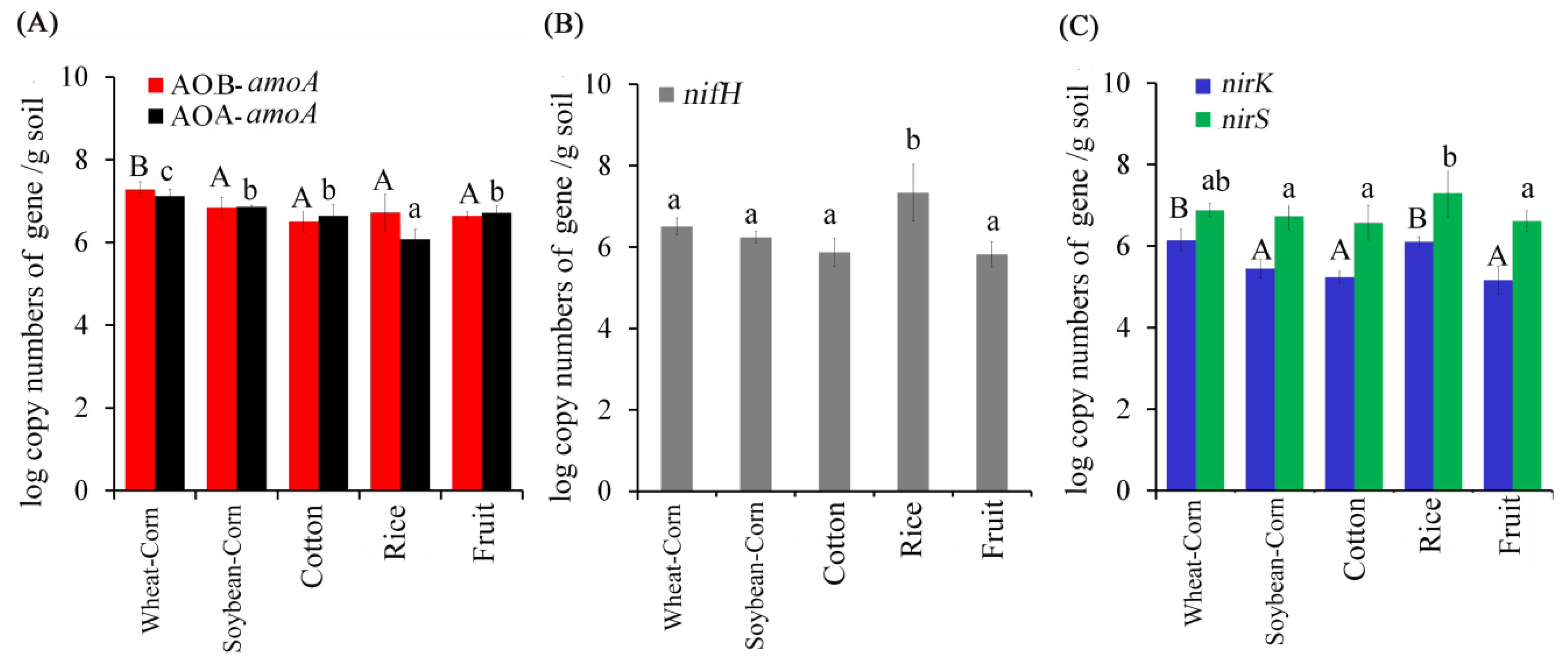
3.4. Abundance of N–Cycling Genes
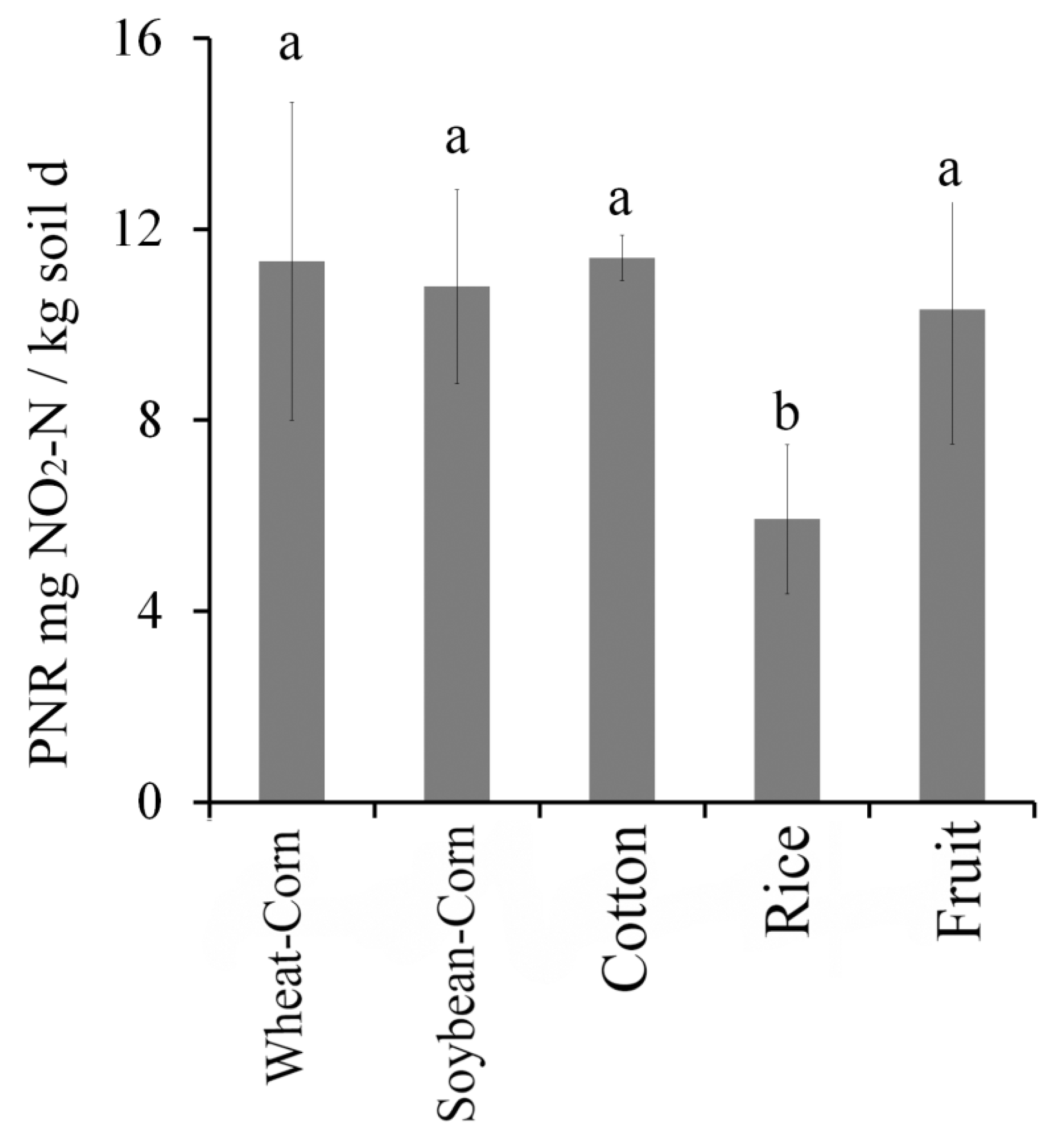
3.5. Potential Nitrification Rate (PNR)
3.6. Composition of N–Cycling Functional Communities
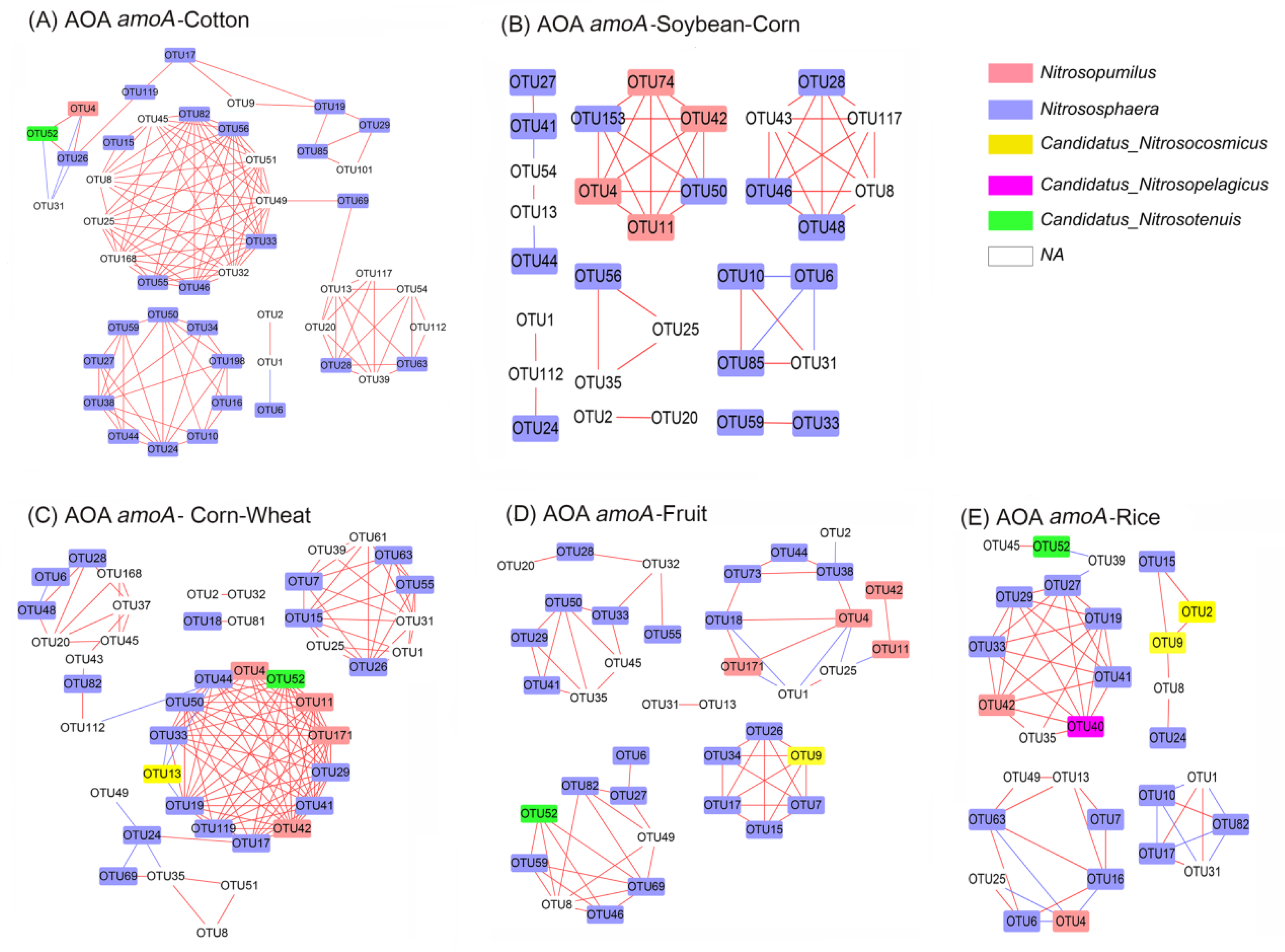
3.7. Co–Ocurrence Network Analysis of amoA–AOA and nirS–Denitrifier
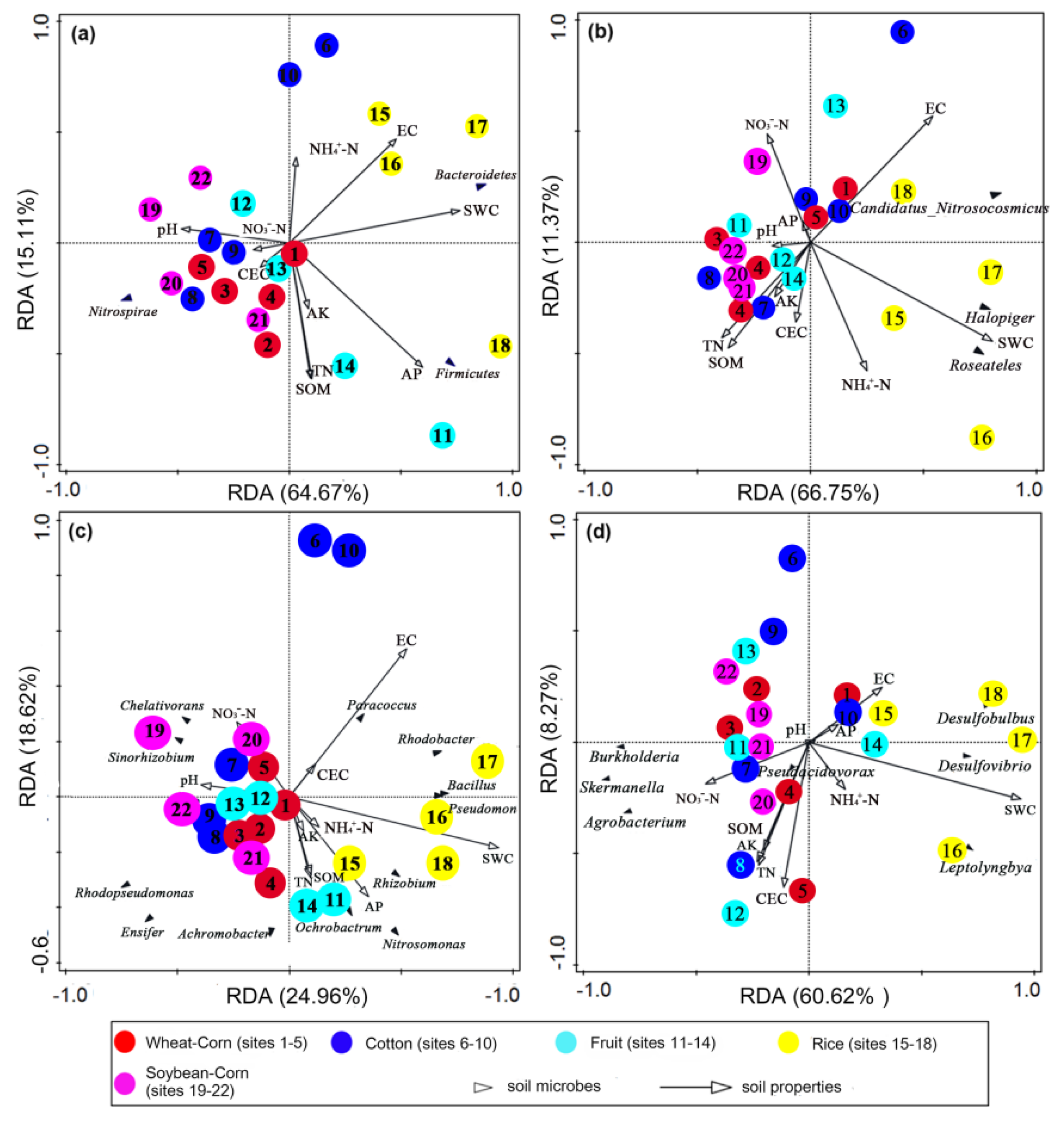
3.8. Key Drivers of Bacterial and N–Cycling Functional Communities
4. Discussion
4.1. Abundance and Diversity of Chosen N–Cycling Functional Genes in Different Cropping Systems
4.2. Factor Affecting the Diversity and Composition of Bacterial and N–Cycling Functional Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, C.; Zhang, X.; Huang, Q. Four Decades of Estuarine Wetland Changes in the Yellow River Delta Based on Landsat Observations between 1973 and 2013. Water 2018, 10, 933. [Google Scholar] [CrossRef]
- Zhang, T.T.; Zeng, S.L.; Gao, Y.; Ouyang, Z.T.; Li, B.; Fang, C.M.; Zhao, B. Assessing impact of land uses on land salinization in the Yellow River Delta, China using an integrated and spatial statistical model. Land Use Policy 2011, 28, 857–866. [Google Scholar] [CrossRef]
- Shan, J.; Chen, X.; Yin, C.; Wen, P.; Yan, K.; Zhang, L.; Zhang, L.; Fu, X.; Sun, H. Comparison of fertilizer-effect models on winter wheat response to nitrogen and phosphorus fertilizers in saline soils in the Yellow River Delta. Chin. J. Eco-Agric. 2017, 25, 1016–1024. (In Chinese) [Google Scholar]
- Yi, X.M.; Yuan, J.; Zhu, Y.H.; Yi, X.J.; Zhao, Q.; Fang, K.K.; Cao, L.K. Comparison of the Abundance and Community Structure of N-Cycling Bacteria in Paddy Rhizosphere Soil under Different Rice Cultivation Patterns. Int. J. Mol. Sci. 2018, 19, 3772. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant Acidification in Major Chinese Croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.S.; Qu, Y.; Kilasara, M.M.; Mmari, W.N.; Funakawa, S. Nitrate leaching from the critical root zone of maize in two tropical highlands of Tanzania: Effects of fertilizer-nitrogen rate and straw incorporation. Soil Tillage Res. 2019, 194, 104295. [Google Scholar] [CrossRef]
- Liu, J.; Yang, W. Water Sustainability for China and Beyond. Science 2016, 337, 649–650. [Google Scholar] [CrossRef]
- Gao, S.; Xu, P.; Zhou, F.; Yang, H.; Zheng, C.; Cao, W.; Tao, S.; Piao, S.; Zhao, Y.; Shang, Z. Quantifying nitrogen leaching response to fertilizer additions in China’s cropland. Environ. Pollut. 2016, 211, 241–251. [Google Scholar] [CrossRef]
- De Boer, W.; Kowalchuk, G.A. Nitrification in acid soils: Micro-organisms and mechanisms. Soil Biol. Biochem. 2001, 33, 853–866. [Google Scholar] [CrossRef]
- Liu, W. Study on N Uptake, N Utilization and Nodule N Fixation Characteristic of Maize-Soyben Relay Strip Intercropping System. Master’s Thesis, Sichuan Agricultural University, Sichuan, China, 2014. [Google Scholar]
- Ding, K.; Zhong, L.; Xin, X.P.; Xu, Z.H.; Kang, X.M.; Liu, W.J.; Rui, Y.C.; Jiang, L.L.; Tang, L.; Wang, Y.F. Effect of grazing on the abundance of functional genes associated with N cycling in three types of grassland in Inner Mongolia. J. Soil Sediment 2015, 15, 683–693. [Google Scholar] [CrossRef]
- Jiang, M.; Zheng, X.; Chen, Y. Enhancement of denitrification performance with reduction of nitrite accumulation and N2O emission by Shewanella oneidensis MR-1 in microbial denitrifying process. Water Res. 2019, 169, 115242. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.D.; Ren, Y.F.; Wang, X.Q.; Hu, Y.G.; Wang, Z.M.; Zeng, Z.H. Ammonia-oxidizing archaea and bacteria responding differently to fertilizer type and irrigation frequency as revealed by Illumina Miseq sequencing. J. Soil Sediment 2018, 18, 1029–1040. [Google Scholar] [CrossRef]
- Ahmed, S.E.; Desoky, E.S.M.; El-Maati, M.F.A.; Elnahal, A.S.; Abdo, A.I.; Raza, S.; Zhou, J.B. Can secondary metabolites extracted from Moringa seeds suppress ammonia oxidizers to increase nitrogen use efficiency and reduce nitrate contamination in potato tubers? Ecotoxicol. Environ. Saf. 2019, 185, 109689. [Google Scholar]
- Hayatsu, M.; Tago, K.; Saito, M. Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 2008, 54, 33–45. [Google Scholar] [CrossRef]
- Liu, R.; Hayden, H.L.; Hu, H.W.; He, J.Z.; Suter, H.; Chen, D.L. Effects of the nitrification inhibitor acetylene on nitrous oxide emissions and ammonia-oxidizing microorganisms of different agricultural soils under laboratory incubation conditions. Appl. Soil Ecol. 2017, 119, 80–90. [Google Scholar] [CrossRef]
- Tago, K.; Ishii, S.; Nishizawa, T.; Otsuka, S.; Senoo, K. Phylogenetic and Functional Diversity of Denitrifying Bacteria Isolated from Various Rice Paddy and Rice-Soybean Rotation Fields. Microbes Environ. 2011, 26, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Munroe, J.W.; Mccormick, I.; Deen, W.; Dunfield, K.E. Effects of 30 Years of Crop Rotation and Tillage on Bacterial and Archaeal Ammonia Oxidizers. J. Environ. Qual. 2016, 45, 940–948. [Google Scholar] [CrossRef]
- Yin, J.; Jiang, Z.; Li, X.; Zhang, S.; Haibara, K. Strategic concept on comprehensive development of Yellow river delta saline and alkaline land. J. Nanjing For. 2000, 24, 61–63. (In Chinese) [Google Scholar]
- Ding, K.; Wen, X.; Li, Y.; Shen, B.; Zhang, B. Ammonia-oxidizing archaea versus bacteria in two soil aquifer treatment systems. Appl. Microbiol. Biotechnol. 2015, 99, 1337–1347. [Google Scholar] [CrossRef]
- Poly, F.; Monrozier, L.J.; Bally, R. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 2001, 152, 95–103. [Google Scholar] [CrossRef]
- Francis, C.A.; Santoro, A.E.; Oakley, B.B.; Beman, J.M.; Roberts, K.J. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microb. 1997, 63, 4704–4712. [Google Scholar] [CrossRef]
- Michotey, V.; Mejean, V.; Bonin, P. Comparison of methods for quantification of cytochrome cd(1)-denitrifying bacteria in environmental marine samples. Appl. Microbiol. Biotechnol. 2000, 66, 1564–1571. [Google Scholar]
- Hallin, S.; Lindgren, P.E. PCR detection of genes encoding nitrile reductase in denitrifying bacteria. Microbiol. Mol. Biol. 1999, 65, 1652–1657. [Google Scholar]
- Ellen, K.; Thomas, B.; Esther, E.; Nicole, D.; Georg, G.; Norbert, L.; Laurent, P. Response of total and nitrate-dissimilating bacteria to reduced N deposition in a spruce forest soil profile. FEMS Microbiol. Ecol. 2009, 67, 444–454. [Google Scholar]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Grobler, D.; Davies, E. The use of the Walkley-Black method for organic carbon determination as a procedure for estimating algal yields. Water 1979, 5, 138–143. [Google Scholar]
- Sparks, D.; Page, A.; Helmke, P.; Loeppert, R.; Soltanpour, P.; Tabatabai, M.; Sumner, M. Methods of Soil Analysis; Soil Science Society of America Inc.: Madison, WI, USA, 1996. [Google Scholar]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; Circular: Washington, DC, USA, 1954. [Google Scholar]
- Gan, Y.; Wang, L.; Yang, G.; Dai, J.; Wang, R.; Wang, W. Multiple factors impact the contents of heavy metals in vegetables in high natural background area of China. Chemosphere 2017, 184, 1388–1395. [Google Scholar] [CrossRef]
- Kurola, J.; Salkinoja-Salonen, M.; Aarnio, T.; Hultman, J.; Romantschuk, M. Activity, diversity and population size of ammonia-oxidising bacteria in oil-contaminated landfarming soil. FEMS Microbiol. Lett. 2005, 250, 33–38. [Google Scholar] [CrossRef]
- Knowles, R. Denitrification. Microbiol. Rev. 1982, 46, 43–49. [Google Scholar] [CrossRef]
- Wei, W.; Isobe, K.; Nishizawa, T.; Zhu, L.; Shiratori, Y.; Ohte, N.; Koba, K.; Otsuka, S.; Senoo, K. Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 2015, 9, 1954–1965. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.; Boumans, L.; Batjes, N. Emissions of N2O and NO from fertilized fields: Summary of available measurement data. Glob. Biogeochem. Cycle 2002, 16, 61–63. [Google Scholar] [CrossRef]
- Torsvik, V.; Ovreas, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.; Hartmann, M. Networking in the plant microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef]
- Verhamme, D.T.; Prosser, J.I.; Nicol, G.W. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 2011, 5, 1067–1071. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, Y.; Zhu-Barker, X.; He, X.; Horwath, W.R.; Luo, H.; Zhao, Y.; Jiang, X. Responses of nitrification and ammonia oxidizers to a range of background and adjusted pH in purple soils. Geoderma 2019, 334, 9–14. [Google Scholar] [CrossRef]
- Prosser, J.I.; Hink, L.; Gubry-Rangin, C.; Nicol, G.W. Nitrous oxide production by ammonia oxidizers: Physiological diversity, niche differentiation and potential mitigation strategies. Glob. Chang. Biol. 2019. [Google Scholar] [CrossRef]
- Dini-Andreote, F.; Silva, M.D.C.P.; Triado-Margarit, X.; Casamayor, E.O.; Van Elsas, J.D.; Salles, J.F. Dynamics of bacterial community succession in a salt marsh chronosequence: Evidences for temporal niche partitioning. ISME J. 2014, 8, 1989–2001. [Google Scholar] [CrossRef]
- Maron, P.-A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microb. 2018, 84, e02738-17. [Google Scholar] [CrossRef]
- Pester, M.; Rattei, T.; Flechl, S.; Gröngröft, A.; Wagner, M. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 2011, 14, 525–539. [Google Scholar] [CrossRef]
- Meinhardt, K.A.; Bertagnolli, A.; Pannu, M.W.; Strand, S.E.; Brown, S.L.; Stahl, D.A. Evaluation of revised polymerase chain reaction primers for more inclusive quantification of ammonia-oxidizing archaea and bacteria. Environ. Microbiol. Rep. 2015, 7, 354–363. [Google Scholar] [CrossRef]
- Throback, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2006, 49, 401–417. [Google Scholar] [CrossRef]
- Bonilla-Rosso, G.; Wittorf, L.; Jones, C.M.; Hallin, S. Design and evaluation of primers targeting genes encoding NO-forming nitrite reductases: Implications for ecological inference of denitrifying communities. Sci. Rep. 2016, 6, 39208. [Google Scholar] [CrossRef]
- Gaby, J.C.; Rishishwar, L.; Valderrama-Aguirre, L.C.; Green, S.J.; Valderrama-Aguirre, A.; Jordan, I.K.; Kostka, J.E. Diazotroph community characterization via a high-throughput nifH amplicon sequencing and analysis pipeline. Appl. Environ. Microbiol. 2017, 84, e01512–e01517. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. The Use of Degenerate Primers in qPCR Analysis of Functional Genes Can Cause Dramatic Quantification Bias as Revealed by Investigation of nifH Primer Performance. Microb. Ecol. 2017, 74, 701–708. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Soil moisture as a factor affecting the microbiological and biochemical activity of soil. Plant Soil Environ. 2016, 62, 250–255. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002, 53, 2401–2410. [Google Scholar] [CrossRef]
- Mmolawa, K.; Or, D. Root zone solute dynamics under drip irrigation: A review. Plant Soil 2000, 222, 163–190. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Błaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Szukics, U.; Abell, G.C.; Hödl, V.; Mitter, B.; Sessitsch, A.; Hackl, E.; Zechmeister-Boltenstern, S. Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol. Lett. 2010, 72, 395–406. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Y.; Xiang, Q.; Yu, X.; Zhao, K.; Zhang, X.; Lindström, K.; Hu, Y.; Liu, S. Implications of wetland degradation for the potential denitrifying activity and bacterial populations with nirS genes as found in a succession in Qinghai-Tibet plateau, China. Eur. J. Soil Biol. 2017, 80, 19–26. [Google Scholar]
- Reed, S.C.; Cleveland, C.C.; Townsend, A.R. Functional Ecology of Free-Living Nitrogen Fixation: A Contemporary Perspective. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 489–512. [Google Scholar] [CrossRef]

| Genes | Premiers | Premier Sequences (5′–3′) | Reference(s) |
|---|---|---|---|
| 16S rRNA | 515F | GTGCCAGCMGCCGCGGTAA | Ding et al. (2015) [20] |
| 926R | CCGTCAATTCMTTTGAGTTT | ||
| nifH | PolF | TGCGAYCCSAARGCBGACTC | Poly et al. (2001) [21] |
| PolR | ATSGCCATCATYTCRCCGGA | ||
| amoA–AOA | Arch amoA F | STAATGGTCTGGCTTAGACG | Francis et al. (2005) [22] |
| Arch amoA R | GCGGCCATCCATCTGTATGT | ||
| amoA–AOB | amoA–1F | GGGGTTTCTACTGGTGGT | Rotthauwe et al. (1997) [23] |
| amoA–2R | CCCCTCKGSAAAGCCTTCTTC | ||
| nirS | cd3aF | GTSAACGTSAAGGARACSGG | Michotey et al. (2000) [24] |
| R3cd | AGTTCTGSGTRGGCTTSAG | Hallin and Lindgren (1999) [25] | |
| nirK | F1aCu | ATCATGGTSCTGCCGCG | Kandeler et al. (2009) [26] |
| R3Cu | GCCTCGATCAGRTTGTGGTT |
| Agroecosystem | SWC (%) | AP (mg kg−1) | AK (g kg−1) | pH | SOM (g kg−1) | TN (g kg−1) | NH4+–N (mg kg−1) | NO3−–N (mg kg−1) | CEC [cmol (+) kg −1] | EC (mS cm−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| Wheat–Corn | 21.3 ± 3.7 a | 14.5 ± 11.1 a | 0.18 ± 0.05 a | 8.0 ± 0.0 a | 14.9 ± 3.3 a | 0.83 ± 0.17 a | 8.0 ± 1.2 a | 36.7 ± 29.4 a | 12.6 ± 2.3 a | 0.34 ± 0.16 a |
| Soybean–Corn | 16.3 ± 2.7 a | 10.4 ± 7.8 a | 0.19 ± 0.04 a | 8.1 ± 0.2 a | 14.1 ± 4.1 a | 0.78 ± 0.23 a | 9.1 ± 0.9 a | 32.9 ± 18.4 a | 10.5 ± 3.0 a | 0.30 ± 0.09 a |
| Cotton | 18.3 ± 1.7 a | 14.0 ± 9.3 a | 0.22 ± 0.06 a | 8.0 ± 0.1 a | 13.2 ± 3.4 a | 0.73 ± 0.21 a | 7.7 ± 0.7 a | 48.1 ± 30.3 a | 11.4 ± 1.7 a | 0.65 ± 0.48 a |
| Rice | 38.1 ± 4.4 b | 31.8 ± 20.0 a | 0.12 ± 0.07 a | 7.9 ± 0.1 a | 13.1 ± 2.6 a | 0.72 ± 0.15 a | 10.9 ± 4.6 a | 8.7 ± 4.8 a | 10.7 ± 3.5 a | 0.63 ± 0.23 a |
| Fruit | 21.0 ± 3.7 a | 71.7 ± 42.9 b | 0.37 ± 0.11 b | 7.8 ± 0.3 a | 16.2 ± 5.5 a | 1.00 ± 0.36 a | 7.8 ± 0.9 a | 75.1 ± 41.1 a | 11.6 ± 1.6 a | 0.60 ± 0.22 a |
| Microbial Community | Gene Type | Samples | Number of OTUs | Coverage (%) | Chao Index | Ace Index | Shannon Index |
|---|---|---|---|---|---|---|---|
| Ammonia-oxidizers | amoA–AOA | Wheat–Corn | 72.40 ± 4.80 a | 99.94 ± 0.02a | 77.21 ± 4.70 a | 77.78 ± 5.90 a | 2.55 ± 0.28 a |
| Soybean–Corn | 72.00 ± 3.00 a | 99.94 ± 0.01a | 79.45 ± 5.31 a | 80.64 ± 3.51 ab | 2.20 ± 0.15 a | ||
| Cotton | 67.00 ± 4.73 a | 99.94 ± 0.02a | 75.07 ± 3.72 a | 77.66 ± 6.15 a | 2.24 ± 0.29 a | ||
| Rice | 87.50 ± 2.87 b | 99.95 ± 0.01a | 94.33 ± 2.90 b | 93.33 ± 3.26 c | 2.55 ± 0.14 a | ||
| Fruit | 75.50 ± 8.67 a | 99.93 ± 0.02a | 90.95 ± 10.49 b | 90.34 ± 6.48 bc | 2.45 ± 0.18 a | ||
| amoA–AOB | Wheat–Corn | 105.0 ± 26.01 ab | 99.78 ± 0.08 ab | 120.1 ± 22.64 ab | 124.0 ± 23.03 ab | 3.08 ± 0.37 a | |
| Soybean–Corn | 91.25 ± 24.03 ab | 99.81 ± 0.07 ab | 105.2 ± 23.30 a | 103.9 ± 23.28 a | 2.58 ± 0.32 a | ||
| Cotton | 89.00 ± 13.60 ab | 99.84 ± 0.02 b | 104.2 ± 20.83 a | 99.9 ± 14.33 a | 2.71 ± 0.084 a | ||
| Rice | 127.5 ± 30.83 b | 99.68 ± 0.09 a | 159.7 ± 31.93 b | 154.7 ± 30.69 b | 2.74 ± 0.33 a | ||
| Fruit | 79.25 ± 17.18 a | 99.78 ± 0.10 ab | 96.9 ± 22.14 a | 98.0 ± 28.40 a | 2.67 ± 0.68 a | ||
| N-fixing bacteria | nifH | Wheat–Corn | 474.2 ± 182.08 a | 99.84 ± 0.04 b | 581.3 ± 195.8 a | 577.0 ± 164.9 a | 4.60 ± 0.38 ab |
| Soybean–Corn | 411.8 ± 91.25 a | 99.84 ± 0.01 b | 509.7 ± 80.04 a | 489.9 ± 69.41 a | 4.37 ± 0.22 a | ||
| Cotton | 412.0 ± 27.18 a | 99.85 ± 0.03 b | 535.1 ± 43.64 a | 520.5 ± 62.26 a | 4.48 ± 0.40 a | ||
| Rice | 998.3 ± 88.85 b | 99.46 ± 0.11 a | 1187 ± 114.0 b | 1163 ± 108.3 b | 5.30 ± 0.19 b | ||
| Fruit | 450.3 ± 253.0 a | 99.84 ± 0.07 b | 562.9 ± 240.8 a | 570.2 ± 219.1 a | 4.33 ± 0.65 a | ||
| Denitrifier | nirK | Wheat–Corn | 434.8 ± 50.92 a | 99.90 ± 0.01 b | 469.2 ± 51.34 a | 455.3 ± 50.21 a | 4.53 ± 0.16 a |
| Soybean–Corn | 391.0 ± 54.54 a | 99.89 ± 0.02 b | 455.4 ± 35.07 a | 423.7 ± 48.51 a | 4.40 ± 0.19 a | ||
| Cotton | 345.0 ± 48.77 a | 99.90 ± 0.00 b | 415.8 ± 59.16 a | 396.6 ± 54.22 a | 4.26 ± 0.42 a | ||
| Rice | 649.0 ± 62.52 b | 99.70 ± 0.02 a | 734.5 ± 82.10 b | 722.1 ± 64.21 b | 4.43 ± 0.18 a | ||
| Fruit | 412.0 ± 114.8 a | 99.91 ± 0.02 b | 452.0 ± 110.8 a | 445.1 ± 105.3 a | 4.26 ± 0.51 a | ||
| nirS | Wheat–Corn | 569.4 ± 107.0 b | 99.82 ± 0.05 b | 663.7 ± 85.82 b | 647.4 ± 92.69 b | 4.69 ± 0.09 b | |
| Soybean–Corn | 399.5 ± 20.93 a | 99.81 ± 0.04 b | 469.4 ± 14.44 a | 459.0 ± 20.46 a | 3.97 ± 0.25 a | ||
| Cotton | 439.0 ± 53.85 ab | 99.84 ± 0.04 b | 506.7 ± 54.16 a | 492.3 ± 53.99 a | 4.41 ± 0.29 ab | ||
| Rice | 830.0 ± 85.79 c | 99.61 ± 0.09 a | 951.8 ± 100.7 c | 931.0 ± 101.0 c | 5.26 ± 0.15 c | ||
| Fruit | 425.3 ± 77.04 ab | 99.81 ± 0.06 b | 515.2 ± 75.98 a | 508.1 ± 67.29 ab | 4.17 ± 0.56 ab | ||
| Bacteria | 16S rRNA | Wheat–Corn | 2930 ± 85.18 a | 98.33 ± 0.28 b | 3725 ± 194.1 a | 3697 ± 157.9 a | 6.49 ± 0.02 a |
| Soybean–Corn | 3050 ± 323.3 a | 98.40 ± 0.13 b | 3832 ± 415.1 a | 3791 ± 367.1 a | 6.50 ± 0.20 a | ||
| Cotton | 2929 ± 278.3 a | 98.54 ± 0.11 b | 3626 ± 314.5 a | 3612 ± 314.3 a | 6.50 ± 0.16 a | ||
| Rice | 3343 ± 279.8 a | 97.85 ± 0.15 a | 4322 ± 356.0 a | 4264 ± 352.4 a | 6.71 ± 0.20 a | ||
| Fruit | 2957 ± 556.5 a | 98.42 ± 0.35 b | 3702 ± 648.3 a | 3686 ± 676.5 a | 6.53 ± 0.25 a |
| Microbial Community | Gene Type | A | Expected δ | p |
|---|---|---|---|---|
| Ammonia–oxidizers | amoA–AOA | 0.185 | 0.426 | 0.007 ** |
| amoA–AOB | 0.018 | 0.664 | 0.177 | |
| N–fixing bacteria | nifH | 0.003 | 0.536 | 0.433 |
| Denitrifier | nirK | 0.025 | 0.525 | 0.154 |
| nirS | 0.063 | 0.401 | 0.027 * | |
| Bacteria | 16S rRNA | 0.532 | 0.533 | 0.001 ** |
| Corn-Wheat | Cotton | Fruit | Rice | Soybean | |||
|---|---|---|---|---|---|---|---|
| Gene | phylum | genus | Relative abundance (%) | ||||
| amoA–AOA | Thaumarchaeota | Candidatus_Nitrosocosmicus | 0.05 | 0.28 | 0.44 | 3.41 | 0.01 |
| Thaumarchaeota | Candidatus_Nitrosopelagicus | 0.29 | 0.03 | 0.03 | 0.28 | 0.04 | |
| Thaumarchaeota | Candidatus_Nitrosotenuis | 0.14 | 0.14 | 0.36 | 0.33 | 0.11 | |
| Thaumarchaeota | Nitrosopumilus | 3.58 | 4.86 | 16.05 | 9.40 | 3.73 | |
| Thaumarchaeota | Nitrososphaera | 25.30 | 27.25 | 29.86 | 28.32 | 29.50 | |
| amoA–AOB | Basidiomycota | Coprinopsis | 0.01 | 0.03 | 0.02 | 0.00 | 0.13 |
| Proteobacteria | Anaeromyxobacter | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Proteobacteria | Roseateles | 0.00 | 0.00 | 0.00 | 1.86 | 0.00 | |
| Proteobacteria | Nitrosococcus | 0.02 | 0.10 | 0.00 | 0.01 | 0.11 | |
| Proteobacteria | Nitrosomonas | 4.81 | 11.57 | 12.16 | 39.62 | 1.77 | |
| Proteobacteria | Nitrospira | 75.65 | 70.71 | 69.53 | 43.19 | 81.65 | |
| Proteobacteria | Nitrosovibrio | 7.47 | 5.62 | 8.12 | 2.42 | 2.14 | |
| nifH | Proteobacteria | Burkholderia | 0.77 | 1.48 | 0.83 | 0.08 | 1.15 |
| Proteobacteria | Dechloromonas | 0.56 | 1.03 | 1.96 | 0.84 | 0.53 | |
| Proteobacteria | Agrobacterium | 1.47 | 2.46 | 6.90 | 0.08 | 5.00 | |
| Cyanobacteria | Anabaena | 0.03 | 0.59 | 0.09 | 0.04 | 1.40 | |
| Proteobacteria | Anaeromyxobacter | 0.00 | 0.00 | 0.00 | 0.53 | 0.01 | |
| Proteobacteria | Azospira | 0.47 | 0.75 | 4.51 | 0.79 | 0.21 | |
| Proteobacteria | Azospirillum | 12.50 | 14.25 | 8.93 | 1.72 | 20.11 | |
| Proteobacteria | Azotobacter | 0.52 | 1.26 | 5.44 | 1.29 | 1.12 | |
| Proteobacteria | Bradyrhizobium | 3.43 | 2.30 | 2.22 | 6.21 | 2.93 | |
| Proteobacteria | Derxia | 0.00 | 0.00 | 0.00 | 1.05 | 0.00 | |
| Proteobacteria | Desulfobulbus | 0.19 | 0.41 | 1.14 | 4.73 | 0.00 | |
| Proteobacteria | Desulfovibrio | 1.13 | 1.67 | 1.59 | 8.98 | 0.60 | |
| Proteobacteria | Desulfuromonas | 3.17 | 5.67 | 1.99 | 7.68 | 1.61 | |
| Proteobacteria | Geobacter | 4.31 | 6.92 | 3.26 | 4.33 | 5.40 | |
| Proteobacteria | Paraburkholderia | 9.06 | 4.25 | 9.18 | 1.19 | 5.25 | |
| Proteobacteria | Pseudacidovorax | 4.16 | 0.44 | 0.43 | 2.44 | 0.46 | |
| Proteobacteria | Pseudomonas | 3.55 | 1.01 | 1.93 | 4.17 | 1.39 | |
| Proteobacteria | Rhizobium | 0.46 | 0.44 | 1.08 | 0.01 | 0.79 | |
| Proteobacteria | Rubrivivax | 0.49 | 0.58 | 0.89 | 0.44 | 0.82 | |
| Proteobacteria | Sideroxydans | 0.94 | 0.55 | 0.05 | 1.41 | 0.04 | |
| Proteobacteria | Stenotrophomonas | 0.01 | 0.80 | 0.04 | 0.06 | 0.26 | |
| Proteobacteria | Skermanella | 4.93 | 5.53 | 4.37 | 0.20 | 7.08 | |
| Cyanobacteria | Leptolyngbya | 0.00 | 0.00 | 0.00 | 1.50 | 0.00 | |
| Cyanobacteria | Nostoc | 0.63 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Firmicutes | Pelosinus | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | |
| nirK | Proteobacteria | Alcaligenes | 4.16 | 0.66 | 0.25 | 3.64 | 0.49 |
| Proteobacteria | Bosea | 2.85 | 1.73 | 0.14 | 0.75 | 0.61 | |
| Proteobacteria | Bradyrhizobium | 15.48 | 13.10 | 10.66 | 11.04 | 15.91 | |
| Proteobacteria | Chelativorans | 4.56 | 7.00 | 0.90 | 1.11 | 5.66 | |
| Proteobacteria | Chelatococcus | 1.08 | 3.66 | 2.87 | 0.77 | 1.97 | |
| Proteobacteria | Enterobacter | 0.51 | 0.18 | 0.50 | 1.62 | 0.26 | |
| Proteobacteria | Mesorhizobium | 13.97 | 20.40 | 19.78 | 19.34 | 12.51 | |
| Proteobacteria | Nitrosomonas | 0.29 | 0.00 | 0.00 | 4.59 | 0.11 | |
| Proteobacteria | Ochrobactrum | 2.71 | 0.00 | 5.90 | 0.95 | 0.05 | |
| Proteobacteria | Rhizobium | 11.56 | 6.47 | 8.76 | 29.13 | 16.84 | |
| Proteobacteria | Rhodobacter | 0.03 | 0.00 | 0.02 | 1.91 | 0.00 | |
| Proteobacteria | Rhodopseudomonas | 13.91 | 6.65 | 11.77 | 2.99 | 14.90 | |
| Proteobacteria | Sinorhizobium | 9.64 | 21.05 | 22.90 | 6.16 | 13.41 | |
| nirS | Proteobacteria | Alicycliphilus | 0.00 | 0.00 | 0.00 | 0.53 | 0.00 |
| Proteobacteria | Aromatoleum | 0.03 | 0.00 | 0.00 | 0.89 | 0.00 | |
| Actinobacteria | Arthrobacter | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | |
| Proteobacteria | Azoarcus | 6.07 | 3.80 | 9.31 | 13.05 | 3.32 | |
| Proteobacteria | Azospira | 0.53 | 0.01 | 0.54 | 0.11 | 0.08 | |
| Proteobacteria | Azospirillum | 3.81 | 4.31 | 4.74 | 0.87 | 7.00 | |
| Firmicutes | Bacillus | 0.97 | 0.22 | 0.72 | 1.58 | 0.04 | |
| Proteobacteria | Cupriavidus | 6.89 | 6.20 | 3.65 | 2.25 | 5.05 | |
| Proteobacteria | Halomonas | 0.08 | 0.58 | 0.89 | 0.03 | 0.04 | |
| Proteobacteria | Magnetospirillum | 2.41 | 1.83 | 3.81 | 0.42 | 5.63 | |
| Proteobacteria | Paracoccus | 0.60 | 1.55 | 0.01 | 0.25 | 0.01 | |
| Proteobacteria | Polymorphum | 0.85 | 0.32 | 1.04 | 0.50 | 0.61 | |
| Proteobacteria | Pseudogulbenkiania | 0.05 | 0.00 | 0.00 | 1.95 | 0.00 | |
| Proteobacteria | Pseudomonas | 3.88 | 2.81 | 3.44 | 9.72 | 0.77 | |
| Proteobacteria | Rhodanobacter | 5.01 | 3.26 | 4.98 | 3.99 | 16.94 | |
| Proteobacteria | Rubrivivax | 5.61 | 4.12 | 4.03 | 1.57 | 5.49 | |
| Proteobacteria | Sulfuritalea | 6.82 | 3.40 | 2.28 | 5.31 | 1.89 | |
| Proteobacteria | Thauera | 0.05 | 0.00 | 0.00 | 0.79 | 0.00 | |
| Proteobacteria | Thiobacillus | 0.75 | 0.02 | 0.00 | 0.66 | 0.00 | |
| Soil Properties | Degrees of Explanation (%) | |||
|---|---|---|---|---|
| AOA–amoA and AOB–amoA | nirS and nirK | nifH | 16S rRNA | |
| SWC | 45.4 ** | 21.6 ** | 54.2 ** | 38.5 ** |
| EC | 15.3 ** | 13.7 ** | NS | 8.9 ** |
| TN | 9.7 * | NS | NS | NS |
| AP | NS | 9.1 * | NS | 21.5 ** |
| CEC | NS | NS | 5.8 * | NS |
| NO3−–N | NS | NS | NS | NS |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, H.; Miao, Y.; Zhang, L.; Chen, Y.; Gan, Y.; Liu, N.; Dong, L.; Dai, J.; Chen, W. The Structure and Diversity of Nitrogen Functional Groups from Different Cropping Systems in Yellow River Delta. Microorganisms 2020, 8, 424. https://doi.org/10.3390/microorganisms8030424
He H, Miao Y, Zhang L, Chen Y, Gan Y, Liu N, Dong L, Dai J, Chen W. The Structure and Diversity of Nitrogen Functional Groups from Different Cropping Systems in Yellow River Delta. Microorganisms. 2020; 8(3):424. https://doi.org/10.3390/microorganisms8030424
Chicago/Turabian StyleHe, Huan, Yongjun Miao, Lvqing Zhang, Yu Chen, Yandong Gan, Na Liu, Liangfeng Dong, Jiulan Dai, and Weifeng Chen. 2020. "The Structure and Diversity of Nitrogen Functional Groups from Different Cropping Systems in Yellow River Delta" Microorganisms 8, no. 3: 424. https://doi.org/10.3390/microorganisms8030424
APA StyleHe, H., Miao, Y., Zhang, L., Chen, Y., Gan, Y., Liu, N., Dong, L., Dai, J., & Chen, W. (2020). The Structure and Diversity of Nitrogen Functional Groups from Different Cropping Systems in Yellow River Delta. Microorganisms, 8(3), 424. https://doi.org/10.3390/microorganisms8030424






