Differences in Gut Microbial Diversity are Driven by Drug Use and Drug Cessation by Either Compulsory Detention or Methadone Maintenance Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects and Screening Conditions
2.2. Dietary Nutrition Intake Survey
2.3. Fecal Sample Collection and DNA Extraction
2.4. Polymerase Chain Reaction (PCR) and High-Throughput Sequencing
2.5. Gut Microbiome Analysis and Statistical Analysis
2.6. Ethic Approval and Consent to Participates
3. Results
3.1. General Demographic Characteristics among All Groups and Drug Abuse Behaviour in CD, MP, and DU Participants
3.2. Dietary Intake Data for Each Group
3.2.1. Higher Nutrient and Energy Intake Reported in MP Group
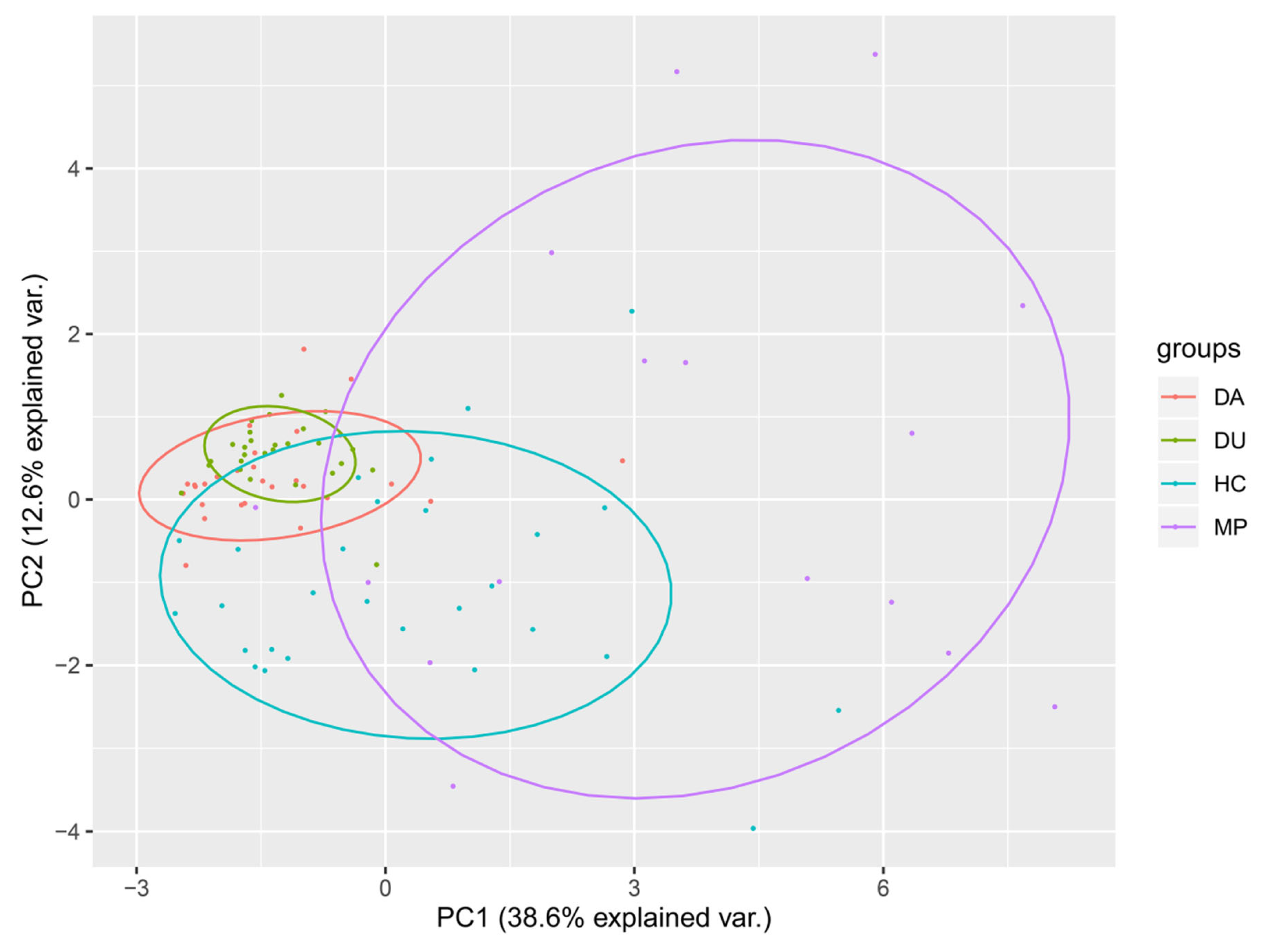
3.2.2. PCA of Dietary Structure Indicates Broad Separation of MP Group Compared to Other Groups
3.3. Fecal Microbiome Data of Four Groups
3.3.1. Alpha Diversity was not Significantly Different among Groups
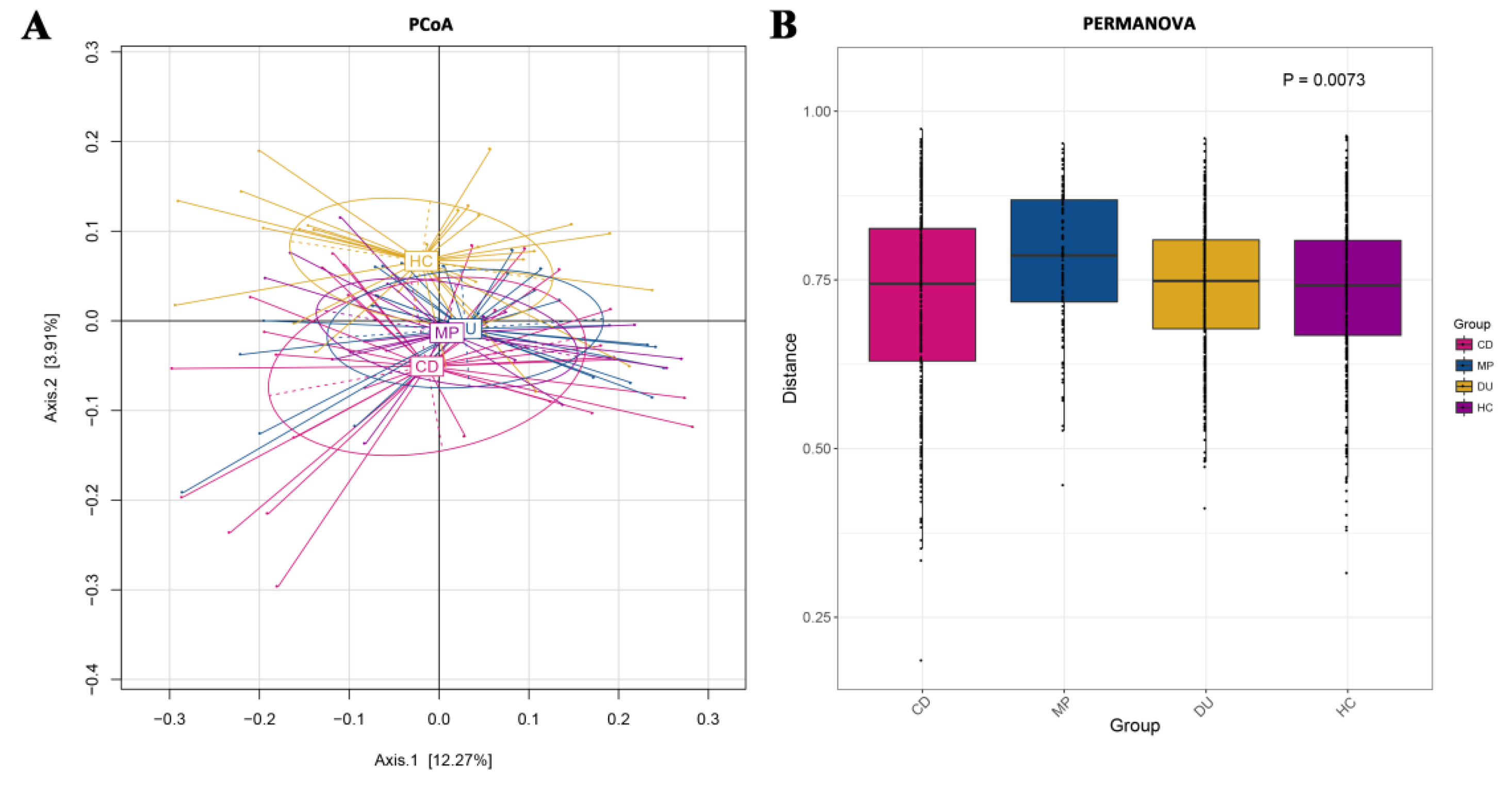
3.3.2. Distribution of Taxa Is Significantly Different across Groups in Beta Diversity
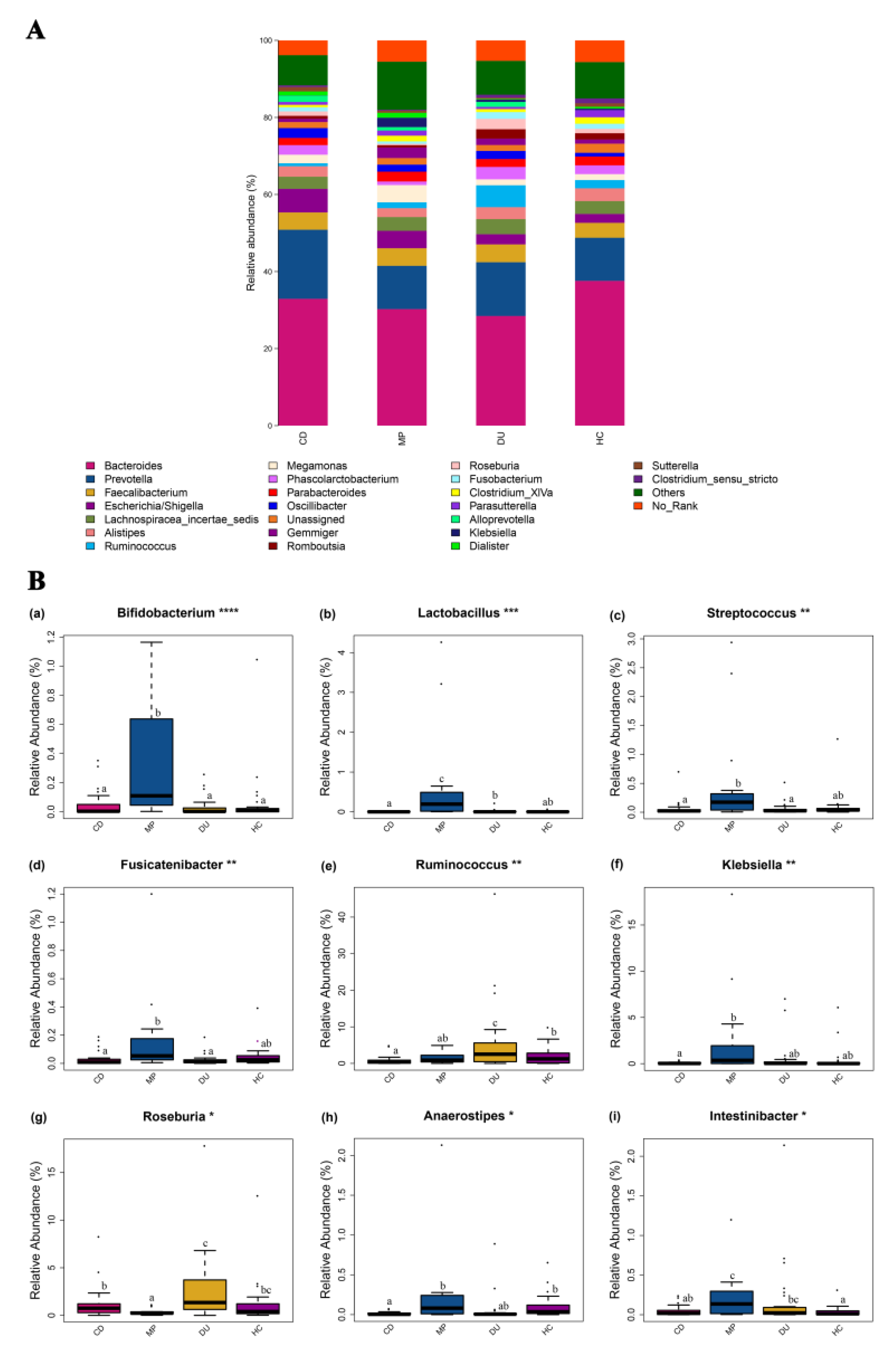
3.3.3. Significant Enrichment for Specific Genera in MP Compared to CD, DU, and HC Groups
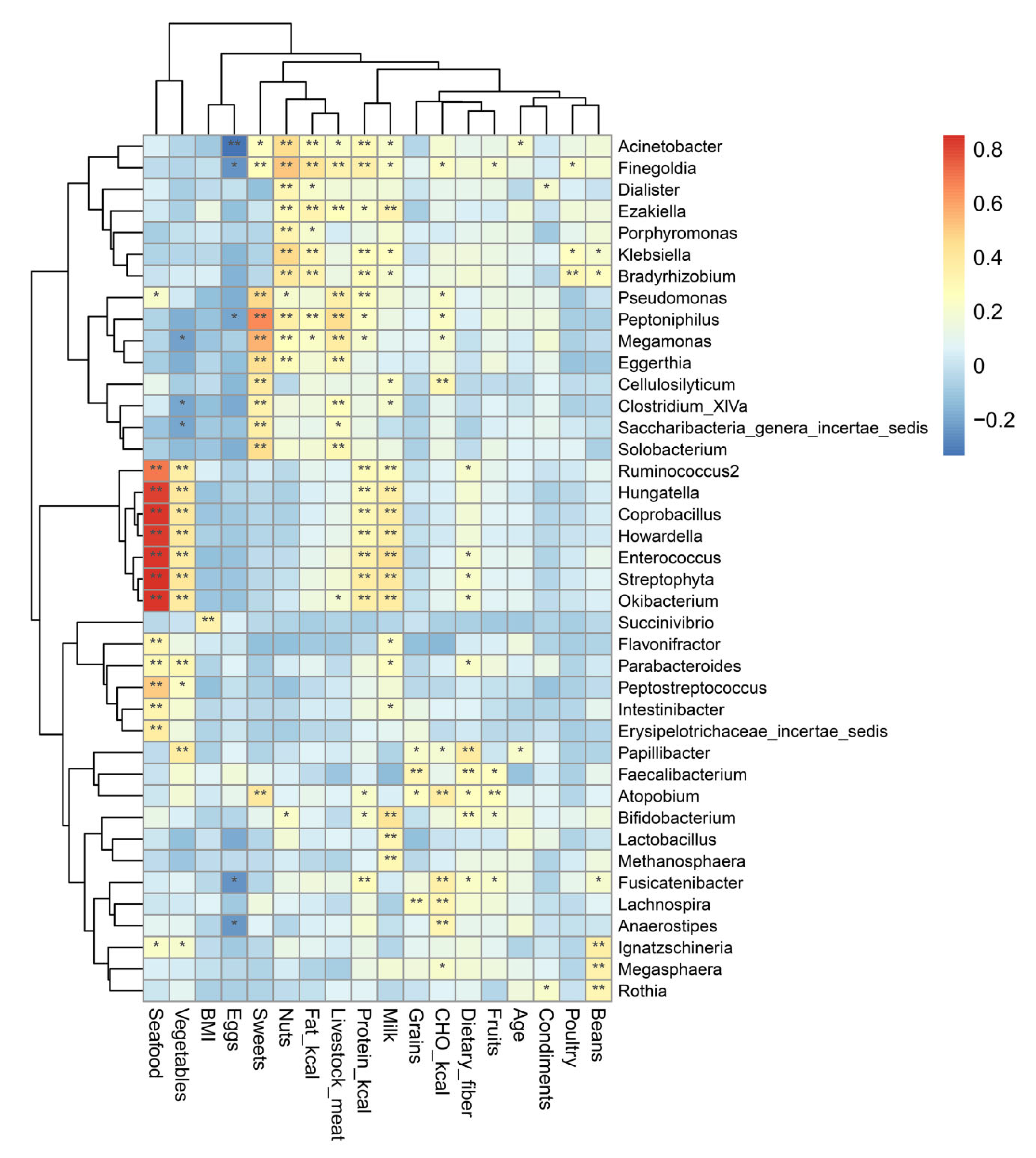
3.3.4. Specific Genera Are Significantly Correlated with Dietary Intake
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MMT | methadone maintenance treatment. |
| PCR | polymerase chain reaction. |
| OTUs | operational taxonomic units. |
| RDP | ribosomal database project. |
| ANOVA | analysis of variance. |
| ACE | abundance-based coverage estimator. |
| PCoA | principal coordinate analysis. |
| PCA | principal component analysis. |
| PERMANOVA | permutational multivariate analysis of variance. |
| LDA | linear discriminant analysis. |
| LEfSe | linear discriminant analysis effect size. |
| BMI | body mass index. |
| CD | compulsory detention (participants). |
| MP | MMT patients. |
| DU | drug users. |
| HC | healthy controls. |
References
- Pence, B.W.; Miller, W.C.; Whetten, K.; Eron, J.J.; Gaynes, B.N. Prevalence of DSM-IV-defined mood, anxiety, and substance use disorders in an HIV clinic in the southeastern United States. Aids J. Acq. Imm. Def. 2006, 42, 298–306. [Google Scholar] [CrossRef]
- Talal, A.H.; Thomas, D.L.; Reynolds, J.L.; Khalsa, J.H. Toward Optimal Control of Hepatitis C Virus Infection in Persons With Substance Use Disorders. Ann. Intern. Med. 2017, 166, 897. [Google Scholar] [CrossRef]
- Azbel, L.; Wickersham, J.A.; Grishaev, Y.; Dvoryak, S.; Altice, F.L. Burden of Infectious Diseases, Substance Use Disorders, and Mental Illness among Ukrainian Prisoners Transitioning to the Community. PLoS ONE 2013, 8, e59643. [Google Scholar] [CrossRef] [PubMed]
- El-Bassel, N.; Gilbert, L.; Wu, E.; Go, H.; Hill, J. Relationship between Drug Abuse and Intimate Partner Violence: A Longitudinal Study Among Women Receiving Methadone. Am. J. Public Health 2005, 95, 465–470. [Google Scholar] [CrossRef]
- Volkow, N.D. The reality of comorbidity: Depression and drug abuse. Biol. Psychiatry 2004, 56, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Swendsen, J.; Burstein, M.; Case, B.; Conway, K.P.; Dierker, L.; He, J.P.; Merikangas, K.R. Use and Abuse of Alcohol and Illicit Drugs in US Adolescents Results of the National Comorbidity Survey-Adolescent Supplement. Arch. Gen. Psychiatry 2012, 69, 390–398. [Google Scholar] [PubMed]
- Elliott, R.; Csete, J.; Palepu, A.; Kerr, T. Reason and rights in global drug control policy. Can Med. Assoc. J. 2005, 172, 655–656. [Google Scholar] [CrossRef]
- Korthuis, P.T.; McCarty, D.; Weimer, M.; Bougatsos, C.; Blazina, I.; Zakher, B.; Grusing, S.; Devine, B.; Chou, R. Primary Care-Based Models for the Treatment of Opioid Use Disorder A Scoping Review. Ann. Intern. Med. 2017, 166, 268. [Google Scholar] [CrossRef]
- Yang, M.; Mamy, J.; Gao, P.C.; Xiao, S.Y. From Abstinence to Relapse: A Preliminary Qualitative Study of Drug Users in a Compulsory Drug Rehabilitation Center in Changsha, China. PLoS ONE 2015, 10, e0130711. [Google Scholar] [CrossRef]
- Alcaraz, S.; Viladrich, C.; Trujols, J.; Sinol, N.; de los Cobos, J.P. Heroin-dependent patient satisfaction with methadone as a medication influences satisfaction with basic interventions delivered by staff to implement methadone maintenance treatment. Patient Prefer Adherence 2018, 12, 1203–1211. [Google Scholar] [CrossRef]
- Gowing, L.R.; Farrell, M.; Bornemann, R.; Sullivan, L.E.; Ali, R.L. Brief report: Methadone treatment of injecting opioid users for prevention of HIV infection. J. Gen. Intern. Med. 2006, 21, 193–195. [Google Scholar] [CrossRef] [PubMed]
- Dole, V.P.; Robinson, J.W.; Orraca, J.; Towns, E.; Searcy, P.; Caine, E. Methadone treatment of randomly selected criminal addicts. N. Engl. J. Med. 1969, 280, 1372–1375. [Google Scholar] [CrossRef] [PubMed]
- Stotts, A.L.; Dodrill, C.L.; Kosten, T.R. Opioid dependence treatment: Options in pharmacotherapy. Expert Opin. Pharm. 2009, 10, 1727–1740. [Google Scholar] [CrossRef] [PubMed]
- Gronbladh, L.; Ohlund, L.S.; Gunne, L.M. Mortality in heroin addiction: Impact of methadone treatment. Acta Psychiatr. Scand. 1990, 82, 223–227. [Google Scholar] [CrossRef]
- Rosenblum, A.; Joseph, H.; Fong, C.; Kipnis, S.; Cleland, C.; Portenoy, R.K. Prevalence and characteristics of chronic pain among chemically dependent patients in methadone maintenance and residential treatment facilities. JAMA J. Am. Med. Assoc. 2003, 289, 2370–2378. [Google Scholar] [CrossRef]
- Sees, K.L.; Delucchi, K.L.; Masson, C.; Rosen, A.; Clark, H.W.; Robillard, H.; Banys, P.; Hall, S.M. Methadone maintenance vs. 180-day psychosocially enriched detoxification for treatment of opioid dependence: A randomized controlled trial. JAMA J. Am. Med. Assoc. 2000, 283, 1303–1310. [Google Scholar] [CrossRef]
- Wang, D.; Teichtahl, H.; Drummer, O.; Goodman, C.; Cherry, G.; Cunnington, D.; Kronborg, I. Central sleep apnea in stable methadone maintenance treatment patients. Chest 2005, 128, 1348–1356. [Google Scholar] [CrossRef]
- Fu, J.J.; Bazazi, A.R.; Altice, F.L.; Mohamed, M.N.; Kamarulzaman, A. Absence of Antiretroviral Therapy and Other Risk Factors for Morbidity and Mortality in Malaysian Compulsory Drug Detention and Rehabilitation Centers. PLoS ONE 2012, 7, e44249. [Google Scholar] [CrossRef][Green Version]
- Poudel, A.; Sharma, C.; Gautam, S. Psychosocial problems among individuals with substance use disorders in drug rehabilitation centers, Nepal. Subst. Abus. Treat. Prev. Pol. 2016, 11, 10. [Google Scholar] [CrossRef][Green Version]
- Harvey-Vera, A.Y.; Gonzalez-Zuniga, P.; Vargas-Ojeda, A.C.; Medina-Mora, M.E.; Magis-Rodriguez, C.L.; Wagner, K.; Strathdee, S.A.; Werb, D. Risk of violence in drug rehabilitation centers: Perceptions of people who inject drugs in Tijuana, Mexico. Subst. Abus. Treat. Prev. Pol. 2016, 11, 9. [Google Scholar] [CrossRef]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Adv. Gastroenter. 2013, 6, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the Microbiota and the Immune System. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.C.; Wisniewski, P.J.; Noji, M.; McGuinness, L.R.; Haggblom, M.M.; Lightfoot, S.A.; Joseph, L.B.; Kerkhof, L.J. The Effect of Diet and Exercise on Intestinal Integrity and Microbial Diversity in Mice. PLoS ONE 2016, 11, e0150502. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Li, M.; Campbell, T.B.; Flores, S.C.; Linderman, D.; Gebert, M.J.; Knight, R.; Fontenot, A.P.; Palmer, B.E. Alterations in the Gut Microbiota Associated with HIV-1 Infection. Cell Host Microbe 2013, 14, 329–339. [Google Scholar] [CrossRef]
- Scorza, C.; Piccini, C.; Martinez Busi, M.; Abin Carriquiry, J.A.; Zunino, P. Alterations in the Gut Microbiota of Rats Chronically Exposed to Volatilized Cocaine and Its Active Adulterants Caffeine and Phenacetin. Neurotox. Res. 2019, 35, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Gibson, G.R. Probiotics, prebiotics, and synbiotics: Approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 1999, 69, S1052–S1057. [Google Scholar] [CrossRef]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 10. [Google Scholar] [CrossRef]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxid. Med. Cell. Longev. 2017, 3831972. [Google Scholar] [CrossRef]
- Cook, M.D.; Allen, J.M.; Pence, B.D.; Wallig, M.A.; Gaskins, H.R.; White, B.A.; Woods, J.A. Exercise and gut immune function: Evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol. Cell Biol. 2016, 94, 158–163. [Google Scholar] [CrossRef]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best Pr. Res. Clin. Gastroenterol. 2017, 31, 579–588. [Google Scholar] [CrossRef]
- Quach, L.A.; Wanke, C.A.; Schmid, C.H.; Gorbach, S.L.; Mkaya Mwamburi, D.; Mayer, K.H.; Spiegelman, D.; Tang, A.M. Drug use and other risk factors related to lower body mass index among HIV-infected individuals. Drug Alcohol. Depend. 2007, 95, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Leppert, W. Emerging therapies for patients with symptoms of opioid-induced bowel dysfunction. Drug Des. Devel. 2015, 9, 2215–2231. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xie, Z.R.; Wang, H.W.; Shen, Z.W.; Guo, Y.B.; Gao, Y.H.; Chen, X.; Wu, Q.; Li, X.J.; Wang, K.H. Bacterial Diversity of Intestinal Microbiota in Patients with Substance Use Disorders Revealed by 16S rRNA Gene Deep Sequencing. Sci. Rep. 2017, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Ning, T.T.; Gong, X.K.; Xie, L.L.; Ma, B.M. Gut Microbiota Analysis in Rats with Methamphetamine-Induced Conditioned Place Preference. Front. Microbiol. 2017, 8, 9. [Google Scholar] [CrossRef]
- Das, B.; Ghosh, T.S.; Kedia, S.; Rampal, R.; Saxena, S.; Bag, S.; Mitra, R.; Dayal, M.; Mehta, O.; Surendranath, A.; et al. Analysis of the Gut Microbiome of Rural and Urban Healthy Indians Living in Sea Level and High Altitude Areas. Sci. Rep. 2018, 8, 15. [Google Scholar] [CrossRef]
- Hu, F.B.; Rimm, E.; Smith-Warner, S.A.; Feskanich, D.; Stampfer, M.J.; Ascherio, A.; Sampson, L.; Willett, W.C. Reproducibility and validity of dietary patterns assessed with a food-frequency questionnaire. Am. J. Clin. Nutr. 1999, 69, 243–249. [Google Scholar] [CrossRef]
- Wang, S.S.; Lay, S.; Yu, H.N.; Shen, S.R. Dietary Guidelines for Chinese Residents (2016): Comments and comparisons. J. Zhejiang. Univ. Sci. B 2016, 17, 649–656. [Google Scholar] [CrossRef]
- Wang, F.Y.; Meng, J.J.; Zhang, L.; Johnson, T.; Chen, C.; Roy, S. Morphine induces changes in the gut microbiome and metabolome in a morphine dependence model. Sci. Rep. 2018, 8, 15. [Google Scholar] [CrossRef]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Dill-McFarland, K.A.; Tang, Z.Z.; Kemis, J.H.; Kerby, R.L.; Chen, G.H.; Palloni, A.; Sorenson, T.; Rey, F.E.; Herd, P. Close social relationships correlate with human gut microbiota composition. Sci. Rep. 2019, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Hakim, J.A.; Schram, J.B.; Galloway, A.W.E.; Morrow, C.D.; Crowley, M.R.; Watts, S.A.; Bej, A.K. The Purple Sea Urchin Strongylocentrotus purpuratus Demonstrates a Compartmentalization of Gut Bacterial Microbiota, Predictive Functional Attributes, and Taxonomic Co-Occurrence. Microorganisms 2019, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Mago, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 17. [Google Scholar] [CrossRef]
- Mazhnaya, A.; Tobin, K.E.; Owczarzak, J. Association between injection in public places and HIV/HCV risk behavior among people who use drugs in Ukraine. Drug Alcohol. Depend. 2018, 189, 125–130. [Google Scholar] [CrossRef]
- Mumtaz, G.R.; Awad, S.F.; Feizzadeh, A.; Weiss, H.A.; Abu-Raddad, L.J. HIV incidence among people who inject drugs in the Middle East and North Africa: Mathematical modelling analysis. J. Int. Aids Soc. 2018, 21, 13. [Google Scholar] [CrossRef]
- Tsai, M.C.; Huang, T.L. Orexin A in men with heroin use disorder undergoing methadone maintenance treatment. Psychiatry Res. 2018, 264, 412–415. [Google Scholar] [CrossRef]
- Sakurai, T. Roles of orexins in the regulation of body weight homeostasis. Obes. Res. Clin. Pr. 2013, 8, e414–e420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.R.; Lian, Z.; Yan, S.Y.; Bao, Y.P.; Liu, Z.M. Different Levels in Orexin Concentrations and Risk Factors Associated with Higher Orexin Levels: Comparison between Detoxified Opiate and Methamphetamine Addicts in 5 Chinese Cities. BioMed Res. Int. 2013, 2013, 282641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Volpe, G.E.; Ward, H.; Mwamburi, M.; Dinh, D.; Bhalchandra, S.; Wanke, C.; Kane, A.V. Associations of Cocaine Use and HIV Infection With the Intestinal Microbiota, Microbial Translocation, and Inflammation. J. Stud. Alcohol. Drugs 2014, 75, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Ding, B.; Feng, C.; Yin, S.; Zhang, T.; Qi, X.; Lv, H.; Guo, X.; Dong, K.; Zhu, Y.; et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J. Affect. Disord. 2017, 207, 300–304. [Google Scholar] [CrossRef]
- Fan, C.Y.; Tan, H.K.L.; Chien, I.C.; Chou, S.Y. Prevalence of psychiatric disorders among heroin users who received methadone maintenance therapy in Taiwan. Am. J. Addict. 2014, 23, 249–256. [Google Scholar] [CrossRef]
- Callaly, T.; Trauer, T.; Munro, L.; Whelan, G. Prevalence of psychiatric disorder in a methadone maintenance population. Aust. N. Z. J. Psychiatry 2001, 35, 601–605. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, S.; Ma, C.; Huo, D.; Peng, Q.; Shao, Y.; Zhang, J. Evaluation of the nutrition and function of cow and goat milk based on intestinal microbiota by metagenomic analysis. Food Funct. 2018, 9, 2320–2327. [Google Scholar] [CrossRef]
- Slavin, J. Fiber and Prebiotics: Mechanisms and Health Benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef]
- Hardy, H.; Harris, J.; Lyon, E.; Beal, J.; Foey, A.D. Probiotics, Prebiotics and Immunomodulation of Gut Mucosal Defences: Homeostasis and Immunopathology. Nutrients 2013, 5, 1869–1912. [Google Scholar] [CrossRef]
- Ze, X.L.; Duncan, S.H.; Louis, P.; Flint, H.J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012, 6, 1535–1543. [Google Scholar] [CrossRef]
- Christopherson, M.R.; Dawson, J.A.; Stevenson, D.M.; Cunningham, A.C.; Bramhacharya, S.; Weimer, P.J.; Kendziorski, C.; Suen, G. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genom. 2014, 15, 1066. [Google Scholar] [CrossRef] [PubMed]
- Beilharz, J.E.; Kaakoush, N.O.; Maniam, J.; Morris, M.J. The effect of short-term exposure to energy-matched diets enriched in fat or sugar on memory, gut microbiota and markers of brain inflammation and plasticity. Brain Behav. Immun. 2016, 57, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Betrapally, N.S.; Gillevet, P.M.; Sterling, R.K.; Akbarali, H.; White, M.B.; Ganapathy, D.; Fagan, A.; Sikaroodi, M.; Bajaj, J.S. Chronic opioid use is associated with altered gut microbiota and predicts readmissions in patients with cirrhosis. Aliment. Pharm. 2017, 45, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.J.; Beiko, R.G. A Phylogenomic View of Ecological Specialization in the Lachnospiraceae, a Family of Digestive Tract-Associated Bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef]
- Takada, T.; Kurakawa, T.; Tsuji, H.; Nomoto, K. Fusicatenibacter saccharivorans gen. nov., sp nova, isolated from human feces. Int. J. Syst. Evol. Microbiol. 2013, 63, 3691–3696. [Google Scholar] [CrossRef]
- Kameue, C.; Tsukahara, T.; Yamada, K.; Koyama, H.; Iwasaki, Y.; Nakayama, K.; Ushida, K. Dietary sodium gluconate protects rats from large bowel cancer by stimulating butyrate production. J. Nutr. 2004, 134, 940–944. [Google Scholar] [CrossRef]
- Mark, A.B.; Kiilerich, P.; Astrup, A.; Kristiansen, K.; Larsen, L.H.; Bendtsen, L.Q.; Holm, J.B.; Blædel, T.; Lorenzen, J.K. High intake of dairy during energy restriction does not affect energy balance or the intestinal microflora compared with low dairy intake in overweight individuals in a randomized controlled trial. Appl. Physiol. Nutr. Metab. 2018, 43, 1–10. [Google Scholar]
- Tanguay, P.; Kamarulzaman, A.; Aramrattana, A.; Wodak, A.; Thomson, N.; Ali, R.; Vumbaca, G.; Lai, G.; Chabungbam, A. Facilitating a transition from compulsory detention of people who use drugs towards voluntary community-based drug dependence treatment and support services in Asia. Harm. Reduct. J. 2015, 12, 5. [Google Scholar] [CrossRef][Green Version]
- Elkana, O.; Adelson, M.; Doniger, G.M.; Sason, A.; Peles, E. Cognitive function is largely intact in methadone maintenance treatment patients. World J. Biol. Psychiatry 2019, 20, 219–229. [Google Scholar] [CrossRef]
- Schuckit, M.A. Treatment of Opioid-Use Disorders. N. Engl. J. Med. 2016, 375, 357–368. [Google Scholar] [CrossRef]
| Food | CD Intake (g) | MP Intake (g) | DU Intake (g) | HC Intake (g) | p-Value |
|---|---|---|---|---|---|
| Grains | 528.50 (521.00, 721.00)ab | 707.50 (498.25, 937.50)b | 629.00 (571.00, 741.00)b | 479.50 (277.50, 686.00)a | 0.013 * |
| Milk | 0.00 (0.00, 0.00)a | 77.00 (7.50, 461.25)b | 0.00 (0.00, 8.00)a | 13.00 (0.80, 167.25)b | <0.001 * |
| Eggs | 35.00 (35.00, 35.00)b | 15.00 (7.00, 33.50)a | 35.00 (35.00, 35.00)b | 14.50 (7.30, 30.00)a | <0.001 * |
| Livestock Meat | 50.00 (50.00, 62.00)a | 127.00 (57.00, 166.50)b | 50.00 (50.00, 52.00)a | 90.00 (55.50, 126.00)b | <0.001 * |
| Poultry | 8.00 (0.00, 21.00) | 11.00 (0.00, 37.30) | 5.00 (0.00, 20.00) | 13.00 (6.50, 60.00) | 0.056 |
| Seafood | 6.00 (2.00, 20.75)b | 10.05 (1.25, 28.00)b | 2.00 (0.00, 3.00)a | 7.50 (3.00, 19.50)b | 0.002 * |
| Beans | 30.00 (30.00, 30.00)a | 102.00 (24.75, 232.25)b | 30.00 (30.00, 30.00)a | 24.00 (11.00, 87.25)a | 0.012 * |
| Vegetables | 283.00 (283.00, 283.00)b | 364.00 (160.50, 565.00)b | 283.00 (283.00, 283.00)b | 186.50 (91.75, 276.75)a | <0.001 * |
| Fruits | 70.00 (40.00, 127.50)a | 215.00 (95.25, 371.75)b | 70.00 (5.00, 137.00)a | 134.00 (42.00, 183.75)ab | 0.003 * |
| Nuts | 0.00 (0.00, 1.00)a | 14.50 (3.50, 31.50)b | 0.00 (0.00, 3.00)a | 3.50 (0.00, 11.75)b | <0.001 * |
| Condiments | 0.00 (0.00, 22.75)a | 87.50 (0.00, 991.25)b | 0.00 (0.00, 250.00)ab | 25.00 (0.00, 179.00)b | 0.046 * |
| Sweets | 0.00 (0.00, 5.25)a | 4.00 (0.00, 179.25)b | 10.00 (0.00, 30.00)b | 8.50 (0.00, 43.50)b | 0.044 * |
| Nutrients | CD | MP | DU | HC | p-Value |
|---|---|---|---|---|---|
| Total energy (kcal) | 1074.00 (928.25, 1358.25)a | 3203.00 (1752.50, 3735.25)c | 1230.00 (1055.00, 1374.00)ab | 1732.50 (1215.25, 2217.75)b | <0.001 * |
| Protein (kcal) | 158.00 (130.50, 189.75)a | 414.50 (284.50, 494.50)c | 160.00 (149.00, 180.00)ab | 235.50 (161.50, 297.00)b | <0.001 * |
| Fat (kcal) | 273.50 (243.75, 365.00)a | 533.00 (373.75, 887.75)b | 297.00 (257.00, 396.00)a | 416.00 (262.50, 555.75)ab | <0.001 * |
| Carbohydrates (kcal) | 607.00 (544.00, 771.50)a | 1849.00 (1178.75, 2688.00)c | 753.00 (646.00, 850.00)ab | 941.50 (747.25, 1334.75)b | <0.001 * |
| Protein (% energy) | 14.00 (14.00, 15.00) | 13.00 (11.00, 15.75) | 13.00 (12.00, 14.00) | 13.00 (12.00, 15.75) | 0.088 |
| Fat (% energy) | 27.00 (25.00, 28.00)b | 20.50 (14.50, 25.00)a | 26.00 (23.00, 28.00)ab | 23.50 (19.25, 32.00)ab | 0.036 * |
| Carbohydrates (% energy) | 59.00 (57.25, 60.75) | 66.50 (55.75, 74.75) | 61.00 (59.00, 64.00) | 61.00 (53.25, 67.00) | 0.079 |
| Dietary fiber | 6.00 (5.00, 8.50)a | 17.00 (6.25, 25.50)b | 6.00 (5.25, 6.75)a | 7.00 (5.00, 11.75)a | <0.001 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Chen, S.; Liu, K.; Long, D.; Liu, D.; Jing, Z.; Huang, X. Differences in Gut Microbial Diversity are Driven by Drug Use and Drug Cessation by Either Compulsory Detention or Methadone Maintenance Treatment. Microorganisms 2020, 8, 411. https://doi.org/10.3390/microorganisms8030411
Li Q, Chen S, Liu K, Long D, Liu D, Jing Z, Huang X. Differences in Gut Microbial Diversity are Driven by Drug Use and Drug Cessation by Either Compulsory Detention or Methadone Maintenance Treatment. Microorganisms. 2020; 8(3):411. https://doi.org/10.3390/microorganisms8030411
Chicago/Turabian StyleLi, Qiaoyan, Siqi Chen, Ke Liu, Danfeng Long, Diru Liu, Zhengchao Jing, and Xiaodan Huang. 2020. "Differences in Gut Microbial Diversity are Driven by Drug Use and Drug Cessation by Either Compulsory Detention or Methadone Maintenance Treatment" Microorganisms 8, no. 3: 411. https://doi.org/10.3390/microorganisms8030411
APA StyleLi, Q., Chen, S., Liu, K., Long, D., Liu, D., Jing, Z., & Huang, X. (2020). Differences in Gut Microbial Diversity are Driven by Drug Use and Drug Cessation by Either Compulsory Detention or Methadone Maintenance Treatment. Microorganisms, 8(3), 411. https://doi.org/10.3390/microorganisms8030411






