Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production
Abstract
1. Introduction
2. Results
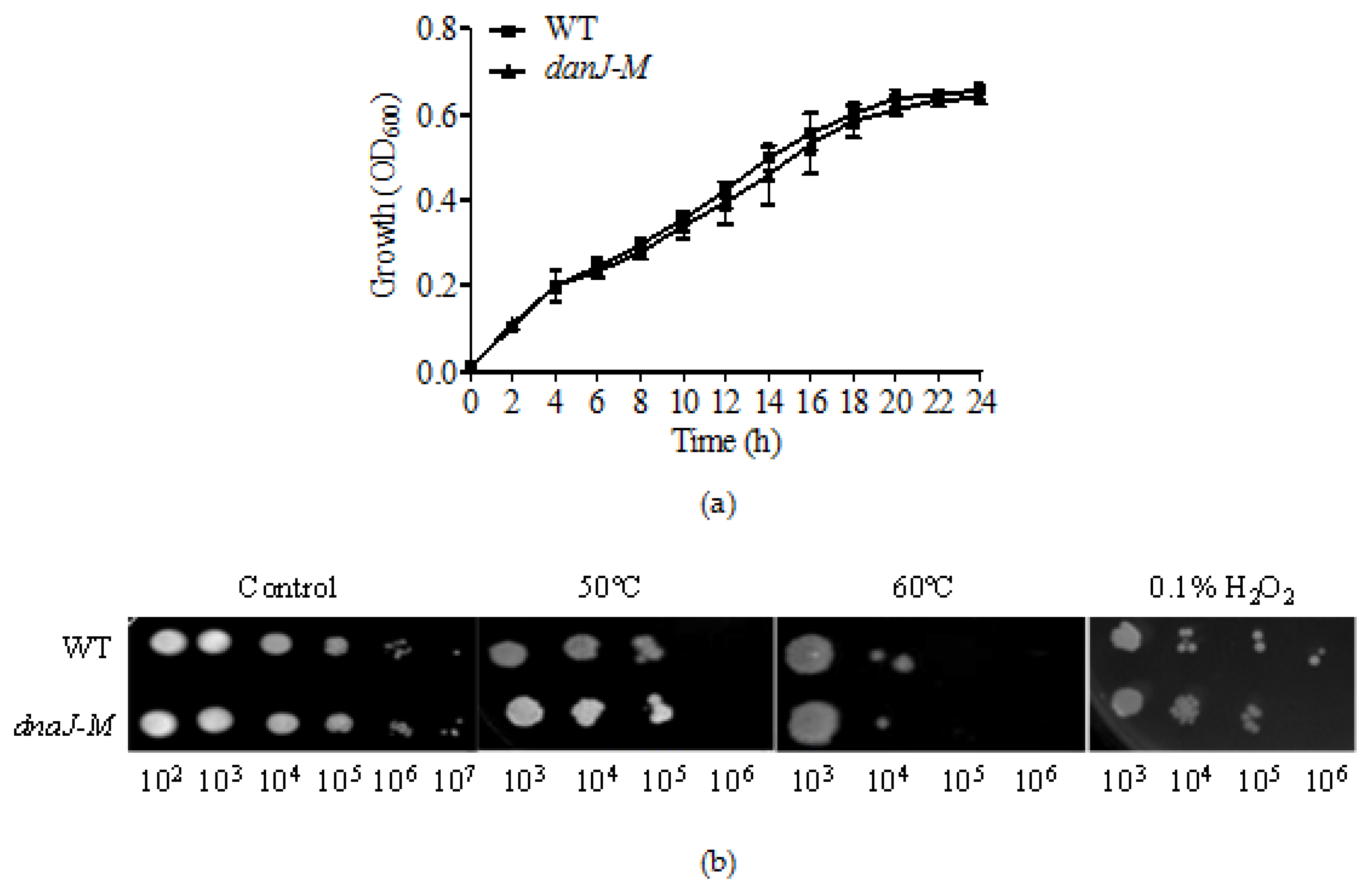
2.1. Disruption of dnaJ Does Not Affect P. aeruginosa Survival under Different Stress Conditions
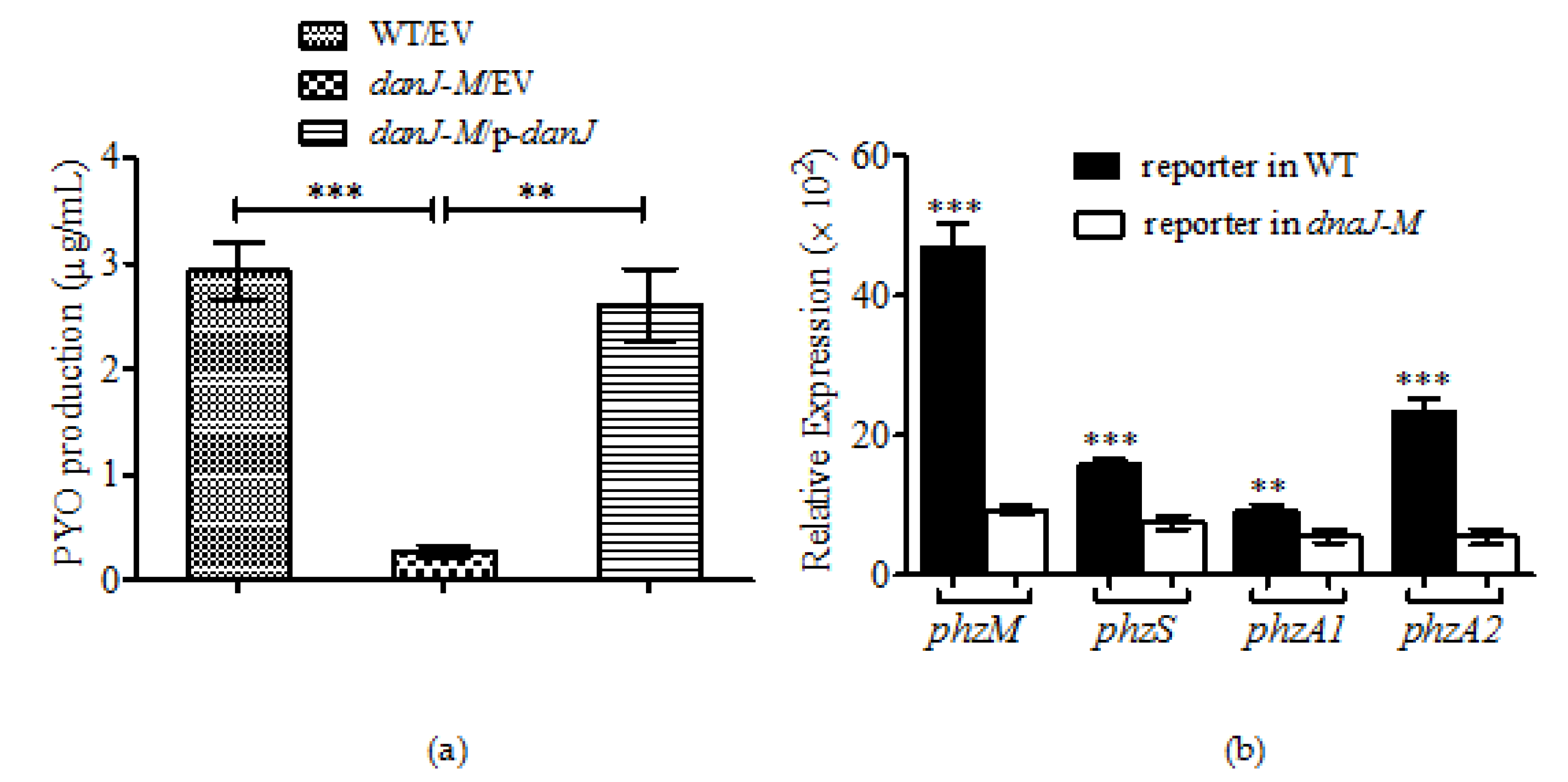
2.2. Production of Pyocyanin Was Drastically Reduced in the Absence of dnaJ
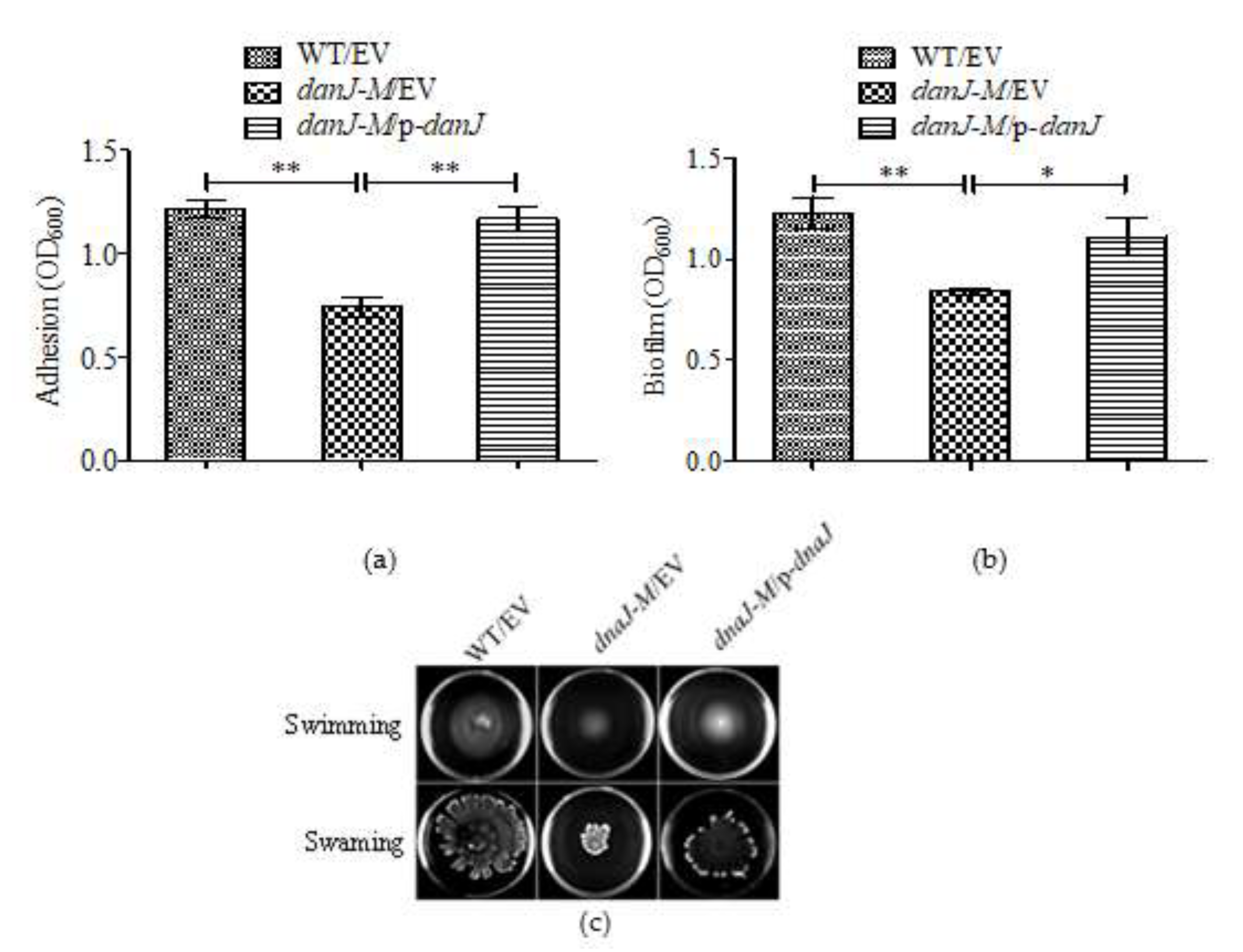
2.3. The dnaJ Mutation Impaired Biofilm Formation
2.4. The Full Pathogenicity of P. aeruginosa Requires Functional dnaJ
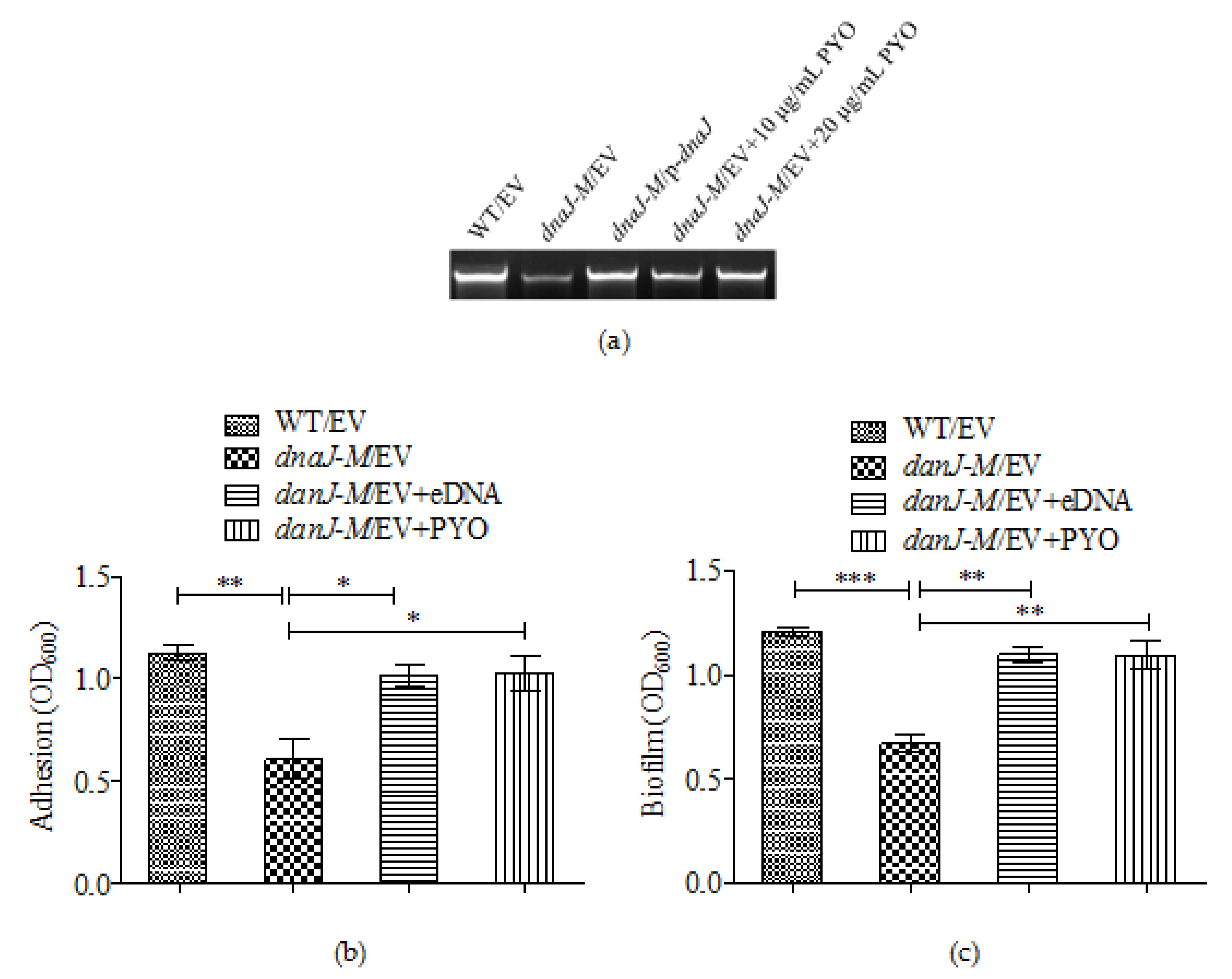
2.5. The Effect of dnaJ on Biofilm Formation Was through PYO and eDNA
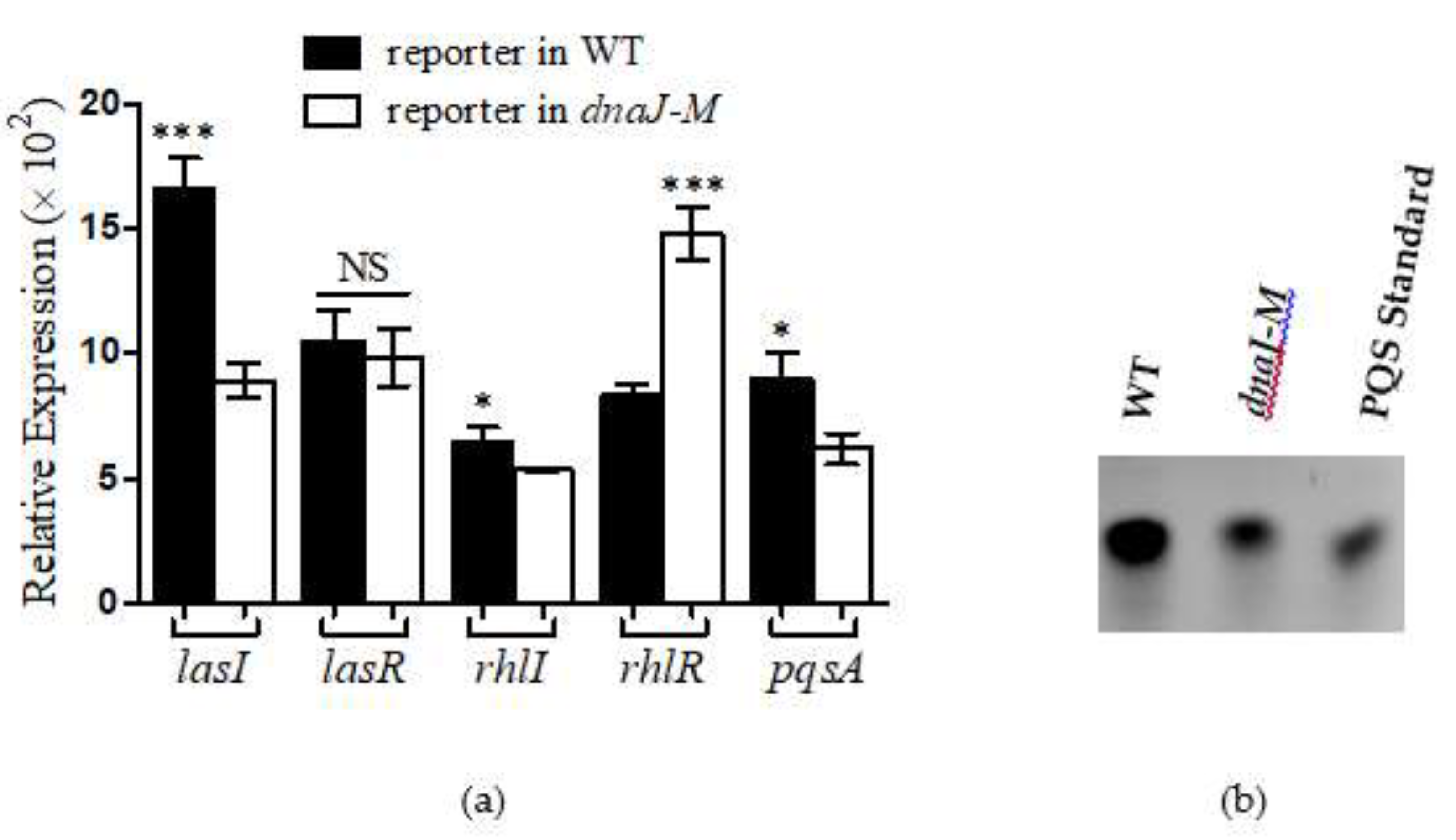
2.6. DnaJ Affected the Production of PYO through QS Pathways
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Culture and Media
4.2. Bacterial Survival Test by Growth Assay
4.3. Motility Assays
4.4. Measurement of PYO Production
4.5. Monitoring Gene Expression by Lux-Based Reporters
4.6. Quantification of Initial Adhesion and Biofilm Formation
4.7. In Vivo Pathogenicity Assay Using Drosophila Melanogaster Feeding Infection Model
4.8. eDNA Assay
4.9. PQS Extraction and Quantification
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ritossa, F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia 1962, 18, 571–573. [Google Scholar] [CrossRef]
- Ghazaei, C. Role and mechanism of the Hsp70 molecular chaperone machines in bacterial pathogens. J. Med. Microbiol. 2017, 66, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.; Morimoto, R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fernandez, M.R.; Valpuesta, J.M. Hsp70 chaperone: A master player in protein homeostasis. F1000Res 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Chenoweth, M.R.; Wickner, S. Complex regulation of the DnaJ homolog CbpA by the global regulators sigmaS and Lrp, by the specific inhibitor CbpM, and by the proteolytic degradation of CbpM. J. Bacteriol. 2008, 190, 5153–5161. [Google Scholar] [CrossRef][Green Version]
- Srinivasan, S.R.; Gillies, A.T.; Chang, L.; Thompson, A.D.; Gestwicki, J.E. Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome. Mol. Biosyst. 2012, 8, 2323–2333. [Google Scholar] [CrossRef]
- Cai, Y.; Yan, W.; Xu, W.; Yin, Y.; He, Y.; Wang, H.; Zhang, X. Screening and identification of DnaJ interaction proteins in Streptococcus pneumoniae. Curr. Microbiol. 2013, 67, 732–741. [Google Scholar] [CrossRef]
- Roncarati, D.; Scarlato, V. Regulation of heat-shock genes in bacteria: From signal sensing to gene expression output. FEMS Microbiol. Rev. 2017, 41, 549–574. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Sandri, A.; Ortombina, A.; Boschi, F.; Cremonini, E.; Boaretti, M.; Sorio, C.; Melotti, P.; Bergamini, G.; Lleo, M. Inhibition of Pseudomonas aeruginosa secreted virulence factors reduces lung inflammation in CF mice. Virulence 2018, 9, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Jayaseelan, S.; Ramaswamy, D.; Dharmaraj, S. Pyocyanin: Production, applications, challenges and new insights. World J. Microbiol. Biotechnol. 2014, 30, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Tolker-Nielsen, T. Pseudomonas aeruginosa biofilm infections: From molecular biofilm biology to new treatment possibilities. APMIS Suppl. 2014, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Camara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Pearson, J.P.; Passador, L.; Iglewski, B.H.; Greenberg, E.P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 1490–1494. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Reiser, J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 6424–6428. [Google Scholar] [CrossRef]
- Cao, H.; Krishnan, G.; Goumnerov, B.; Tsongalis, J.; Tompkins, R.; Rahme, L.G. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 2001, 98, 14613–14618. [Google Scholar] [CrossRef]
- Schuster, M.; Lostroh, C.P.; Ogi, T.; Greenberg, E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J. Bacteriol. 2003, 185, 2066–2079. [Google Scholar] [CrossRef]
- Diggle, S.P.; Winzer, K.; Chhabra, S.R.; Worrall, K.E.; Camara, M.; Williams, P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 2003, 50, 29–43. [Google Scholar] [CrossRef]
- Arenas, J.; Tommassen, J. Meningococcal Biofilm Formation: Let’s Stick Together. Trends Microbiol. 2017, 25, 113–124. [Google Scholar] [CrossRef]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Yu, Y.; Yan, R.; Zhang, H.; Shen, L.; Chen, L.; Duan, K. Identification of Genes that Regulate rsmA and Phenotypes Affected. Sci. Sin. Vitae 2016, 46, 861. [Google Scholar] [CrossRef]
- Dang, W.; Zhang, M.; Sun, L. Edwardsiella tarda DnaJ is a virulence-associated molecular chaperone with immunoprotective potential. Fish. Shellfish Immunol. 2011, 31, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Kutty, S.K.; Tavallaie, R.; Ibugo, A.I.; Panchompoo, J.; Sehar, S.; Aldous, L.; Yeung, A.W.; Thomas, S.R.; Kumar, N.; et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci. Rep. 2015, 5, 8398. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef]
- Das, T.; Manefield, M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e46718. [Google Scholar] [CrossRef]
- Price-Whelan, A.; Dietrich, L.E.; Newman, D.K. Rethinking ‘secondary’ metabolism: Physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef]
- Ohnishi, H.; Mizunoe, Y.; Takade, A.; Tanaka, Y.; Miyamoto, H.; Harada, M.; Yoshida, S. Legionella dumoffii DjlA, a member of the DnaJ family, is required for intracellular growth. Infect. Immun. 2004, 72, 3592–3603. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, X.; Yang, L.; Fang, L.; Shen, H.; Lu, X.; Fang, W. DnaJ of Streptococcus suis Type 2 Contributes to Cell Adhesion and Thermotolerance. J. Microbiol. Biotechnol. 2015, 25, 771–781. [Google Scholar] [CrossRef]
- Caldwell, C.C.; Chen, Y.; Goetzmann, H.S.; Hao, Y.; Borchers, M.T.; Hassett, D.J.; Young, L.R.; Mavrodi, D.; Thomashow, L.; Lau, G.W. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am. J. Pathol. 2009, 175, 2473–2488. [Google Scholar] [CrossRef]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Drake, D.; Montie, T.C. Flagella, motility and invasive virulence of Pseudomonas aeruginosa. J. Gen. Microbiol. 1988, 134, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, M.; Nishigaki, T.; Ihara, K.; Nishioka, N.; Kojima, S.; Homma, M. A novel dnaJ family gene, sflA, encodes an inhibitor of flagellation in marine Vibrio species. J. Bacteriol. 2013, 195, 816–822. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef]
- Duan, K.; Dammel, C.; Stein, J.; Rabin, H.; Surette, M.G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003, 50, 1477–1491. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Chugani, S.A.; Whiteley, M.; Lee, K.M.; D’Argenio, D.; Manoil, C.; Greenberg, E.P. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2001, 98, 2752–2757. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, B.; Wang, C.; Zhang, P.; Guo, Z.; Chen, L.; Duan, K. Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production. Microorganisms 2020, 8, 395. https://doi.org/10.3390/microorganisms8030395
Zeng B, Wang C, Zhang P, Guo Z, Chen L, Duan K. Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production. Microorganisms. 2020; 8(3):395. https://doi.org/10.3390/microorganisms8030395
Chicago/Turabian StyleZeng, Bo, Chong Wang, Pansong Zhang, Zisheng Guo, Lin Chen, and Kangmin Duan. 2020. "Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production" Microorganisms 8, no. 3: 395. https://doi.org/10.3390/microorganisms8030395
APA StyleZeng, B., Wang, C., Zhang, P., Guo, Z., Chen, L., & Duan, K. (2020). Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production. Microorganisms, 8(3), 395. https://doi.org/10.3390/microorganisms8030395




