Yeast Associated with Rice Phylloplane and Their Contribution to Control of Rice Sheath Blight Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Rice Leaf Collection and Phylloplane Yeast Isolation
2.2. Yeast Identification
2.3. Selection of Antagonistic Yeasts Capable of Antagonize Fungi Cause Rice Diseases
2.4. Determination of Antagonistic Mechanisms of Antagonistic Yeasts In Vitro
2.4.1. Production of Antifungal Volatile Organic Compounds
2.4.2. Production of β-Glucanase and Chitinase
2.4.3. Competition of Nutrients
2.4.4. Phosphate and Zinc Oxide Solubilization
2.4.5. Siderophore Production
2.4.6. Biofilm Formation
2.5. Controlling of Rice Sheath Blight Disease in Rice Plants in the Greenhouse by the Selected Antagonistic Yeasts
2.6. Yeast Population and Development of Sheath Blight Lesion on Rice Plants
2.7. Statistical Analysis
3. Results
3.1. Rice Phylloplane Yeast Isolation and Identification
3.2. Selection of Antagonistic Yeasts Capable of Antagonizing Fungi Causing Rice Diseases
3.3. Antagonistic Mechanisms of Antagonistic Yeasts
3.3.1. Production of Antifungal Volatile Organic Compounds
3.3.2. Production of β-Glucanase and Chitinase
3.3.3. Competition for Nutrients and Space
3.3.4. Phosphate and Zinc Oxide Solubilization
3.3.5. Siderophore Production
3.3.6. Biofilm Formation
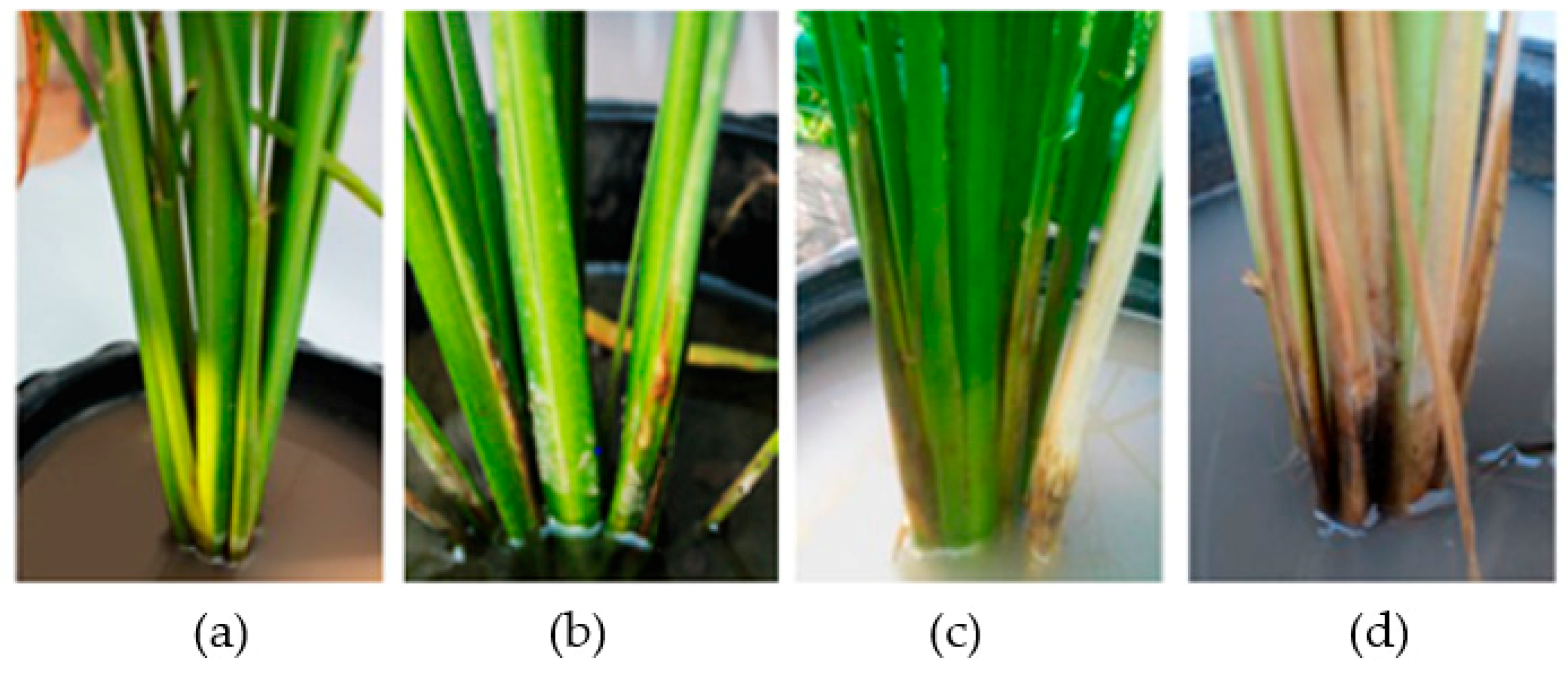
3.4. Controlling of Rice Sheath Blight Disease in Rice Plants in the Greenhouse by the Selected Antagonistic Yeasts
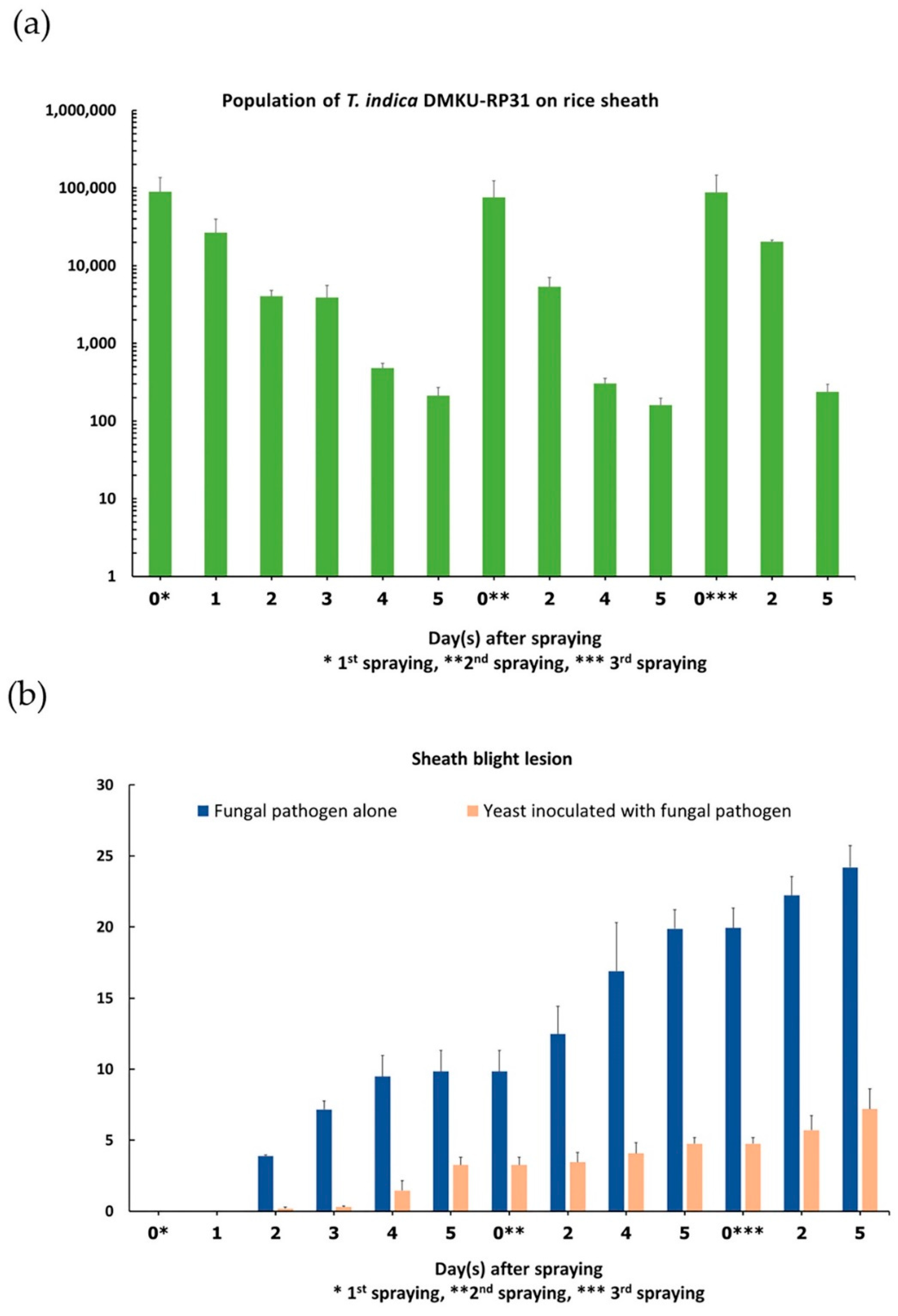
3.5. Yeast Population and Development of Sheath Blight Lesion on Rice Plant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Phaff, H.J.; Starmer, W.T. Yeasts associated with plants, insects and soil. In The Yeasts, 2nd ed.; Rose, A.H., Harrison, J.S., Eds.; Academic Press: London, UK, 1987; pp. 123–180. [Google Scholar]
- Fiala, V.; Glad, C.; Martin, M.; Jolivet, E.; Derridj, S. Occurrence of soluble carbohydrates on thephylloplane of maize (Zea mays L.): Variations in relation to leaf heterogeneity and position on the plant. New Phytol. 1990, 115, 609–615. [Google Scholar] [CrossRef]
- Xin, G.; Glawe, D.; Doty, S.L. Characterization of three endophytic, indole-3-acetic acidproducing yeasts occurring in Populus trees. Mycol. Res. 2009, 113, 973–980. [Google Scholar] [CrossRef] [PubMed]
- de Azeredo, L.A.I.; Gomes, E.A.T.; Mendonca-Hagler, L.C.; Hagler, A.N. Yeast communities associated with sugarcane in Campos, Rio de Janeiro, Brazil. Int. Microbiol. 1998, 1, 205–208. [Google Scholar]
- Inácio, J.; Portugal, L.; Spencer-Martins, I.; Fonseca, Á. Phylloplane yeasts from Portugal: Seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Res. 2005, 5, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Nasanit, R.; Jaibangyang, S.; Tantirungkij, M.; Limtong, S. Yeast diversity and novel yeast D1/D2 sequences from corn phylloplane obtained by a culture-independent approach. Antonie Leeuwenhoek 2016, 109, 1615–1634. [Google Scholar] [CrossRef] [PubMed]
- Nasanit, R.; Krataithong, K.; Tantirungkij, M.; Limtong, S. Assessment of epiphytic yeast diversity in rice (Oryza sativa) phyllosphere in Thailand by a culture-independent approach. Antonie Leeuwenhoek 2015, 107, 1475–1490. [Google Scholar] [CrossRef]
- Nasanit, R.; Tangwong-o-thai, A.; Tantirungkij, M.; Limtong, S. The assessment of epiphytic yeast diversity in sugarcane phyllosphere in Thailand by culture-independent method. Fungal Biol. 2015, 119, 1145–1157. [Google Scholar] [CrossRef]
- Plodpai, P.; Chuenchitt, S.; Petcharat, V.; Chakthong, S.; Voravuthikunchai, S.P. Anti-Rhizoctonia solani activity by Desmos chinensis extracts and its mechanism of action. Crop Prot. 2012, 43, 65–71. [Google Scholar] [CrossRef]
- Boukaew, S.; Prasertsan, P. Suppression of rice sheath blight disease using a heat stable culture filtrate from Streptomyces philanthi RM-1-138. Crop Prot. 2014, 61, 1–10. [Google Scholar] [CrossRef]
- Groth, D.E. Azoxystrobin rate and timing effects on rice sheath blight incidence and severity and rice grain and milling yields. Plant Dis. 2005, 89, 1171–1174. [Google Scholar] [CrossRef]
- Nandakumar, R.; Babu, S.; Viswanathan, R.; Raguchander, T.; Samiyappan, R. Induction of systemic resistance in rice against sheath blight disease by Pseudomonas fluorescens. Soil Biol. Biochem. 2001, 33, 603–612. [Google Scholar] [CrossRef]
- Groth, D.E.; Bond, J.A. Effects of cultivars and fungicides on rice sheath blight, yield, and quality. Plant Dis. 2007, 91, 1647–1650. [Google Scholar] [CrossRef] [PubMed]
- Boukaew, S.; Klinmanee, C.; Prasertsan, P. Potential for the integration of biological and chemical control of sheath blight disease caused by Rhizoctonia solani on rice. World J. Microbiol. Biotechnol. 2013, 29, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.A.; Fokkema, N.J. Phyllosphere yeasts antagonize penetration from appressoria and subsequent infection of maize leaves by Colletotrichum graminicola. Neth. J. Plant Pathol. 1985, 91, 265–276. [Google Scholar] [CrossRef]
- Ziedan, E.S.H.E.; Farrag, E.S. Application of yeasts as biocontrol agents for controlling foliar diseases on sugar beet plants. J. Agric. Technol. 2011, 7, 1789–1799. [Google Scholar]
- Curtis, F.D.; Cicco, V.D.; Lima, G. Efficacy of biocontrol yeasts combined with calcium silicate or Sulphur for controling durum wheat powdery mildew and increasing grain yield components. Field Crops Res. 2012, 134, 36–46. [Google Scholar] [CrossRef]
- Suzzi, G.; Romano, P.; Ponti, I.; Montuschi, C. Natural wine yeasts as biocontrol agents. J. Appl. Bacteriol. 1995, 78, 304–308. [Google Scholar] [CrossRef]
- Bar-Shimon, M.; Yehuda, H.; Cohen, L.; Weiss, B.; Kobeshnikov, A.; Duas, A.; Goldway, M.; Wisniewski, M.; Droby, S. Characterization of extracellular lytic enzymes produced by the yeast biological agent Candida oleophila. Curr. Genet. 2004, 45, 140–148. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]
- Rosa, M.M.; Tauk-Tornisielo, S.M.; Rampazzo, P.E.; Ceccato-Antonini, S.R. Evaluation of the biological control by the yeast Torulaspora globosa against Colletotrichum sublineolum in sorghum. World J. Microbiol. Biotechnol. 2010, 26, 1491–1502. [Google Scholar] [CrossRef]
- Schisler, D.A.; Janisiewicz, W.J.; Boekhout, T.; Kurtzman, C.P. Agriculturally important yeasts: Biological control of field and postharvest diseases using yeast antagonists, and yeasts as pathogens of plants. In The Yeasts, A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: New York, NY, USA, 2011; pp. 45–52. [Google Scholar]
- Bautista-Rosales, P.U.; Calderon-Santoyo, M.; Servín-Villegas, R.; Ochoa-Álvarez, N.A.; Ragazzo-Sánchez, J.A. Action mechanisms of the yeast Meyerozyma caribbica for the control of the phytopathogen Colletotrichum gloeosporioides in mangoes. Biol. Control 2013, 65, 293–301. [Google Scholar] [CrossRef]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Cordero-Bueso, G.; Mangieri, N.; Maghradez, D.; Foschino, R.; Valdetara, F.; Cantoral, J.M.; Vigentini, I. Wild grape-associated yeasts as promising biological agents against Vitis vinifera fungal pathogen. Front. Microbiol. 2017, 8, 2025. [Google Scholar] [CrossRef] [PubMed]
- Vespermann, A.; Kai, M.; Piechulla, B. Rhizobacterial volatiles affect the growth of fungi and Arabidopsis thaliana. Appl. Environ. Microbiol. 2007, 73, 5639–5641. [Google Scholar] [CrossRef]
- Huang, R.; Li, G.Q.; Zhang, J.; Yang, L.; Che, H.J.; Jiang, D.H.; Huang, H.C. Control of postharvest Botrytis fruit rot of strawberry by volatile organic compounds of Candida intermedia. Phytopathology 2011, 101, 859–869. [Google Scholar] [CrossRef]
- Ando, H.; Hatanaka, K.; Ohata, I.; Yamashita-Kitaguchi, Y.; Kurata, A.; Kishimoto, N. Antifungal activities of volatile substances generated by yeast isolated from Iranian commercial cheese. Food Control 2012, 26, 472–478. [Google Scholar] [CrossRef]
- Huang, R.; Che, H.J.; Zhang, J.; Yang, L.; Jiang, D.H.; Li, G.Q. Evaluation of Sporidiobolus pararoseus strain YCXT3 as biocontrol agent of Botrytis cinerea on post-harvest strawberry fruits. Biol. Control 2012, 62, 53–63. [Google Scholar] [CrossRef]
- Hua, S.S.T.; Beck, J.J.; Sarreal, S.B.L.; Gee, W. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014, 30, 71–78. [Google Scholar] [CrossRef]
- Chanchaichaovivat, A.; Panijpan, B.; Ruenwongsa, P. Putative modes of action of Pichia guilliermondii strain R13 in controlling chilli anthracnose after harvest. Biol. Control 2008, 47, 207–215. [Google Scholar] [CrossRef]
- Masih, E.I.; Paul, B. Secretion of β-1,3-glucanases by the yeast Pichia membranifaciens and its possible role in the biocontrol of Botrytis cinerea causing grey mold disease of the grapevine. Curr. Microbiol. 2002, 44, 391–395. [Google Scholar] [CrossRef]
- Dikin, A.; Sijam, K.; Kadir, J.; Seman, I.A. Mode of Action of Antimicrobial Substances from Burkholderia multivorans and Microbacterium testaceum Against Scizophyllum commune. J. Agric. Biol. 2007, 9, 311–314. [Google Scholar]
- Calvente, V.; De Orellano, M.E.; Sansone, G.; Benuzzi, D.; De Tosetti, M.S. Effect of nitrogen source and pH on siderophore production by Rhodotorula strains and their application to biocontrol of phytopathogenic moulds. J. Ind. Microbiol. Biot. 2001, 26, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Sansone, G.; Rezza, I.; Calvente, V.; Benuzzi, D.; de Tosetti, M.I.S. Control of Botrytis cinerea strains resistant to iprodione in apple with rhodotorulic acid and yeasts. Postharvest Biol. Technol. 2005, 35, 245–251. [Google Scholar] [CrossRef]
- Sharma, R.R.; Singh, D.; Singh, R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol. Control 2009, 50, 205–221. [Google Scholar] [CrossRef]
- Zhang, D.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Efficacy of the antagonist Aureobasidium pullulans PL5 against postharvest pathogens of peach, apple and plum and its modes of action. Biol. Control 2010, 54, 172–180. [Google Scholar] [CrossRef]
- Platania, C.; Restuccia, C.; Muccilli, S.; Cirvilleri, G. Efficacy of killer yeasts in the biological control of Penicillium digitatum on Tarocco orange fruits (Citrus sinensis). Food Microbiol. 2012, 30, 219–225. [Google Scholar] [CrossRef]
- Tian, Y.Q.; Li, W.; Jiang, Z.T.; Jing, M.M.; Shao, Y.Z. The preservation effect of Metschnikowia pulcherrima yeast on anthracnose of postharvest mango fruits and the possible mechanism. Food Sci. Biotechnol. 2018, 27, 95–105. [Google Scholar] [CrossRef]
- Surussawadee, J.; Jindamorakot, S.; Nakase, T.; Lee, C.F.; Limtong, S. Hannaella phyllophila sp. nov., a novel basidiomycetous yeast species associated with plants in Thailand and Taiwan. Int. J. Syst. Evolut. Microbiol. 2015, 65, 2135–2140. [Google Scholar] [CrossRef]
- Limtong, S.; Koowadjanakul, N.; Jindamorakot, S.; Yongmanitchai, W.; Nakase, T. Candida sirachaensis sp. nov. and Candida sakaeoensis sp. nov. two anamorphic yeast species from phylloplane in Thailand. Antonie Leeuwenhoek 2012, 102, 221–229. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycete yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evolut. Microbiol. 2000, 50, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evolut. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Ugolini, L.; Lazzeri, L.; Mari, M. Production of volatile organic compounds by Aureobasidium pullulans as a potential mechanism of action against postharvest fruit pathogens. Biol. Control 2015, 81, 8–14. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Khan, M.A.; Hamid, R.; Ahmad, M.; Abdin, M.Z.; Javed, S. Optimization of culture media for enhanced chitinase production from a novel strain of Stenotrophomonas maltophilia using response surface methodology. J. Microbiol. Biotechnol. 2010, 20, 1597–1602. [Google Scholar] [CrossRef]
- Zaidi, S.; Usmani, S.; Singh, B.R.; Musarrat, J. Significance of Bacillus subtilis strain SJ-101 as a bioinoculant for concurrent plant growth promotion and nickel accumulation in Brassica juncea. Chemosphere 2006, 64, 991–997. [Google Scholar] [CrossRef]
- Rokhbakhsh-Zamin, F.; Sachdev, D.; Kazemi-Pour, N.; Engineer, A.; Pardesi, K.R.; Zinjarde, S.; Dhakephalka, P.K.; Chopade, B.A. Characterization of plant-growth-promoting traits of Acinetobacter species isolated from rhizosphere of Pennisetum glaucum. J. Microbiol. Biotechnol. 2011, 21, 556–566. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Růžička, F.; Holá, V.; Votava, M.; Tekkalov, R. Importance of biofilm in Candida parapsilosis and evaluation of its susceptibility to antifungal agents by colorimetric method. Folia Microbiol. 2007, 52, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, D.; Ciavorella, A.; Zhang, D.; Garibaldi, A.; Gullino, M.L. Effect of culture media and pH on the biomass production and biocontrol efficacy of a Metschnikowia pulcherrima strain to be used as a biofungicide for postharvest disease control. Can. J. Microbiol. 2010, 56, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Sayler, R.; Hong, Y.; Nam, M.; Yang, Y. A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 2008, 92, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakashvel, M.; Selvakumar, M.; Srinivasan, K.; Ramesh, S.; Mathivanan, N. Control of sheath blight disease in rice by thermostable secondary metabolites of Trichoderma roseum MML003. Eur. J. Plant Pathol. 2010, 126, 229–239. [Google Scholar] [CrossRef]
- Nix, S.; Burpee, L.L.; Buck, J.W. Responses of 2 epiphytic yeasts to foliar infection by Rhizoctonia solani or mechanical wounding on the phylloplane of tall fescue. Can. J. Microbiol. 2009, 55, 1160–1165. [Google Scholar] [CrossRef]
- Jindamorakot, S.; Am-In, S.; Kaewwichian, R.; Limtong, S. Yamadazyma insecticola fa, sp. nov. and Yamadazyma epiphylla fa, sp. nov., two novel yeast species. Int. J. Syst. Evolut. Microbiol. 2015, 65, 1290–1296. [Google Scholar] [CrossRef]
- Kaewwichian, R.; Jindamorakot, S.; Am-In, S.; Sipiczki, M.; Limtong, S. Hannaella siamensis sp. nov. and Hannaella phetchabunensis sp. nov., two new anamorphic basidiomycetous yeast species isolated from plants. Int. J. Syst. Evolut. Microbiol. 2015, 65, 1297–1303. [Google Scholar] [CrossRef]
- Srisuk, N.; Nutaratat, P.; Surussawadee, J.; Limtong, S. Yeast communities in sugarcane phylloplane. Microbiology 2019, 88, 353–369. [Google Scholar] [CrossRef]
- Limtong, S.; Kaewwichian, R. The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann. Microbiol. 2015, 65, 667–675. [Google Scholar] [CrossRef]
- Coelho, A.R.; Celli, M.G.; Ono, E.Y.S.; Wosiacki, G.; Hoffmann, F.L.; Pagnocca, F.C.; Hirooka, Y.E. Penicillium expansum versus antagonist yeasts and patulin degradation in vitro. Braz. Arch. Biol. Technol. 2007, 50, 725–733. [Google Scholar] [CrossRef]
- Sundh, I.; Melin, P. Safety and regulation of yeasts used for biocontrol or biopreservation in the food or feed chain. Antonie Leeuwenhoek 2011, 99, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol yeasts: Mechanisms and applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef] [PubMed]
- Khunnamwong, P.; Lertwattanasakul, N.; Jindamorakot, S.; Suwannarach, N.; Matsui, K.; Limtong, S. Evaluation of antagonistic activity and mechanisms of endophytic yeasts against pathogenic fungi causing economic crop diseases. Folia Microbiol. 2019, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Feliziani, E.; Ciania, M.; Romanazzib, G.; Comitinia, F. Volatile organic compounds from Wickerhamomyces anomalus, Metschnikowia pulcherrima and Saccharomyces cerevisiae inhibit growth of decay causing fungi and control postharvest diseases of strawberries. Int. J. Food Microbiol. 2018, 265, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, E.M.; do Vale, H.M.M.; Moreira, G.A.M. Yeasts from native Brazilian Cerrado plants: Occurrence, diversity and use in the biocontrol of citrus green mould. Fungal Boil. 2015, 119, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Fokkema, N.J.; den Houter, J.G.; Kosterman, Y.J.C.; Nelis, A.L. Manipulation of yeasts on field-grown wheat leaves and their antagonistic effect on Cochliobolus sativus and Septoria nodorum. Trans. Br. Mycol. Soc. 1979, 72, 19–29. [Google Scholar] [CrossRef]
- Elead, Y.; Köhl, J.; Fokkema, N.J. Control of infection and sporulation of Botrytis cinerea on bean and tomato by saprophytic bacteria and fungi. Eur. J. Plant Pathol. 1994, 100, 315–336. [Google Scholar] [CrossRef]
- Dik, A.J.; Fokkema, N.J.; van Pelt, J.A. Influence of climatic and nutritional factors on yeast population dynamics in the phyllosphere of wheat. Microb. Ecol. 1992, 23, 41–52. [Google Scholar] [CrossRef]
- Shahjahan, A.K.M.; Rush, M.C.; Groth, D.E. Phylloplane yeasts as potential biocontrol agents for rice sheath blight disease. In Major Fungal Diseases of Rice; Sreenivasaprasad, S., Johnson, R., Eds.; Kluwer Academic: New York, NY, USA, 2001; pp. 235–252. [Google Scholar]
| Province | District | Location | Sampling Month and Year | No. of Samples | No. of Strains |
|---|---|---|---|---|---|
| Chachoengsao | Ban Pho | 13°35′46.0″N 01°04′56.8″E | Dec 2011 | 1 | 3 |
| Bang Pakong | 13°29′46.3″N 00°57′14.5″E | Dec 2011 | 1 | 1 | |
| Bang Khla | 13°41′11.5″N 01°04′13.3″E | Dec 2011 | 2 | 8 | |
| Mueang Chachoengsao | 13°43′53.0″N 00°59′22.8″E | Dec 2011 | 1 | 6 | |
| Phanom Sarakham | 13°45′54.1″N 01°19′42.5″E | Dec 2011 | 2 | 7 | |
| Ratchasan | 13°48′45.8″N 01°16′54.9″E | Dec 2011 | 1 | 2 | |
| Chai Nat | Manorom | 15°20′37.9″N 00°08′52.0″E | Mar 2012 | 3 | 7 |
| Mueang Chai Nat | 15°13′28.7″N 00°05′45.7″E | Mar 2012 | 3 | 7 | |
| Kanchanaburi | Phanom Thuan | 14°09′08.7″N 99°40′48.2″E | Jan 2012 | 4 | 14 |
| Nakhon Nayok | Ban Na | 14°15′34.0″N 01°01′51.4″E | Dec 2011 | 4 | 8 |
| Mueang Nakhon Nayok | 14°15′29.3″N 01°13′04.8″E | Dec 2011 | 1 | 2 | |
| Pak Phli | 14°19′34.6″N 01°21′47.7″E | Feb 2012 | 4 | 6 | |
| Nakhon Pathom | Bang Len | 14°01′55.8″N 00°09′08.6″E | Jan 2012 | 3 | 7 |
| Don Tum | 13°58′00.6″N 00°04′13.7″E | Jan 2012 | 1 | 3 | |
| Kamphaeng Saen | 14°04′53.0″N 99°57′02.3″E | Jan 2012 | 8 | 29 | |
| Nakhon Sawan | Mueang Nakhon Sawan | 15°45′10.4″N 00°07′38.4″E | Mar 2012 | 2 | 11 |
| Phayuha Khiri | 15°30′33.9″N 00°09′51.0″E | Mar 2012 | 1 | 2 | |
| Nonthaburi | Bang Bua Thong | 13°55′56.9″N 00°24′37.1″E | Feb 2012 | 1 | 10 |
| Sai Noi | 14°01′04.3″N 00°18′55.7″E | Jan 2012 | 8 | 36 | |
| Prachin Buri | Mueang Prachin Buri | 14°08′44.1″N 01°22′55.1″E | Dec 2011 | 2 | 6 |
| Si Mahosot | 13°55′01.6″N 01°24′23.2″E | Dec 2011 | 2 | 5 | |
| Suphan Buri | Bang Pla Ma | 14°23′02.7″N 00°08′46.8″E | Mar 2012 | 7 | 20 |
| Doem Bang Nang Buat | 14°52′00.9″N 00°09′29.9″E | Mar 2012 | 1 | 1 | |
| Don Chedi | 14°39′53.3″N 99°57′59.0″E | Jan 2012 | 3 | 9 | |
| Mueang Suphan Buri | 14°26′57.4″N 00°03′44.0″E | Jan 2012 | 7 | 23 | |
| Song Phi Nong | 14°13′02.3″N 99°58′44.9″E | Jan 2012 | 4 | 12 | |
| U Thong | 14°26′27.5″N 99°52′39.5″E | Jan 2012 | 12 | 37 |
| Taxa | No. of Strains | Frequency of Occurrence (%) c | No. of Strains Evaluated for Antagonistic Activity |
|---|---|---|---|
| Phylum Ascomycota, Subphylum Saccharomycotina | |||
| Blastobotrys arbuscular | 1 | 1.1 | |
| Candida diddensiae | 1 | 1.1 | |
| Candida maltosa | 2 | 2.2 | 2 |
| Candida parapsilosis | 6 | 6.7 | |
| Candida tropicalis | 4 | 4.5 | 2 |
| Candida wangnamkhiaoensis | 1 | 1.1 | |
| Debaryomyces nepalensis | 1 | 1.1 | 1 |
| Hyphopichia burtonii | 1 | 1.1 | |
| Kodamaea ohmeri | 7 | 7.9 | 7 |
| Meyerozyma caribbica | 11 | 12.4 | 11 |
| Meyerozyma guilliermondii | 2 | 2.2 | 1 |
| Torulaspora indica | 2 | 2.2 | 2 |
| Wickerhamomyces anomalus | 3 | 3.4 | 2 |
| Wickerhamomyces edaphicus | 1 | 1.1 | |
| Yamadazyma epiphyllaa | 1 | 1.1 | |
| Phylum Basidiomycota, Subphylum Agaricomycotina | |||
| Hannaella sinensis | 4 | 4.5 | |
| Hannaella siamensisb | 7 | 7.9 | 2 |
| Hannaella pagnoccae | 1 | 1.1 | 1 |
| Hannaella phetchabunensis | 2 | 2.2 | |
| Papiliotrema aspenensis | 4 | 4.5 | |
| Papiliotrema flavescens | 7 | 7.9 | |
| Papiliotrema japonica | 15 | 16.9 | 5 |
| Papiliotrema laurentii | 2 | 2.2 | |
| Papiliotrema nemorosus | 1 | 1.1 | 1 |
| Papiliotrema rajasthanensis | 5 | 5.6 | |
| Papiliotrema siamense | 3 | 3.4 | |
| Saitozyma flava | 2 | 2.2 | 2 |
| Trichosporon asahii | 1 | 1.1 | |
| Trichosporon asteroides | 1 | 1.1 | |
| Trichosporon insectorum | 1 | 1.1 | |
| Potential new species closest to Vishniacozyma taibaiensis | 1 | 1.1 | |
| Phylum Basidiomycota, Subphylum Pucciniomycotina | |||
| Occultifur plantarum | 2 | 2.2 | 2 |
| Rhodotorula mucilaginosa | 9 | 10.1 | 4 |
| Rhodotorula paludigena | 7 | 7.9 | 1 |
| Rhodotorula taiwanensis | 24 | 27.0 | 12 |
| Rhodotorula toruloides | 2 | 2.2 | 2 |
| Potential new species closest to Rhodotorula toruloides | 2 | 2.2 | 2 |
| Sakaguchia oryzae | 5 | 5.6 | 3 |
| Sporobolomyces blumeae | 14 | 15.7 | 1 |
| Sporobolomyces carnicolor | 4 | 4.5 | |
| Sporobolomyces nakasei | 1 | 1.1 | |
| Sporidiobolus pararoseus | 7 | 7.9 | 3 |
| Symmetrospora vermiculata | 2 | 2.2 | 1 |
| Phylum Basidiomycota, Subphylum Ustilaginomycotina | |||
| Dirkmeia churashimaensis | 36 | 40.4 | 7 |
| Jaminaea angkoriensis | 3 | 3.4 | |
| Kalmanozyma vetiver | 1 | 1.1 | |
| Moesziomyces antarcticus | 55 | 61.8 | 5 |
| Moesziomyces aphidis | 1 | 1.1 | |
| Moesziomyces parantarcticus | 1 | 1.1 | 1 |
| Pseudozyma alboarmeniaca | 2 | 2.2 | |
| Pseudozyma hubeiensis | 1 | 1.1 | |
| Ustilago siamensis | 2 | 2.2 | |
| Yeast | Growth Inhibition by Yeast (%) a | ||||
|---|---|---|---|---|---|
| Cu. lunata DOAC 2313 | F. moniliforme DOAC 1224 | H. oryzae DOAC 2293 | R. solani DOAC 1406 | P. oryzae | |
| Kodamaea ohmeri | |||||
| DMKU-RP06 | 48.3 ± 1.4d | 23.3 ± 5.73cd | 0 | 0 | 0 |
| DMKU-RP18 | 0 | 20.0 ± 3.57e | 0 | 0 | 0 |
| DMKU-RP24 | 0 | 16.7 ± 4.72f | 0 | 0 | 45.9 ± 6.5c |
| DMKU-RP34 | 0 | 25.1 ± 2.31c | 0 | 0 | 0 |
| DMKU-RP44 | 0 | 23.2 ± 3.89cd | 0 | 0 | 0 |
| DMKU-RP57 | 44.7 ± 1.5e | 46.6 ± 2.53a | 0 | 0 | 0 |
| DMKU-RP233 | 63.1 ± 0.5a | 23.3 ± 4.75cd | 0 | 0 | 38.8 ± 7.8d |
| Meyerozyma caribbica | |||||
| DMKU-RP07 | 38.0 ± 0.6 | 23.3 ± 3.27cd | 0 | 0 | 0 |
| DMKU-RP55 | 0 | 25.0 ± 2.9c | 59.8 ± 0.86bc | 0 | 33.5 ± 7.9d |
| Meyerozyma guilliermondii | |||||
| DMKU-RP26 | 0 | 15.2 ± 2.6g | 0 | 0 | 43.4 ± 8.7c |
| Torulaspora indica | |||||
| DMKU-RP31 | 62.0 ± 1.7ab | 46.6 ± 3.2a | 64.1 ± 0.7a | 86.3 ± 0.9a | 62.6 ± 4.4a |
| DMKU-RP35 | 61.0 ± 0.9ab | 46.6 ± 3.2a | 64.9 ± 1.5a | 85.4 ± 0.8a | 62.2 ± 4.6a |
| Wickerhamomyces anomalus | |||||
| DMKU-RP04 | 50.3 ± 1.7c | 30.0 ± 2.0b | 48.5 ± 2.9d | 0 | 0 |
| DMKU-RP25 | 59.1 ± 1.0b | 29.9 ± 2.1b | 60.2 ± 1.1b | 79.7 ± 0.5b | 55.7 ± 6.6b |
| Rice Pathogenic Fungus and Antagonistic Yeast | Growth Inhibition by VOCs (%) a | Growth Inhibition in Different Nutrient Competition b | ||||
|---|---|---|---|---|---|---|
| A c | B d | C e | D f | Sum | ||
| Cu. lunata DOAC 2313 | ||||||
| K. ohmeri DMKU-RP06 | 15.2 ± 2.0d | 48.3a | 18.8b | 6.3c | 0d | - |
| K. ohmeri DMKU-RP57 | 35.1 ± 0.5b | 44.7a | 15.1b | 0c | 0c | - |
| K. ohmeri DMKU-RP233 | 19.4 ± 0.3cd | 31.4a | 7.5b | 0c | 0c | - |
| M. caribbica DMKU-RP07 | 32.1 ± 0.6bc | 38.0a | 17.2b | 0c | 0c | - |
| T. indica DMKU-RP31 | 60.2 ± 0.3a | 62.1a | 38.4b | 6.9c | 0d | - |
| T. indica DMKU-RP35 | 59.6 ± 0.9a | 61.0a | 38.3b | 4.4c | 0d | - |
| W. anomalus DMKU-RP04 | 15.7 ± 1.1d | 50.3a | 22.8b | 6.1c | 0d | - |
| W. anomalus DMKU-RP25 | 23.8 ± 1.2c | 59.1a | 21.1b | 6.1c | 0d | - |
| F. moniliforme DOAC 1224 | ||||||
| K. ohmeri DMKU-RP06 | 6.8 ± 1.3e | 23.3a | 0b | 0b | 0b | - |
| K. ohmeri DMKU-RP18 | 25.3 ± 1.2c | 20.0a | 6.9b | 0c | 0c | - |
| K. ohmeri DMKU-RP24 | 17.6 ± 6.9d | 16.7a | 10.1b | 0c | 0c | - |
| K. ohmeri DMKU-RP34 | 20.2 ± 0.5cd | 25.1a | 8.7b | 0c | 0c | - |
| K. ohmeri DMKU-RP44 | 23.3 ± 0.7cd | 23.2a | 8.6b | 2.3c | 0d | - |
| K. ohmeri DMKU-RP57 | 0f | 46.6a | 25.6b | 11.9c | 0d | - |
| K. ohmeri DMKU-RP233 | 22.1 ± 1.1cd | 23.3a | 5.8b | 0c | 0c | - |
| M. caribbica DMKU-RP07 | 6.4 ± 1.9e | 23.3a | 6.8b | 0c | 0d | - |
| M. caribbica DMKU-RP55 | 0f | 25.0a | 11.6b | 1.6c | 0d | - |
| M. guilliermondii DMKU-RP26 | 0f | 15.2a | 3.8b | 0c | 0c | - |
| T. indica DMKU-RP31 | 50.9 ± 0.5a | 46.6a | 28.1b | 11.1c | 0d | |
| T. indica DMKU-RP35 | 51.2 ± 1.2a | 46.6a | 24.7b | 5.9c | 0d | - |
| W. anomalus DMKU-RP04 | 41.1 ± 1.4b | 30.0a | 10.7b | 0c | 0c | - |
| W. anomalus DMKU-RP25 | 35.2 ± 2.5b | 30.0a | 20.0b | 5.6c | 0d | - |
| H. oryzae DOAC 2293 | ||||||
| M. caribbica DMKU-RP55 | 0d | 59.8a | 44.4b | 23.5c | 0d | - |
| T. indica DMKU-RP31 | 49.3 ± 0.5a | 64.1a | 47.3b | 25.8c | 0d | - |
| T. indica DMKU-RP35 | 31.5 ± 0.6b | 64.9a | 47.2b | 24.6c | 0d | - |
| W. anomalus DMKU-RP04 | 21.5 ± 0.4c | 48.5a | 37.9b | 23.8c | 0d | - |
| W. anomalus DMKU-RP25 | 49.3 ± 0.5a | 60.2a | 46.6b | 25.6c | 0d | - |
| R. solani DOAC 1406 | ||||||
| T. indica DMKU-RP31 | 94.1 ± 0.0a | 86.3a | 80.7b | 0c | 0c | - |
| T. indica DMKU-RP35 | 94.1 ± 0.0a | 85.4a | 80.3b | 0c | 0c | - |
| W. anomalus DMKU-RP25 | 94.1 ± 0.0a | 79.7a | 73.3b | 0c | 0c | - |
| P. oryzae | ||||||
| K. ohmeri DMKU-RP24 | 9.8 ± 1.0d | 36.0a | 10.6b | 0c | 0c | - |
| K. ohmeri DMKU-RP233 | 73.3 ± 1.1b | 27.8a | 13.8b | 0c | 0c | - |
| M. caribbica DMKU-RP55 | 8.1 ± 0.4d | 21.4a | 0b | 0b | 0b | - |
| M. guilliermondii DMKU-RP26 | 7.2 ± 0.7d | 33.3a | 10.3b | 0c | 0c | - |
| T. indica DMKU-RP31 | 91.9 ± 0.0a | 55.8a | 26.7b | 0c | 0c | - |
| T. indica DMKU-RP35 | 91.9 ± 0.0a | 55.3a | 26.7b | 0c | 0c | - |
| W. anomalus DMKU-RP25 | 52.2 ± 1.0c | 47.7a | 17.0b | 0c | 0c | - |
| Antagonistic Yeast | Enzyme Activities (mU/mL) | SE a | Siderophore Production b | Biofilm Formation | ||||
|---|---|---|---|---|---|---|---|---|
| Glucanase | Chitinase | Ca3(PO)4 | ZnO | A c | A value d | Sum e | ||
| K. ohmeri DMKU-RP06 | 0 | 25.1 ± 0.5 | 0 | 0 | 0 | 0.1664 ± 0.03 | 2.2 | + |
| K. ohmeri DMKU-RP18 | 0.2 ± 0.0 | 0 | 0 | 0 | 0 | 0.4848 ± 0.06 | 6.5 | + |
| K. ohmeri DMKU-RP24 | 4.6 ± 0.9 | 0 | 0 | 0 | 0 | 0.5614 ± 0.07 | 7.5 | + |
| K. ohmeri DMKU-RP34 | 27.1 ± 2.5 | 2.0 ± 0.4 | 0 | 0 | 0 | 0.3535 ± 0.02 | 4.7 | + |
| K. ohmeri DMKU-RP44 | 0 | 0 | 0 | 0 | 0 | 0.4905 ± 0.03 | 6.6 | + |
| K. ohmeri DMKU-RP57 | 11.0 ± 2.1 | 249.2 ± 39.6 | 0 | 0 | 0 | 0.1505 ± 0.02 | 2.0 | + |
| K. ohmeri DMKU-RP233 | 17.8 ± 2.2 | 0 | 0 | 0 | 0 | 0.1188 ± 0.01 | 1.6 | + |
| M. caribbica DMKU-RP07 | 4.7 ± 0.3 | 88.4 ± 5.9 | 0 | 0 | 0 | 0.1616 ± 0.01 | 2.2 | + |
| M. caribbica DMKU-RP55 | 0.6 ± 0.2 | 0 | 0 | 0 | 0 | 0.1351 ± 0.02 | 1.8 | + |
| M. guilliermondii DMKU-RP26 | 4.6 ± 1.2 | 0 | 0 | 0 | 0 | 0.0746 ± 0.07 | 1.0 | - |
| T. indica DMKU-RP31 | 1.7 ± 0.4 | 35.2 ± 3.5 | 1.2 | 1.2 | 0 | 0.5407 ± 0.06 | 7.3 | + |
| T. indica DMKU-RP35 | 2.2 ± 0.3 | 166.8 ± 5.7 | 1.2 | 1.2 | 0 | 0.4252 ± 0.03 | 5.7 | + |
| W. anomalus DMKU-RP04 | 4.2 ± 0.3 | 109.8 ± 9.4 | 1.0 | 1.0 | 3.0 | 0.2967 ± 0.01 | 4.0 | + |
| W. anomalus DMKU-RP25 | 1.8 ± 0.2 | 107.5 ± 7.0 | 1.0 | 1.2 | 2.9 | 0.2121 ± 0.02 | 2.8 | + |
| Treatment | Plant Height (cm) a | Lesion Height (cm) b | Disease Incidence (%) | Disease Suppression (%) |
|---|---|---|---|---|
| Control (negative control) | 94.0 ± 2.9a | 0d | 0d | 0 |
| R. solani (positive control) | 92.0 ± 2.6a | 23.8 ± 1.6a | 25.9 ± 2.3a | 0 |
| R. solani + T. indica DMKU-RP31 | 93.0 ± 4.1a | 7.2 ± 1.4b | 7.7 ± 1.4b | 70.3 |
| R. solani + T. indica DMKU-RP35 | 93.2 ± 2.3a | 8.3 ± 1.5b | 8.8 ± 1.4b | 66.0 |
| R. solani + W. anomalus DMKU-RP25 | 92.6 ± 3.0a | 8.1 ± 1.0b | 8.7 ± 0.8b | 66.4 |
| R. solani + 3% Validamycin | 93.4 ± 3.5a | 3.9 ± 0.5c | 4.2 ± 0.6c | 83.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Into, P.; Khunnamwong, P.; Jindamoragot, S.; Am-in, S.; Intanoo, W.; Limtong, S. Yeast Associated with Rice Phylloplane and Their Contribution to Control of Rice Sheath Blight Disease. Microorganisms 2020, 8, 362. https://doi.org/10.3390/microorganisms8030362
Into P, Khunnamwong P, Jindamoragot S, Am-in S, Intanoo W, Limtong S. Yeast Associated with Rice Phylloplane and Their Contribution to Control of Rice Sheath Blight Disease. Microorganisms. 2020; 8(3):362. https://doi.org/10.3390/microorganisms8030362
Chicago/Turabian StyleInto, Parichat, Pannida Khunnamwong, Sasitorn Jindamoragot, Somjit Am-in, Wanwilai Intanoo, and Savitree Limtong. 2020. "Yeast Associated with Rice Phylloplane and Their Contribution to Control of Rice Sheath Blight Disease" Microorganisms 8, no. 3: 362. https://doi.org/10.3390/microorganisms8030362
APA StyleInto, P., Khunnamwong, P., Jindamoragot, S., Am-in, S., Intanoo, W., & Limtong, S. (2020). Yeast Associated with Rice Phylloplane and Their Contribution to Control of Rice Sheath Blight Disease. Microorganisms, 8(3), 362. https://doi.org/10.3390/microorganisms8030362





