Indoor Microbiome: Quantification of Exposure and Association with Geographical Location, Meteorological Factors, and Land Use in France
Abstract
1. Introduction
2. Material and Methods
2.1. Samples and qPCR Method
2.2. qPCR Data
2.3. Climate Data and Land Use
2.4. Statistical Analyses
3. Results
3.1. qPCR Analysis
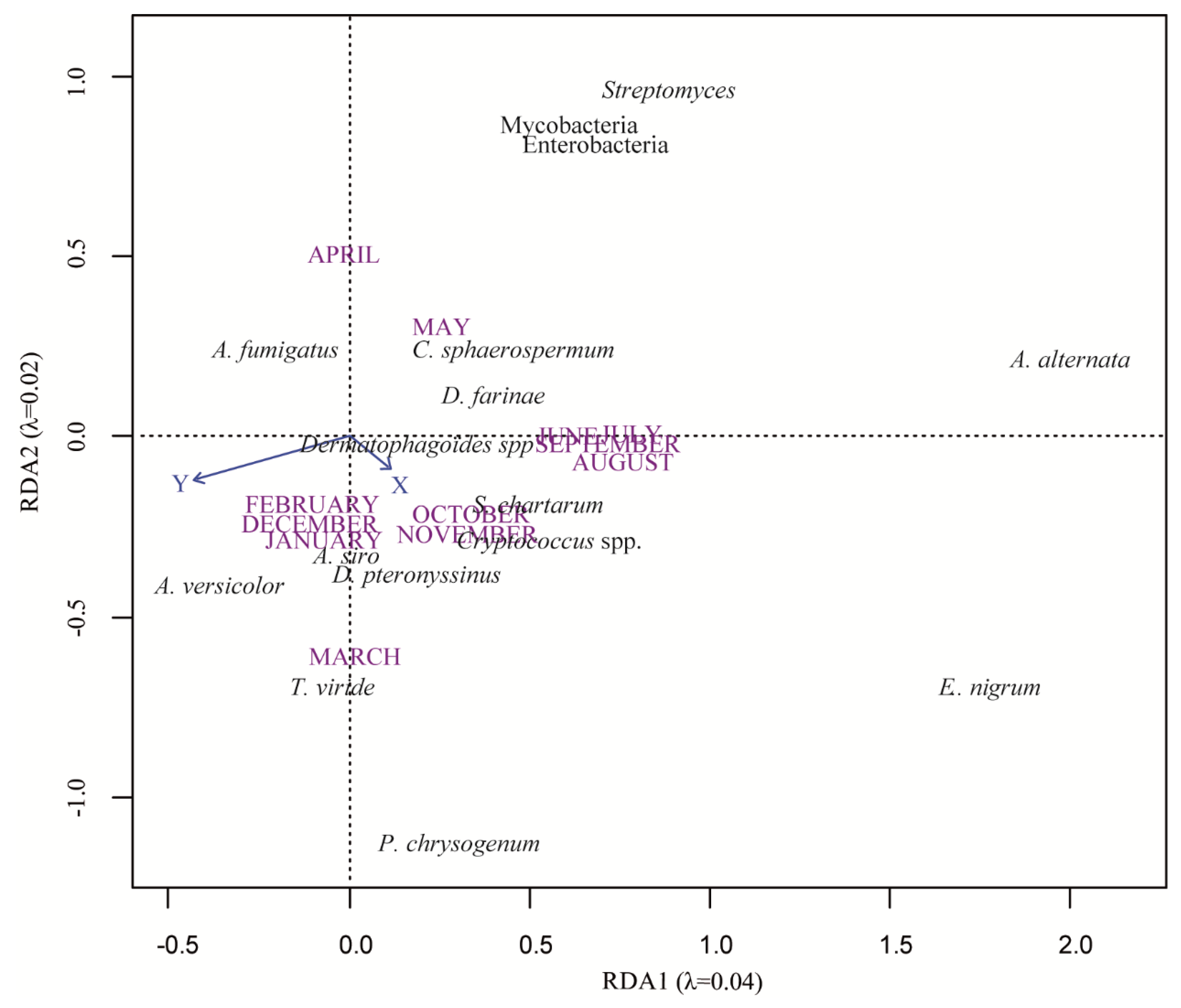
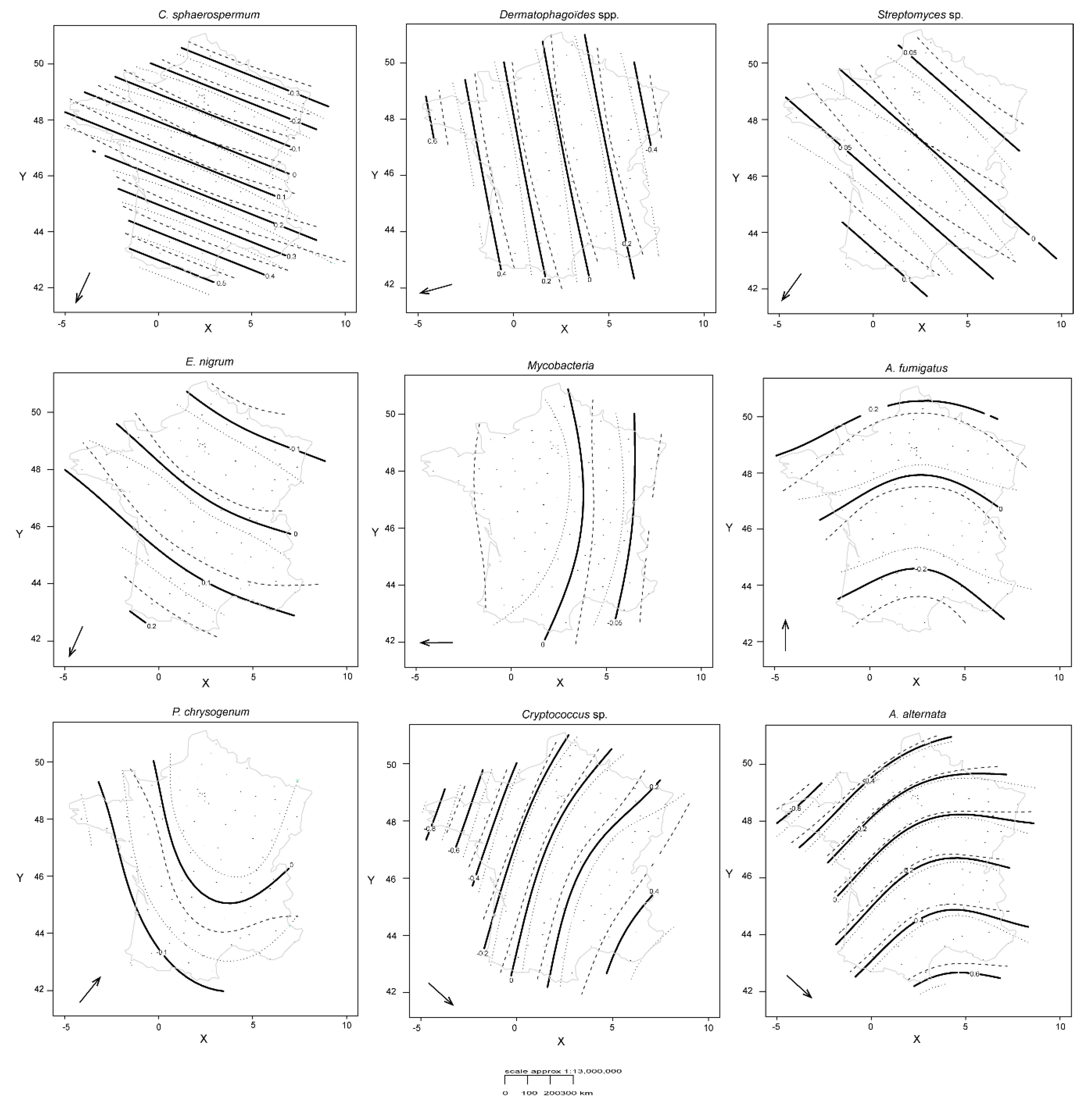
3.2. Spatio-Temporal Distribution of Microorganisms
3.3. Climate and Biophysical Land Use for Each Individual Target
3.4. Microorganism Community
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kirjavainen, P.V.; Hyytiäinen, H.; Täubel, M. The environmental microbiota and asthma. In The Lung Microbiome (ERS Monograph); Cox, M.J., Ege, M.J., von Mutius, E., Eds.; European Respiratory Society: Lausanne, Switzerland, 2019; pp. 216–239. [Google Scholar] [CrossRef]
- von Mutius, E. Allergies, infections and the hygiene hypothesis—The epidemiological evidence. Immunobiology 2007, 212, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Tischer, C.G.; Hohmann, C.; Thiering, E.; Herbarth, O.; Muller, A.; Henderson, J.; Granell, R.; Fantini, M.P.; Luciano, L.; Bergstrom, A.; et al. Meta-analysis of mould and dampness exposure on asthma and allergy in eight European birth cohorts: An ENRIECO initiative. Allergy 2011, 66, 1570–1579. [Google Scholar] [CrossRef] [PubMed]
- Douwes, J.; Thorne, P.; Pearce, N.; Heederik, D. Bioaerosol Health Effects and Exposure Assessment: Progress and Prospects. Ann. Occup. Hyg. 2003, 47, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Platts-Mills, T.A.; Vervloet, D.; Thomas, W.R.; Aalberse, R.C.; Chapman, M.D. Indoor allergens and asthma: Report of the Third International Workshop. J. Allergy Clin. Immunol. 1997, 100, S2–S24. [Google Scholar] [CrossRef]
- Kanchongkittiphon, W.; Mendell, M.J.; Gaffin, J.M.; Wang, G.; Phipatanakul, W. Indoor environmental exposures and exacerbation of asthma: An update to the 2000 review by the Institute of Medicine. Environ. Health Perspect. 2015, 123, 6–20. [Google Scholar] [CrossRef]
- Martin, L.J.; Adams, R.I.; Bateman, A.; Bik, H.M.; Hawks, J.; Hird, S.M.; Hughes, D.; Kembel, S.W.; Kinney, K.; Kolokotronis, S.-O.; et al. Evolution of the indoor biome. Trends Ecol. Evol. 2015, 30, 223–232. [Google Scholar] [CrossRef]
- Gibbons, S.M. The Built Environment Is a Microbial Wasteland. mSystems 2016, 1, e00033-16. [Google Scholar] [CrossRef]
- Horve, P.F.; Lloyd, S.; Mhuireach, G.A.; Dietz, L.; Fretz, M.; MacCrone, G.; Van Den Wymelenberg, K.; Ishaq, S.L. Building upon current knowledge and techniques of indoor microbiology to construct the next era of theory into microorganisms, health, and the built environment. J. Expo. Sci. Environ. Epidemiol. 2019, 30, 219–235. [Google Scholar] [CrossRef]
- Amato, P.; Joly, M.; Besaury, L.; Oudart, A.; Taib, N.; Mone, A.I.; Deguillaume, L.; Delort, A.M.; Debroas, D. Active microorganisms thrive among extremely diverse communities in cloud water. PLoS ONE 2017, 12, e0182869. [Google Scholar] [CrossRef]
- Golan, J.J.; Pringle, A. Long-Distance Dispersal of Fungi. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Prospero, J.M.; Blades, E.; Mathison, G.; Naidu, R. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 2005, 21, 1–19. [Google Scholar] [CrossRef]
- Brown, J.K.; Hovmoller, M.S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.H.; Lee, P.K. The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: A review. Microbiome 2016, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Pitkäranta, M.; Meklin, T.; Hyvärinen, A.; Paulin, L.; Auvinen, P.; Nevalainen, A.; Rintala, H. Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl. Environ. Microbiol. 2008, 74, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; McLetchie, S.; Knight, R.; Fierer, N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2010, 5, 601. [Google Scholar] [CrossRef] [PubMed]
- Weikl, F.; Tischer, C.; Probst, A.J.; Heinrich, J.; Markevych, I.; Jochner, S.; Pritsch, K. Fungal and bacterial communities in indoor dust follow different environmental determinants. PLoS ONE 2016, 11, e0154131. [Google Scholar] [CrossRef] [PubMed]
- Grantham, N.S.; Reich, B.J.; Pacifici, K.; Laber, E.B.; Menninger, H.L.; Henley, J.B.; Barberan, A.; Leff, J.W.; Fierer, N.; Dunn, R.R. Fungi identify the geographic origin of dust samples. PLoS ONE 2015, 10, e0122605. [Google Scholar] [CrossRef]
- Barberan, A.; Ladau, J.; Leff, J.W.; Pollard, K.S.; Menninger, H.L.; Dunn, R.R.; Fierer, N. Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl. Acad. Sci. USA 2015, 112, 5756–5761. [Google Scholar] [CrossRef]
- Vesper, S.; Wakefield, J.; Ashley, P.; Cox, D.; Dewalt, G.; Friedman, W. Geographic distribution of Environmental Relative Moldiness Index molds in USA homes. J. Environ. Public Health 2011, 2011, 242457. [Google Scholar] [CrossRef]
- Shelton, B.G.; Kirkland, K.H.; Flanders, W.D.; Morris, G.K. Profiles of airborne fungi in buildings and outdoor environments in the United States. Appl. Environ. Microbiol. 2002, 68, 1743–1753. [Google Scholar] [CrossRef]
- Pakpour, S.; Li, D.-W.; Klironomos, J. Relationships of fungal spore concentrations in the air and meteorological factors. Fungal Ecol. 2015, 13, 130–134. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Ye, Z.; Li, J.; Feng, Y.; Lu, Q. Relationships between fungal and plant communities differ between desert and grassland in a typical dryland region of northwest China. Front. Microbiol. 2018, 9, 2327. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Villarreal Ruiz, L.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Fungal biogeography. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Knorr, K.; Jorgensen, L.N.; Nicolaisen, M. Fungicides have complex effects on the wheat phyllosphere mycobiome. PLoS ONE 2019, 14, e0213176. [Google Scholar] [CrossRef] [PubMed]
- Pellissier, L.; Oppliger, A.; Hirzel, A.H.; Savova-Bianchi, D.; Mbayo, G.; Mascher, F.; Kellenberger, S.; Niculita-Hirzel, H. Airborne and Grain Dust Fungal Community Compositions Are Shaped Regionally by Plant Genotypes and Farming Practices. Appl. Environ. Microbiol. 2016, 82, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Grinn-Gofron, A.; Nowosad, J.; Bosiacka, B.; Camacho, I.; Pashley, C.; Belmonte, J.; De Linares, C.; Ianovici, N.; Manzano, J.M.M.; Sadys, M.; et al. Airborne Alternaria and Cladosporium fungal spores in Europe: Forecasting possibilities and relationships with meteorological parameters. Sci. Total Environ. 2019, 653, 938–946. [Google Scholar] [CrossRef]
- Crawford, J.A.; Rosenbaum, P.F.; Anagnost, S.E.; Hunt, A.; Abraham, J.L. Indicators of airborne fungal concentrations in urban homes: Understanding the conditions that affect indoor fungal exposures. Sci. Total Environ. 2015, 517, 113–124. [Google Scholar] [CrossRef]
- Salonen, H.; Duchaine, C.; Mazaheri, M.; Clifford, S.; Lappalainen, S.; Reijula, K.; Morawska, L. Airborne viable fungi in school environments in different climatic regions—A review. Atmos. Environ. 2015, 104, 186–194. [Google Scholar] [CrossRef]
- Adams, R.I.; Miletto, M.; Lindow, S.E.; Taylor, J.W.; Bruns, T.D. Airborne bacterial communities in residences: Similarities and differences with fungi. PLoS ONE 2014, 9, e91283. [Google Scholar] [CrossRef]
- Caillaud, D.; Cheriaux, M.; Martin, S.; Segala, C.; Dupuy, N.; Evrard, B.; Thibaudon, M. Short-term effect of outdoor mould spore exposure on prescribed allergy medication sales in Central France. Clin. Exp. Allergy 2018, 48, 837–845. [Google Scholar] [CrossRef]
- Grewling, Ł.; Nowak, M.; Szymańska, A.; Kostecki, Ł.; Bogawski, P. Temporal variability in the allergenicity of airborne Alternaria spores. Med. Mycol. 2019, 57, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, S.; Reboux, G.; Frossard, V.; Scherer, E.; Valot, B.; Laboissiere, A.; Zaros, C.; Vacheyrou, M.; Gillet, F.; Roussel, S.; et al. Microbiological characterization of 3193 French dwellings of Elfe cohort children. Sci. Total Environ. 2015, 505, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Reboux, G.; Valot, B.; Rocchi, S.; Scherer, E.; Roussel, S.; Millon, L. Storage mite concentrations are underestimated compared to house dust mite concentrations. Exp. Appl. Acarol. 2019, 77, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Dannemiller, K.C.; Mendell, M.J.; Macher, J.M.; Kumagai, K.; Bradman, A.; Holland, N.; Harley, K.; Eskenazi, B.; Peccia, J. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air 2014, 24, 236–247. [Google Scholar] [CrossRef]
- Nevalainen, A.; Täubel, M.; Hyvärinen, A. Indoor fungi: Companions and contaminants. Indoor Air 2015, 25, 125–156. [Google Scholar] [CrossRef]
- Amend, A.S.; Seifert, K.A.; Samson, R.; Bruns, T.D. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. USA 2010, 107, 13748–13753. [Google Scholar] [CrossRef]
- Chase, J.; Fouquier, J.; Zare, M.; Sonderegger, D.L.; Knight, R.; Kelley, S.T.; Siegel, J.; Caporaso, J.G. Geography and location are the primary drivers of office microbiome composition. mSystems 2016, 1, e00022-16. [Google Scholar] [CrossRef]
- Barrera, C.; Rocchi, S.; Degano, B.; Soumagne, T.; Laurent, L.; Bellanger, A.P.; Laplante, J.J.; Millon, L.; Dalphin, J.C.; Reboux, G. Microbial exposure to dairy farmers’ dwellings and COPD occurrence. Int. J. Environ. Health Res. 2019, 29, 387–399. [Google Scholar] [CrossRef]
- Rocchi, S.; Valot, B.; Reboux, G.; Millon, L. DNA metabarcoding to assess indoor fungal communities: Electrostatic dust collectors and Illumina sequencing. J. Microbiol. Methods 2017, 139, 107–112. [Google Scholar] [CrossRef]
- EPA. Microbiological and Chemical Exposure Assessment, EPA Technology for Mold Identification and Enumeration. Available online: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=2ahUKEwiCpMX5tvXjAhURfxoKHa4fCJwQFjAAegQIARAC&url=https%3A%2F%2Firp-cdn.multiscreensite.com%2Fc4e267ab%2Ffiles%2Fuploaded%2FgCQnkBNWQuSD96fPIikY_EPA_Technology%2520for%2520Mold%2520Identification%2520and%2520Enumeration.pdf&usg=AOvVaw0hz56KTQnKxxabVWqzeW3t (accessed on 20 August 2019).
- Sen, K.; Asher, D.M. Multiplex PCR for detection of Enterobacteriaceae in blood. Transfusion 2001, 41, 1356–1364. [Google Scholar] [CrossRef]
- Torvinen, E.; Torkko, P.; Rintala, A.N. Real-time PCR detection of environmental mycobacteria in house dust. J. Microbiol. Methods 2010, 82, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Rintala, H.; Nevalainen, A. Quantitative measurement of streptomycetes using real-time PCR. J. Environ/ Monitor. 2006, 8, 745–749. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Heisterkamp, S.; van Willigen, B.; R-Core Team. Nlme: Linear and Nonlinear Mixed Effects Models; R Package Version 3.1-122. Available online: https://cran.r-project.org/web/packages/nlme/nlme.pdf (accessed on 2 February 2020).
- Wood, S. Generalized Additive Models: An Introduction with R; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Becker, R.A.; Wilks, A.R. Mapdata: Extra Map Databases; R Package Version 2.2-2. Available online: https://cran.r-project.org/web/packages/mapdata/index.html (accessed on 20 August 2018).
- Zuur, A.F.; Ieno, E.N.; Walker, N.J.; Saveliev, A.A.; Smith, G.M. Mixed Effects Models and Extensions in Ecology with R; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Adams, R.I.; Miletto, M.; Taylor, J.W.; Bruns, T.D. Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013, 7, 1262–1273. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.I.; Amend, A.S.; Taylor, J.W.; Bruns, T.D. A unique signal distorts the perception of species richness and composition in high-throughput sequencing surveys of microbial communities: A case study of fungi in indoor dust. Microb. Ecol. 2013, 66, 735–741. [Google Scholar] [CrossRef]
- Amend, A.S.; Seifert, K.A.; Bruns, T.D. Quantifying microbial communities with 454 pyrosequencing: Does read abundance count? Mol. Ecol. 2010, 19, 5555–5565. [Google Scholar] [CrossRef]
- Adhikari, A.; Reponen, T.; Rylander, R. Airborne fungal cell fragments in homes in relation to total fungal biomass. Indoor Air 2013, 23, 142–147. [Google Scholar] [CrossRef]
- Reponen, T.; Seo, S.C.; Grimsley, F.; Lee, T.; Crawford, C.; Grinshpun, S.A. Fungal fragments in moldy houses: A field study in homes in New Orleans and Southern Ohio. Atmos. Environ. 2007, 41, 8140–8149. [Google Scholar] [CrossRef]
- Madsen, A.M.; Matthiesen, C.B.; Frederiksen, M.W.; Frederiksen, M.; Frankel, M.; Spilak, M.; Gunnarsen, L.; Timm, M. Sampling, extraction and measurement of bacteria, endotoxin, fungi and inflammatory potential of settling indoor dust. J. Environ. Monitor. 2012, 14, 3230–3239. [Google Scholar] [CrossRef]
- Sordillo, J.E.; Alwis, U.K.; Hoffman, E.; Gold, D.R.; Milton, D.K. Home characteristics as predictors of bacterial and fungal microbial biomarkers in house dust. Environ. Health Perspect. 2011, 119, 189–195. [Google Scholar] [CrossRef]
- Adams, R.I.; Bhangar, S.; Dannemiller, K.C.; Eisen, J.A.; Fierer, N.; Gilbert, J.A.; Green, J.L.; Marr, L.C.; Miller, S.L.; Siegel, J.A.; et al. Ten questions concerning the microbiomes of buildings. Build. Environ. 2016, 109, 224–234. [Google Scholar] [CrossRef]
- Fog Nielsen, K. Mycotoxin production by indoor molds. Fungal Genet. Biol. 2003, 39, 103–117. [Google Scholar] [CrossRef]
- Thind, B.B.; Clarke, P.G. The occurrence of mites in cereal-based foods destined for human consumption and possible consequences of infestation. Exp. Appl. Acarol. 2001, 25, 203–215. [Google Scholar] [CrossRef] [PubMed]

| Targets and Designers [Reference] | (5’–3’) Sequences | ||
|---|---|---|---|
| Molds | Alternaria alternata designed by EPA [41] | Forward primer | GGCGGGCTGGAACCTC |
| Reverse primer | GCAATTACAAAAGGTTTATGTTTGTCGTA | ||
| Probe | TTACAGCCTTGCTGAATTATTCACCCTTGTCTTT | ||
| Aspergillus fumigatus designed by EPA [41] | Forward primer | GCCCGCCGTTTCGAC | |
| Reverse primer | CCGTTGTTGAAAGTTTTAACTGATTAC | ||
| Probe | CCCGCCGAAGACCCCAACATG | ||
| Aspergillus versicolor designed by EPA [41] | Forward primer | CGGCGGGGAGCCCT | |
| Reverse primer | CCATTGTTGAAAGTTTTGAcTGATCTTA | ||
| Probe | AGACTGCATCACTCTCAGGCATGAAGTTCAG | ||
| Cladosporium sphaerospermum designed by EPA [41] | Forward primer | ACCGGCTGGGTCTTTCG | |
| Reverse primer | GGGGTTGTTTTACGGCGTG | ||
| Probe | CCCGCGGCACCCTTTAGCGA | ||
| Epicoccum nigrum designed by EPA [41] # | Forward primer | TTGTAGACTTCGGTCTGCTACCTCTT | |
| Reverse primer | TGCAACTGCAAAGGGTTTGAAT | ||
| Probe | CATGTCTTTTGAGTACCTTCGTTTCCTCGGC | ||
| Penicillium chrysogenum Modified from EPA [40] | Forward primer | TGCCTGTCCGAGCGTCATT | |
| Reverse primer | CCCCCGGGATCGGAG | ||
| Probe | CCAACACACAAGCCGTGCTTGAGG | ||
| Stachybotrys chartarum designed by EPA [41] | Forward primer | TCCCAAACCCTTATGTGAACC | |
| Reverse primer | GTTTGCCACTCAGAGAATACTGAAA | ||
| Probe | CTGCGCCCGGATCCAGGC | ||
| Trichoderma viride designed by EPA [41] # | Forward primer | CCCAAACCCAATGTGAACCA | |
| Reverse primer | TCCGCGAGGGGACTACAG | ||
| Probe | CCAAACTGTTGCCTCGGCGGG | ||
| Chaetomium globosum designed by EPA [41] # | Forward primer | CCGCAGGCCCTGAAAAG | |
| Reverse primer | CGCGGCGCGACCA | ||
| Probe | AGATGTATGCTACTACGCTCGGTGCGACAG | ||
| Yeasts | Cryptococcus spp. designed for this study # | Forward primer | CCTGCGGAAGGATCATTAATG |
| Reverse primer | GCACAGGTGTTATGGATATGATGTG | ||
| Probe | TTGACCGTCTGTCGAGCTTGCTCACA | ||
| Mites | Acarus siro Designed by our team [34] # | Forward primer | CGCAAACTGTGGTGCGAGTA |
| Reverse primer | GCTCCTTGGTCCGTGTTTCA | ||
| Probe | TCGGTCTCCACCCGACCCGTC | ||
| Dermatophagoïdes spp. Designed by our team [34] | Forward primer | TGTTGTGGTTAAAAAGCTCGTAGTTG | |
| Reverse primer | ATGCGATAATCTGCTCAGTATGACA | ||
| Probe | CAGCTCATGTATGGCGGTCCACCTG | ||
| Dermatophagoïdes farinea Designed by our team [34] # | Forward primer | CACACATTCAACCAGAGTGGTACTT | |
| Reverse primer | GGCTAACACTCCCCCTAGTTTAGA | ||
| Probe | CGCTTACGCGATCCTACGAGCCATT | ||
| Dermatophagoïdes pteronyssinus Designed by our team [34] # | Forward primer | CATCCAACCAGAGTGGTATTTCC | |
| Reverse primer | GCTATTGCGCATACTCCACCTA | ||
| Probe | TATGCAATCCTTCGGGCTATCCCATCA | ||
| Bacteria | Enterobacteriaceae designed by Sen and Asher [42] | Forward primer | GGCGGCAGGCCTAAC |
| Reverse primer | CAGGCAGTTTCCCAGACATTACT | ||
| Probe | AGCAAGCTCTCTGTGCTACCGCTCGA | ||
| Mycobacteria designed by Torvinen et al. [43] | Forward primer | GATGCAACGCGAAGAACCTT | |
| Reverse primer | TGCACCACCTGCACACAGG | ||
| Probe | CCTGGGTTTGACATGCACAGGACG | ||
| Streptomyces spp. designed by Rintala and Nevalainen [44] | Forward primer | GCCGATTGTGGTGAAGTGGA | |
| Reverse primer | GTACGGGCCGCCATGAAA | ||
| Probe | ATCCTATGCTGTCGAGAAAAGCCTCTAGCG | ||
| qPCR Targets | Number of Positive EDCs | Median Value (copy/µL) | Max Value (copy/µL) | |
|---|---|---|---|---|
| Fungi (molds and yeasts) | E. nigrum | 2763 | 76 | 39106 |
| A. alternata | 2752 | 66 | 32634 | |
| P. chrysogenum | 2660 | 9 | 19169 | |
| C. sphaerospermum | 2195 | 8 | 12162 | |
| A. versicolor | 2307 | 2 | 5671 | |
| A. fumigatus | 1468 | 0 | 5138 | |
| T. viride | 1390 | 0 | 1536 | |
| S. chartarum | 848 | 0 | 333 | |
| Cryptococcus spp. | 2314 | <1 | 7 | |
| Bacteria | Enterobacteriaceae | 3086 | 134 | 78163 |
| Mycobacteria | 3053 | 235 | 12511 | |
| Streptomyces | 2879 | 51 | 5887 | |
| Dust mites | Dermatophagoïdes spp. | 1505 | <1 | 97673 |
| D. pteronyssinus | 1908 | 0 | 9657 | |
| D. farinae | 2192 | <1 | 1330 | |
| A. siro | 2300 | <1 | 348 | |
| qPCR Targets | p | edf | F | adjR2 | |
|---|---|---|---|---|---|
| Molds | A. alternata | <0.001 * | 2.954 | 162.6 | 0.12 |
| A. fumigatus | <0.001 * | 2.7 | 10.54 | 0.00909 | |
| E. nigrum | <0.001 * | 2.345 | 10.85 | 0.0069 | |
| C. sphaerospermum | <0.001 * | 2 | 59.59 | 0.0418 | |
| P. chrysogenum | 0.028 * | 2.738 | 4.472 | 0.00212 | |
| S. chartarum | 0.122 | 2.853 | 1.808 | 0.00119 | |
| A. versicolor | 0.126 | 2 | 2.071 | 0.000853 | |
| T. viride | 0.794 | 2 | 0.231 | −0.000452 | |
| Yeasts | Cryptococcus spp. | <0.001 * | 2.852 | 42.88 | 0.0391 |
| Dust mites | Dermatophagoïdes spp. | <0.001 * | 2.122 | 39.51 | 0.0286 |
| D. pteronyssinus | <0.001 * | 2 | 15.77 | 0.00918 | |
| D. farinea | 0.672 | 2.248 | 0.44 | −0.000307 | |
| A. siro | 0.779 | 2 | 0.249 | −0.000525 | |
| Bacteria | Enterobacteriaceae | 0.12 | 2.25 | 1.823 | 0.00139 |
| Mycobacteria | 0.009 * | 2.694 | 3.544 | 0.00456 | |
| Streptomyces | <0.001 * | 2 | 11.71 | 0.00851 | |
| C. sphaerospermum | Dermatophagoïdes spp. | Streptomyces | E. nigrum | Mycobacteria | A. fumigatus | P. chrysogenum | Cryptococcus spp. | A. alternata | |
|---|---|---|---|---|---|---|---|---|---|
| Annual average temperature | + | − | + | − | + | ||||
| Number of days temperature < −5 °C | − | − | − | + | + | ||||
| Number of days temperature >30 °C | + | + | + | ||||||
| Annual temperature range * | − | − | + | + | |||||
| Total annual precipitation | + | − | − | ||||||
| Number of precipitation days in January | + | + | + | + | + | − | − | ||
| Number of precipitation days in July | − | ||||||||
| Ratio between autumn ** and July precipitations | − | − | − | − | − | ||||
| Urbanized areas | − | − | − | − | + | − | − | ||
| Industrial or commercial areas | + | − | − | ||||||
| Mines, landfills, and construction sites | + | ||||||||
| Artificial, non-agricultural green spaces | + | − | |||||||
| Arable land | + | − | − | − | − | − | − | ||
| Permanent crops | + | − | − | − | − | − | − | ||
| Prairies | + | − | − | − | − | + | − | − | − |
| Heterogeneous agricultural areas | − | − | − | + | − | − | − | ||
| Forests | + | − | − | − | + | − | − | ||
| Scrub and/or herbaceous vegetation associations | + | − | − | + | |||||
| Open spaces, with little or no vegetation | − | − | + | − | − | ||||
| Inland wetlands | + | − | |||||||
| Inland waters | + | − | − | − | − | − | |||
| Maritime waters | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocchi, S.; Reboux, G.; Scherer, E.; Laboissière, A.; Zaros, C.; Rouzet, A.; Valot, B.; Khan, S.; Dufourg, M.-N.; Leynaert, B.; et al. Indoor Microbiome: Quantification of Exposure and Association with Geographical Location, Meteorological Factors, and Land Use in France. Microorganisms 2020, 8, 341. https://doi.org/10.3390/microorganisms8030341
Rocchi S, Reboux G, Scherer E, Laboissière A, Zaros C, Rouzet A, Valot B, Khan S, Dufourg M-N, Leynaert B, et al. Indoor Microbiome: Quantification of Exposure and Association with Geographical Location, Meteorological Factors, and Land Use in France. Microorganisms. 2020; 8(3):341. https://doi.org/10.3390/microorganisms8030341
Chicago/Turabian StyleRocchi, Steffi, Gabriel Reboux, Emeline Scherer, Audrey Laboissière, Cécile Zaros, Adeline Rouzet, Benoit Valot, Sadia Khan, Marie-Noëlle Dufourg, Bénédicte Leynaert, and et al. 2020. "Indoor Microbiome: Quantification of Exposure and Association with Geographical Location, Meteorological Factors, and Land Use in France" Microorganisms 8, no. 3: 341. https://doi.org/10.3390/microorganisms8030341
APA StyleRocchi, S., Reboux, G., Scherer, E., Laboissière, A., Zaros, C., Rouzet, A., Valot, B., Khan, S., Dufourg, M.-N., Leynaert, B., Raherison, C., & Millon, L. (2020). Indoor Microbiome: Quantification of Exposure and Association with Geographical Location, Meteorological Factors, and Land Use in France. Microorganisms, 8(3), 341. https://doi.org/10.3390/microorganisms8030341







