Pesticides Decrease Bacterial Diversity and Abundance of Irrigated Rice Fields
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling Procedure for Soil
2.3. Bacteria Culture
2.4. DNA Extraction and 16S Ribosomal RNA (rRNA) Gene Sequencing
2.5. Data Analysis
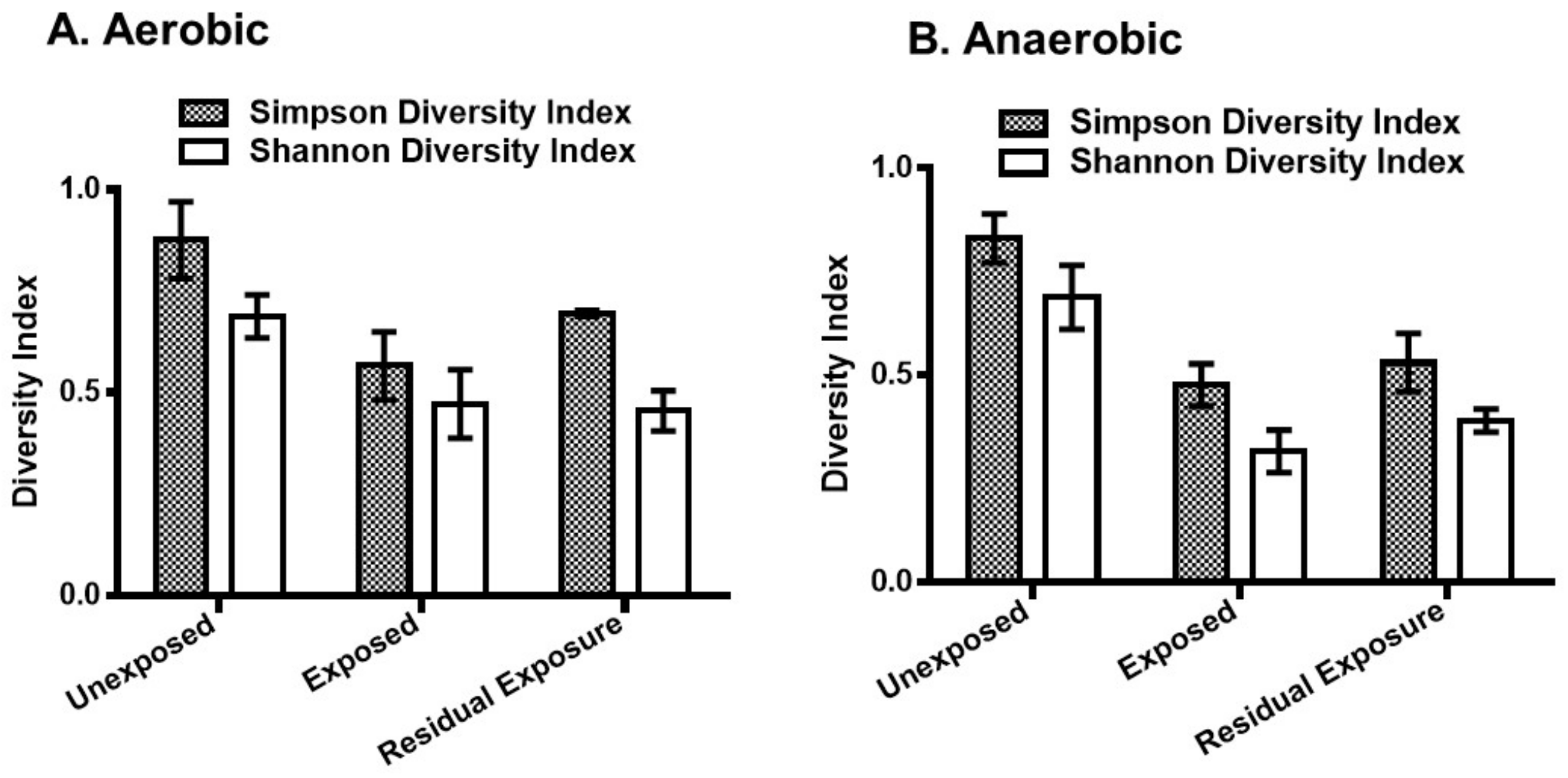
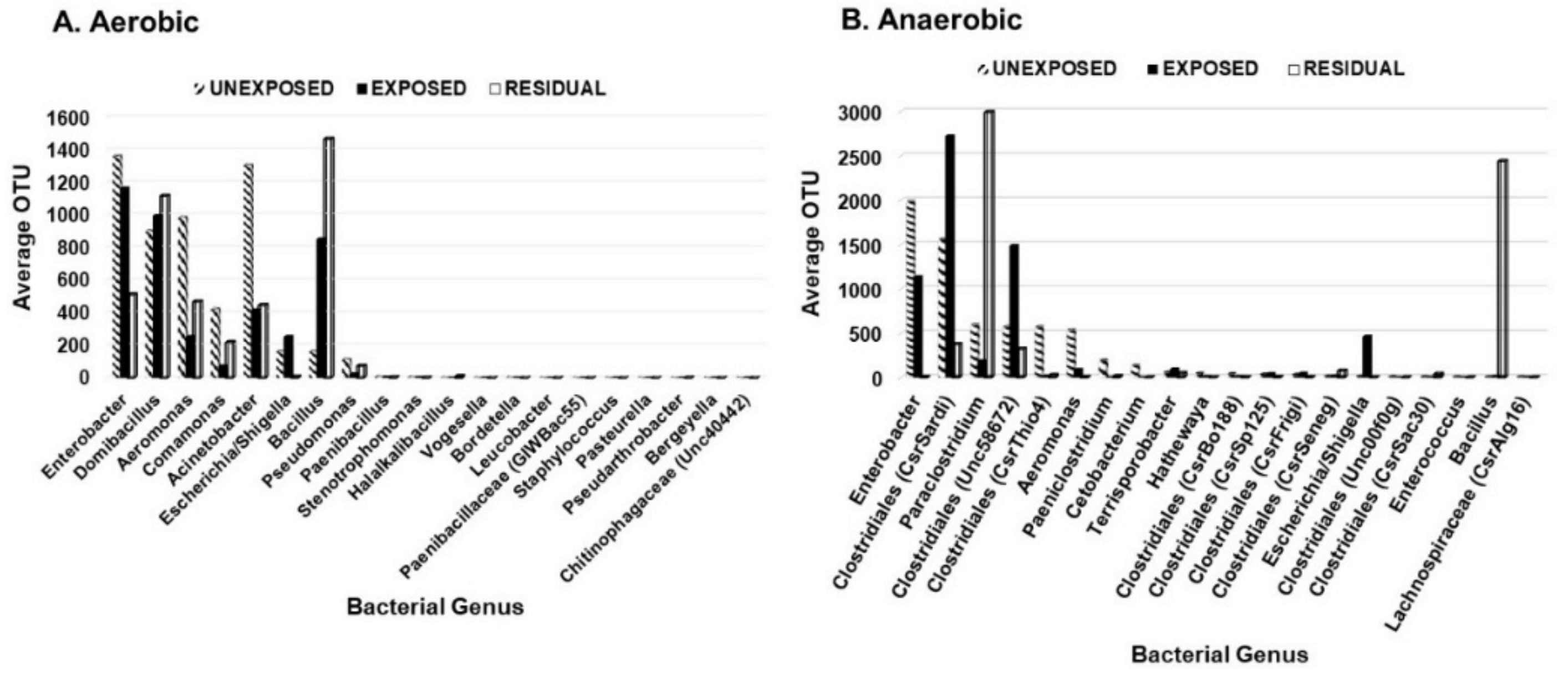
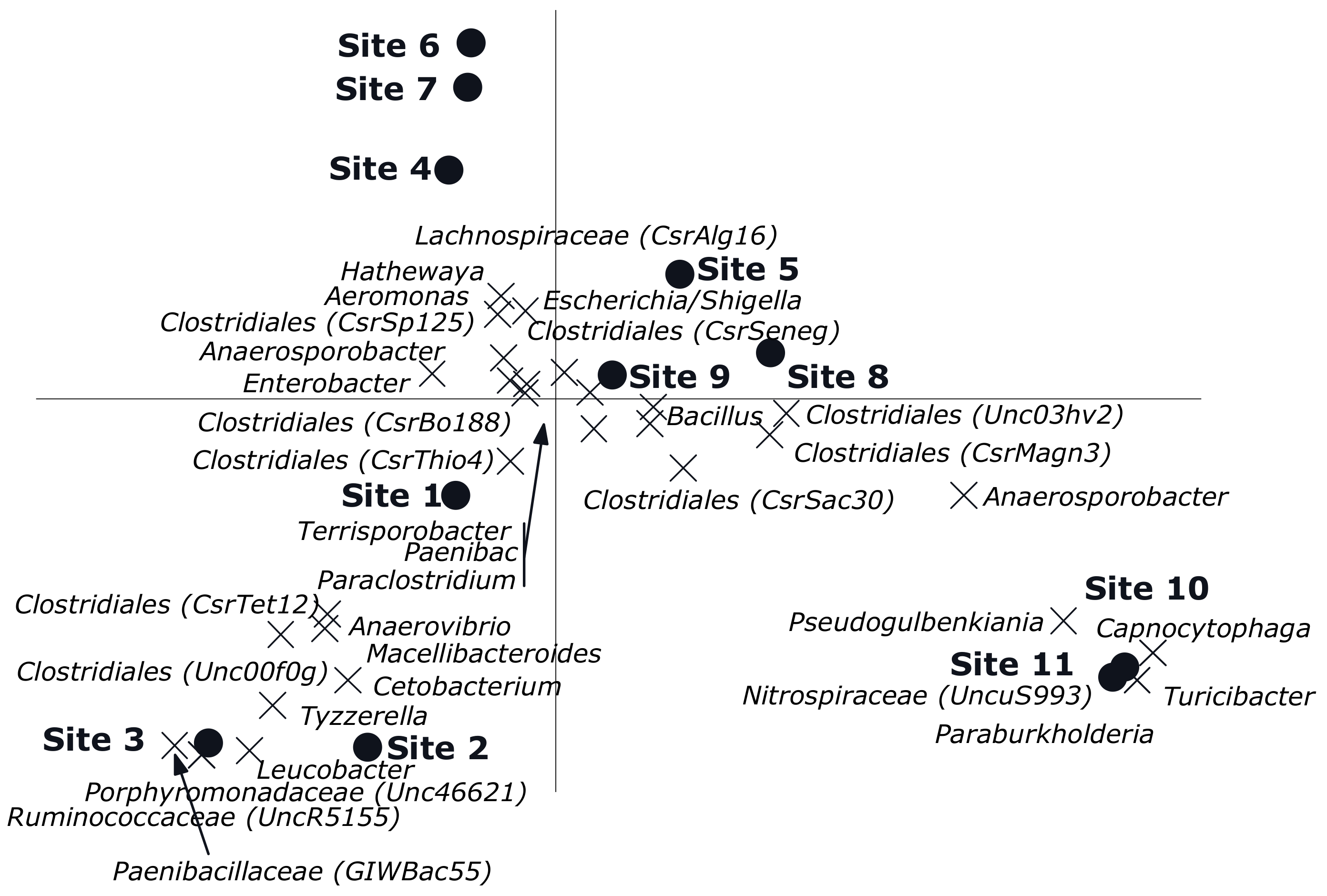
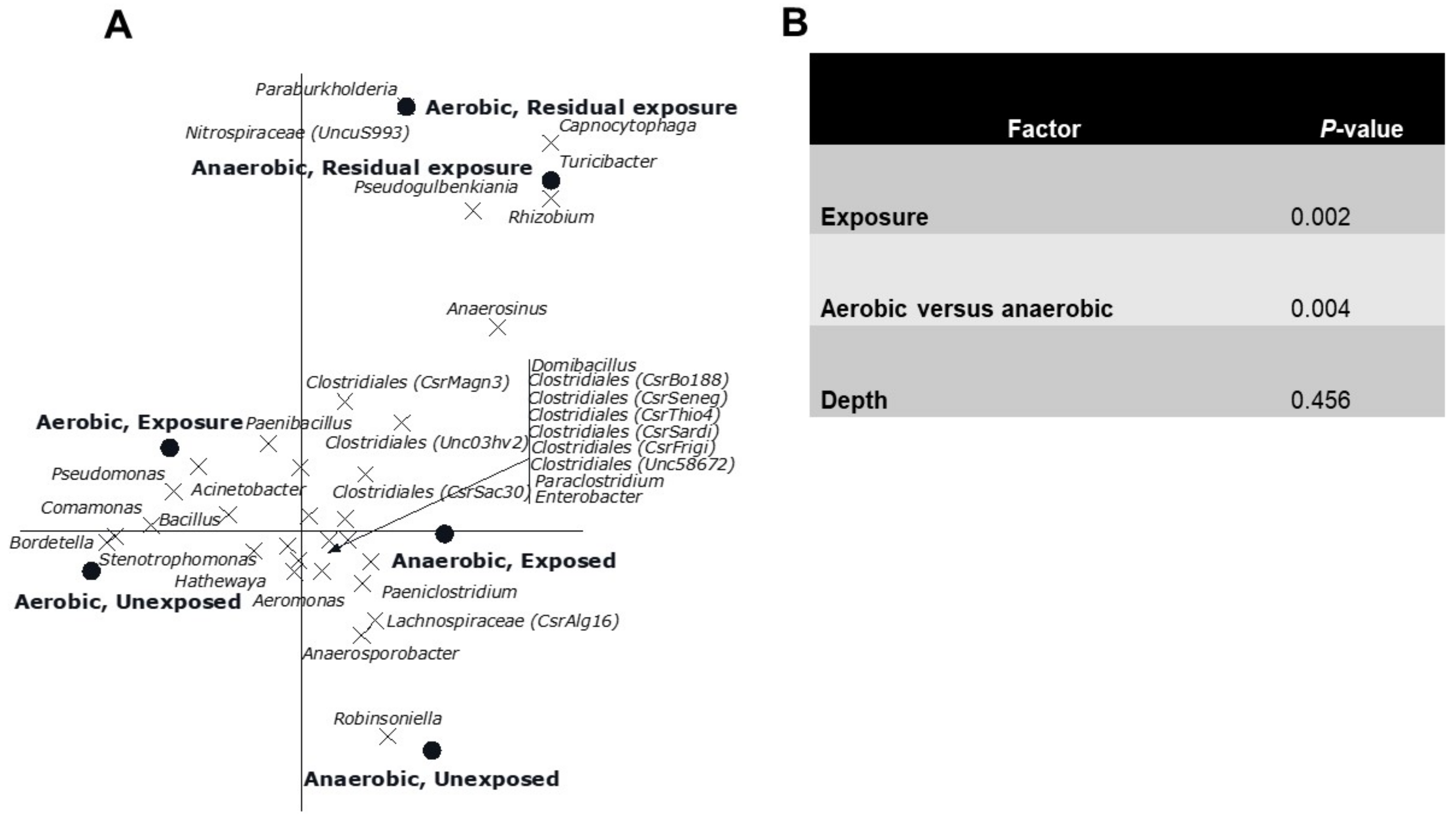
3. Results
3.1. Effect of Pesticides on Soil Bacterial Abundance and Diversity
3.2. Multivariate Analyses
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- DeLorenzo, M.E.; Scott, G.I.; Ross, P.E. Toxicity of pesticides to aquatic microorganisms: A review. Environ. Toxicol. Chem. 2001, 20, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Cao, Q.; Hesham, A.B.; Xia, Y.; He, J. Soil enzymatic activities and microbial community structure with different application rates of Cd and Pb. J. Environ. Sci. 2007, 19, 834–840. [Google Scholar] [CrossRef]
- Khan, S.; Hesham, A.L.; Qiao, M.; Rehman, S.; He, J.Z. Effects of Cd and Pb on soil microbial community structure and activities. Environ. Sci. Pollut. Res. Int. 2010, 17, 288–296. [Google Scholar] [CrossRef] [PubMed]
- United Nations Department of Economic and Social Affairs. The World Population Prospects: 2015 Revision. Available online: https://www.un.org/en/development/desa/publications/world-population-prospects-2015-revision.html (accessed on 12 July 2019).
- Munoz-Leoz, B.; Garbisu, C.; Charcosset, J.Y.; Sanchez-Perez, J.M.; Antiguedad, I.; Ruiz-Romera, E. Non-target effects of three formulated pesticides on microbially-mediated processes in a clay-loam soil. Sci. Total Environ. 2013, 449, 345–354. [Google Scholar] [CrossRef]
- Prado, A.G.; Airoldi, C. Toxic effect caused on microflora of soil by pesticide picloram application. J. Environ. Monit. 2001, 3, 394–397. [Google Scholar] [CrossRef]
- De, A.; Bose, R.; Kumar, A.; Mozumdar, S. Worldwide Pesticide Use. In Targeted Delivery of Pesticides Using Biodegradable Polymeric Nanoparticles; Springer: New Delhi, India, 2014. [Google Scholar]
- Atwood, D.; Paisley-Jones, C. Pesticides Industry Sales and Usage 2008–2012. Available online: https://www.epa.gov/sites/production/files/2017-01/documents/pesticides-industry-sales-usage-2016_0.pdf (accessed on 8 September 2018).
- Geiger, F.; Bengtsson, J.; Berendse, F.; Weisser, W.W.; Emmerson, M.; Morales, M.B.; Ceryngier, P.; Liira, J.; Tscharntke, T.; Winqvist, C.; et al. Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl. Ecol. 2010, 11, 97–105. [Google Scholar] [CrossRef]
- Zhao, C.; Xie, H.; Zhang, J.; Xu, J.; Liang, S. Spatial distribution of organochlorine pesticides (OCPs) and effect of soil characters: A case study of a pesticide producing factory. Chemosphere 2013, 90, 2381–2387. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition-Current Knowledge and Future Directions. Front. Plant. Sci. 2017, 8, 1617. [Google Scholar] [CrossRef]
- Bohlen, P.J.; Edwards, C.A.; Zhang, Q.; Parmelee, R.W.; Allen, M. Indirect effects of earthworms on microbial assimilation of labile carbon. Appl. Soil Ecol. 2002, 20, 255–261. [Google Scholar] [CrossRef]
- Kent, A.D.; Triplett, E.W. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 2002, 56, 211–236. [Google Scholar] [CrossRef]
- Abdullah, A.R.; Bajet, C.M.; Matin, M.A.; Nhan, D.D.; Sulaiman, A.H. Ecotoxicology of pesticides in the tropical paddy field ecosystem. Environ. Toxicol. Chem. 1997, 16, 59–70. [Google Scholar] [CrossRef]
- Tejada, A.W. Pesticide residues in foods and the environment as a consequence of crop protection. Philipp. J. Agric. 1995, 78, 63–79. [Google Scholar]
- Sally, H.; Abernethy, C.L. Private irrigation in Sub-Saharan Africa: Regional Seminar on Private Sector Participation and Irrigation Expansion in Sub-Saharan Africa, Accra, Ghana, 22–26 October 2001. Available online: https://cgspace.cgiar.org/handle/10568/38800 (accessed on 8 September 2018).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Mansbach, J.M.; Ajami, N.J.; Espinola, J.A.; Henke, D.M.; Petrosino, J.F.; Piedra, P.A.; Shaw, C.A.; Sullivan, A.F.; Camargo, C.A., Jr. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur. Respir. J. 2016, 48, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Meng, R.; He, L.S.; Guo, L.G.; Xi, B.D.; Li, Z.Q.; Shu, J.M.; Diao, X.J.; Li, B.C. Canonical correspondence analysis between phytoplankton community and environmental factors in macrophtic lakes of the middle and lower reaches of Yangtze River. Huan Jing Ke Xue 2013, 34, 2588–2596. [Google Scholar] [PubMed]
- Braak, C.J.F.; Smilauer, P. CANOCO Reference Manual and CanoDraw for Windows User’s Guide: Software for Canonical Community Ordination, version 4.5; Microcomputer Power: Ithaca, NY, USA, 2002. [Google Scholar]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef]
- Inbar, J.; Chet, I. Evidence that chitinase produced by Aeromonas caviae is involved in the biological control of soil-borne plant pathogens by this bacterium. Soil Biol. Biochem. 1991, 23, 973–978. [Google Scholar] [CrossRef]
- Jeong, H.; Jeong, D.E.; Kim, S.H.; Song, G.C.; Park, S.Y.; Ryu, C.M.; Park, S.H.; Choi, S.K. Draft genome sequence of the plant growth-promoting bacterium Bacillus siamensis KCTC 13613T. J. Bacteriol. 2012, 194, 4148–4149. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, L.; Wang, G.; Zheng, S. Domibacillus antri sp. nov., isolated from the soil of a cave. Int. J. Syst. Evol. Microbiol. 2016, 66, 2502–2508. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Q.; Liao, S.; Zhi, H.; Xing, D.; Xiao, Y.; Yang, Q. Characterization and sequence analysis of potential biofertilizer and biocontrol agent Bacillus subtilis strain SEM-9 from silkworm excrement. Can. J. Microbiol. 2019, 65, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Fira, D.; Dimkic, I.; Beric, T.; Lozo, J.; Stankovic, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Soumana, I.H.; Linz, B.; Harvill, E.T. Environmental Origin of the Genus Bordetella. Front. Microbiol. 2017, 8, 28. [Google Scholar]
- Rosenberg, E.T. The family Chitinophagaceae. In The Prokaryotes: Applied Bacteriology and Biotechnology; Springer: Berlin, Germany, 2014; pp. 493–495. [Google Scholar]
- Willems, A.; De Vos, P. Comamonas. In The Prokaryotes; Springer: New York, NY, USA, 2006. [Google Scholar]
- Garcia-Gonzalez, T.; Saenz-Hidalgo, H.K.; Silva-Rojas, H.V.; Morales-Nieto, C.; Vancheva, T.; Koebnik, R.; Avila-Quezada, G.D. Enterobacter cloacae, an Emerging Plant-Pathogenic Bacterium Affecting Chili Pepper Seedlings. Plant. Pathol. J. 2018, 34, 1–10. [Google Scholar]
- Zhu, B.; Wang, G.; Xie, G.; Zhou, Q.; Zhao, M.; Praphat, K.; Li, B.; Tian, W. Enterobacter spp.: A new evidence causing bacterial wilt on mulberry. Sci. China Life Sci. 2010, 53, 292–300. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Lee, J.S.; Saravanan, V.S.; Lee, K.C.; Santhanakrishnan, P. Enterobacter arachidis sp. nov., a plant-growth-promoting diazotrophic bacterium isolated from rhizosphere soil of groundnut. Int. J. Syst. Evol. Microbiol. 2010, 60, 1559–1564. [Google Scholar] [CrossRef]
- Ge, S.; Ai, W.; Dong, X. High-Quality Draft Genome Sequence of Leucobacter sp. Strain G161, a Distinct and Effective Chromium Reducer. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microbial. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Busse, H.J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 9–37. [Google Scholar] [PubMed]
- Peix, A.; Ramirez-Bahena, M.H.; Velazquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infection, genetics and evolution. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2009, 9, 1132–1147. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.J.; Fonseca, S.; Meena, R.M.; Ramaiah, N. Improved Sprouting and Growth of Mung Plants in Chromate Contaminated Soils Treated with Marine Strains of Staphylococcus Species. Indian J. Microbiol. 2017, 57, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Reche, M.H.L.R.; Fiuza, L.M. Bacterial diversity in rice-field water in Rio Grande do Sul. Braz. J. Microbiol. 2005, 36, 253–257. [Google Scholar] [CrossRef]
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S.; Rehman, A.; Chauhan, P.S. Environmental Escherichia coli occur as natural plant growth-promoting soil bacterium. Arch. Microbiol. 2010, 192, 185–193. [Google Scholar] [CrossRef]
- Echigo, A.; Fukushima, T.; Mizuki, T.; Kamekura, M.; Usami, R. Halalkalibacillus halophilus gen. nov., sp. nov., a novel moderately halophilic and alkaliphilic bacterium isolated from a non-saline soil sample in Japan. Int. J. Syst. Evol. Microbiol. 2007, 57, 1081–1085. [Google Scholar] [CrossRef]
- Sheu, S.Y.; Chen, J.C.; Young, C.C.; Chen, W.M. Vogesella fluminis sp. nov., isolated from a freshwater river, and emended description of the genus Vogesella. Int. J. Syst. Evol. Microbiol. 2013, 63, 3043–3049. [Google Scholar] [CrossRef]
- Subhash, Y.; Tushar, L.; Sasikala, C.; Ramana Ch, V. Vogesella alkaliphila sp. nov., isolated from an alkaline soil, and emended description of the genus Vogesella. Int. J. Syst. Evol. Microbiol. 2013, 63, 2338–2343. [Google Scholar] [CrossRef]
- Backstrand, J.M.; Botzler, R.G. Survival of Pasteurella multocida in soil and water in an area where avian cholera is enzootic. J. Wildl. Dis. 1986, 22, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Hugo, C.J.; Bruun, B.; Jooste, P.J. The Genera Bergeyella and Weeksella. In The Prokaryotes; Springer: New York, NY, USA, 2006. [Google Scholar]
- Sasi Jyothsna, T.S.; Tushar, L.; Sasikala, C.; Ramana, C.V. Paraclostridium benzoelyticum gen. nov., sp. nov., isolated from marine sediment and reclassification of Clostridium bifermentans as Paraclostridium bifermentans comb. nov. Proposal of a new genus Paeniclostridium gen. nov. to accommodate Clostridium sordellii and Clostridium ghonii. Int. J. Syst. Evol. Microbiol. 2016, 66, 1268–1274. [Google Scholar] [PubMed]
- Lawson, P.A.; Rainey, F.A. Proposal to restrict the genus Clostridium Prazmowski to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 2016, 66, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Khalifa, A.Y.; Alsyeeh, A.M.; Almalki, M.A.; Saleh, F.A. Characterization of the plant growth promoting bacterium, Enterobacter cloacae MSR1, isolated from roots of non-nodulating Medicago sativa. Saudi J. Biol. Sci. 2016, 23, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.C.; Fegan, N.; Fegan, M.; Stirling, G.R. Stenotrophomonas and Lysobacter: Ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 2010, 108, 756–770. [Google Scholar] [CrossRef]
- Lu, Z.; Sang, L.; Li, Z.; Min, H. Catalase and superoxide dismutase activities in a Stenotrophomonas maltophilia WZ2 resistant to herbicide pollution. Ecotoxicol. Environ. Saf. 2009, 72, 136–143. [Google Scholar] [CrossRef]
- Saeki, M.T.K. Effect of bensulfuron-methyl (a sulfonylurea herbicide) on the soil bacterial community of a paddy soil microcosm. Biol. Fertil. Soils 2004, 40, 110–118. [Google Scholar] [CrossRef]
- Kumar, U.; Behera, S.; Saha, S.; Das, D.; Guru, P.K.; Kaviraj, M.; Munda, S.; Adak, T.; Nayak, A.K. Non-target effect of bispyribac sodium on soil microbial community in paddy soil. Ecotoxicol. Environ. Saf. 2019, 189, 110019. [Google Scholar] [CrossRef]
- Boivin, A.; Amellal, S.; Schiavon, M.; van Genuchten, M.T. 2,4-Dichlorophenoxyacetic acid (2,4-D) sorption and degradation dynamics in three agricultural soils. Environ. Pollut. 2005, 138, 92–99. [Google Scholar] [CrossRef]
- Gardner, J.M.; Feldman, A.W.; Zablotowicz, R.M. Identity and behavior of xylem-residing bacteria in rough lemon roots of Florida citrus trees. Appl. Environ. Microbiol. 1982, 43, 1335–1342. [Google Scholar] [CrossRef]
- Gilani, R.A.; Rafique, M.; Rehman, A.; Munis, M.F.; Rehman, S.U.; Chaudhary, H.J. Biodegradation of chlorpyrifos by bacterial genus Pseudomonas. J. Basic Microbiol. 2016, 56, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Seenivasan, S.; Premkumar, R. Biodegradation of propiconazole by Pseudomonas putida isolated from tea rhizosphere. Plant. Soil Environ. 2009, 55, 196–201. [Google Scholar] [CrossRef]
- Hughes, S.M.; Cooper, D.G. Biodegradation of phenol using the self-cycling fermentation (SCF) process. Biotechnol. Bioeng. 1996, 51, 112–119. [Google Scholar] [CrossRef]

| Pesticide Name | Active Ingredient Conc. | Application Rate/ha | Target Pest/Disease | Application Method | Frequency |
|---|---|---|---|---|---|
| Condex | Bensylfuron methyl (30%) | 0.42 Kg | Selective herbicide | spraying | 3 |
| Kilsect 2.5 EC | Lambda Cyhalothrin (25 g/L) | 1.0 L | Grasshoppers, Worms, Thrips | spraying | 5 |
| Bounty/Nakitse | Bispyribac sodium (400 g/L) | 62.5-75 Ml | Selective herbicide | spraying | 3 |
| Nativo | Tebuconazole (200 g/L) | 1.0 L | Blast | spraying | 5 |
| Trifloxystrobin (100 g/L) | |||||
| Orizo plus | Propanil (360 g/L) | 2.0 L | Selective herbicide | spraying | 3 |
| 2,4,D amine (200 g/L) | |||||
| Dursban/Sunpyrifos | Chlorpyrifos (480 g/L) | 1.0 L | Grasshoppers, Worms, Thrips | spraying | 5 |
| Allogator | Pendimethalin (400 g/L) | 3.0 L | Selective herbicide | spraying | 3 |
| Genus | Respiration | Habitat | Role in Soil | Potential Pathogen | Open Literature References | * Zone of Highest OTU |
|---|---|---|---|---|---|---|
| Acinetobacter | Obligate aerobes | Soil, water | Mineralization | Pathogenic | [25] | Residual |
| Aeromonas | Facultative anaerobes | Soil, water | Microbial equilibrium | Pathogenic | [26] | Unexposed |
| Bacillus | Obligate aerobes Facultative anaerobes | Ubiquitous | Plant protection from plants and insects | Pathogenic | [27,28,29,30] | Residual |
| Bordetella | Aerobes | Soil, water, sediment, plants | Possible degradation of organic compounds | Pathogenic | [31] | Unexposed |
| Chitinophagaceae (Unc40442) | Facultative anaerobes | Soil | Chitin degradation | Not Pathogenic | [32] | Exposed |
| Comamonas | Facultative anaerobes, Aerobes | Soil, water | Possible degradation of organic compounds | Pathogenic | [33] | Unexposed |
| Enterobacter | Facultative anaerobes | Ubiquitous | Plant growth regulator | Pathogenic | [34,35,36] | Unexposed |
| Leucobacter | Aerobes | Soil, sediment, water | Possible bioremediation | Pathogenic | [37] | Unexposed |
| Paenibacillaceae (GIWBac55) | Facultative anaerobes | Soil, water, plants | Nitrogen fixation, plant growth, plant protection from microbes and insects | Pathogenic | [38] | Unexposed |
| Paenibacillus | Facultative anaerobes | Soil, water, plants | Nitrogen fixation, plant growth, plant protection from microbes and insects | Pathogenic | [38] | Unexposed |
| Pseudarthrobacter | Obligate aerobes | Soil | Possible biodegradation of organic compounds | Pathogenic | [39] | Residual |
| Pseudomonas | Facultative anaerobes, aerobes | Ubiquitous, Soil, water, plants, rhizosphere | Biocontrol, Plant Growth promotion, nutrient mobilization, soil bioremediation | Pathogenic | [40] | Residual |
| Staphylococcus | Facultative anaerobes | Ubiquitous, Soil, water | Possible degradation of organic compounds, Plant Growth promotion | Pathogenic | [41] | Unexposed |
| Stenotrophomonas | Aerobes | Soil | Plant protection from plants and insects | Pathogenic | [33,42] | Unexposed |
| Escherichia/Shigella | Aerobes | Ubiquitous | Plant growth promoter | Pathogenic | [43,44] | Exposed |
| Domibacillus | Aerobes | Ubiquitous, Soil, water | Unknown | Unknown | [28] | Residual |
| Halalkalibacillus | Aerobes | Soil, water | Unknown | Pathogenic | [45] | Exposed |
| Vogesella | Aerobes | Soil, water sediments | Unknown | Not Pathogenic | [46,47] | Exposed |
| Pasteurella | Facultative Aerobes | Soil, water | Biocontrol | Pathogenic | [48] | Exposed |
| Bergeyella | Aerobes | Soil, water | Unknown | Pathogenic | [49] | Exposed |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onwona-Kwakye, M.; Plants-Paris, K.; Keita, K.; Lee, J.; Brink, P.J.V.d.; Hogarh, J.N.; Darkoh, C. Pesticides Decrease Bacterial Diversity and Abundance of Irrigated Rice Fields. Microorganisms 2020, 8, 318. https://doi.org/10.3390/microorganisms8030318
Onwona-Kwakye M, Plants-Paris K, Keita K, Lee J, Brink PJVd, Hogarh JN, Darkoh C. Pesticides Decrease Bacterial Diversity and Abundance of Irrigated Rice Fields. Microorganisms. 2020; 8(3):318. https://doi.org/10.3390/microorganisms8030318
Chicago/Turabian StyleOnwona-Kwakye, Michael, Kimberly Plants-Paris, Kadiatou Keita, Jessica Lee, Paul J. Van den Brink, Jonathan N. Hogarh, and Charles Darkoh. 2020. "Pesticides Decrease Bacterial Diversity and Abundance of Irrigated Rice Fields" Microorganisms 8, no. 3: 318. https://doi.org/10.3390/microorganisms8030318
APA StyleOnwona-Kwakye, M., Plants-Paris, K., Keita, K., Lee, J., Brink, P. J. V. d., Hogarh, J. N., & Darkoh, C. (2020). Pesticides Decrease Bacterial Diversity and Abundance of Irrigated Rice Fields. Microorganisms, 8(3), 318. https://doi.org/10.3390/microorganisms8030318





