Molecular Mechanisms Underpinning Aggregation in Acidiphilium sp. C61 Isolated from Iron-Rich Pelagic Aggregates
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions, and Microscopic Characterization of Aggregate Formation in Acidophilic Bacteria
2.2. Genomic DNA Extraction and Whole Genome Sequencing of Acidiphilium sp. C61
2.3. Genome Assembly and Annotation
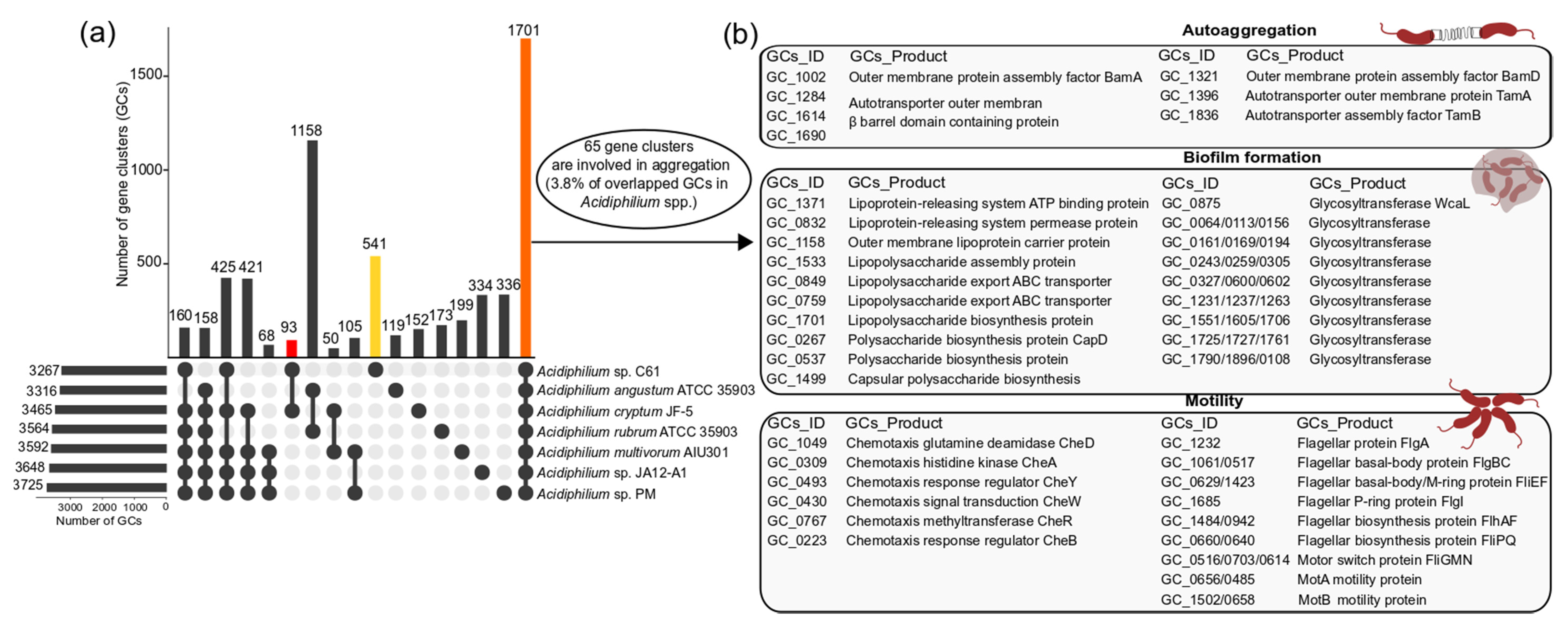
2.4. Pangenomic Analysis
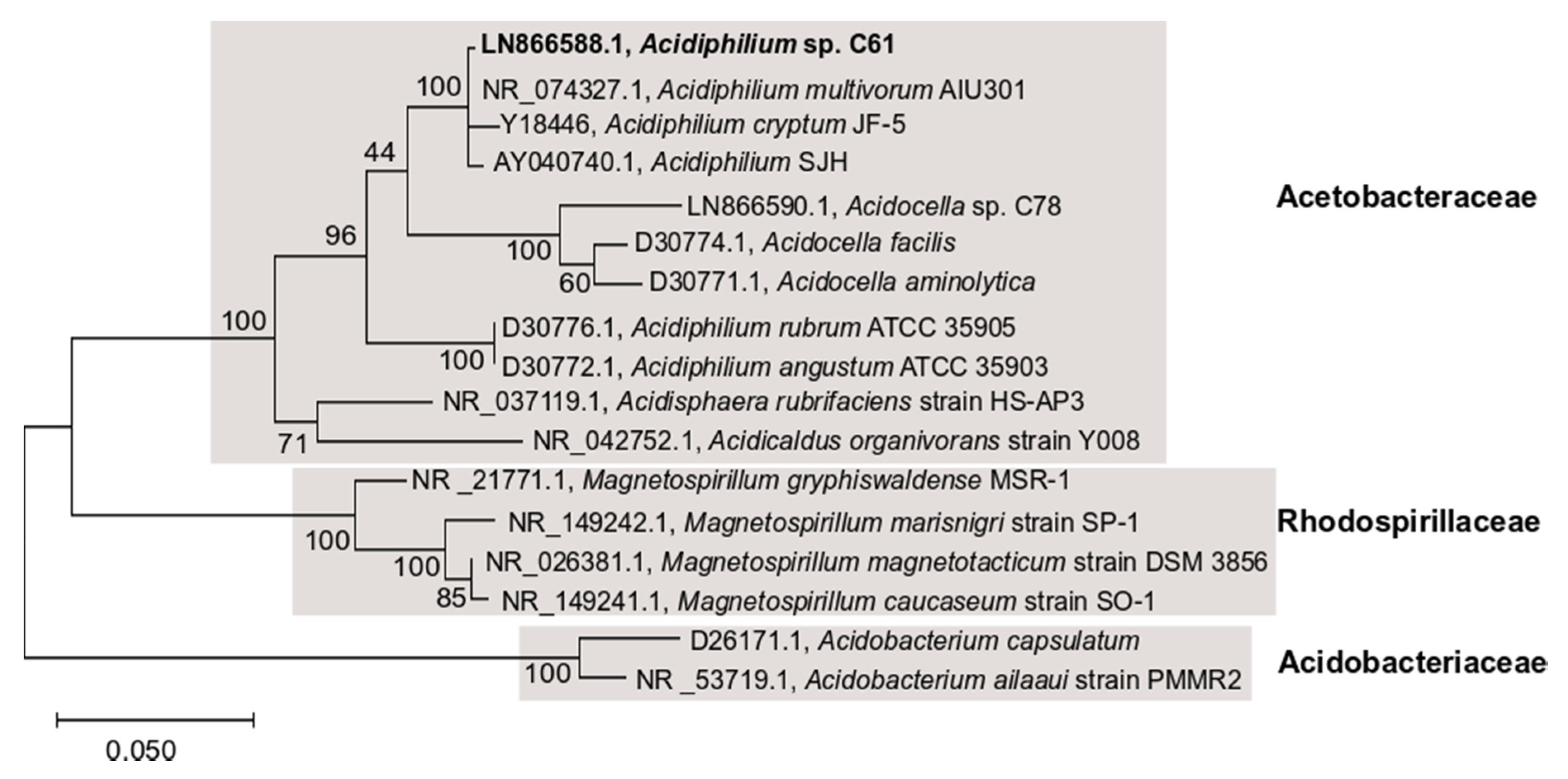
2.5. 16S rRNA Gene Phylogenetic Analysis
2.6. RNA Extraction
2.7. RNA-seq Library Preparation and Sequencing
2.8. RNA-seq Data Pre-Processing
2.9. Differential Gene Expression Analysis
2.10. Quantitative PCR
2.11. Quantification of eDNA Concentrations
2.12. Data Deposition
3. Results
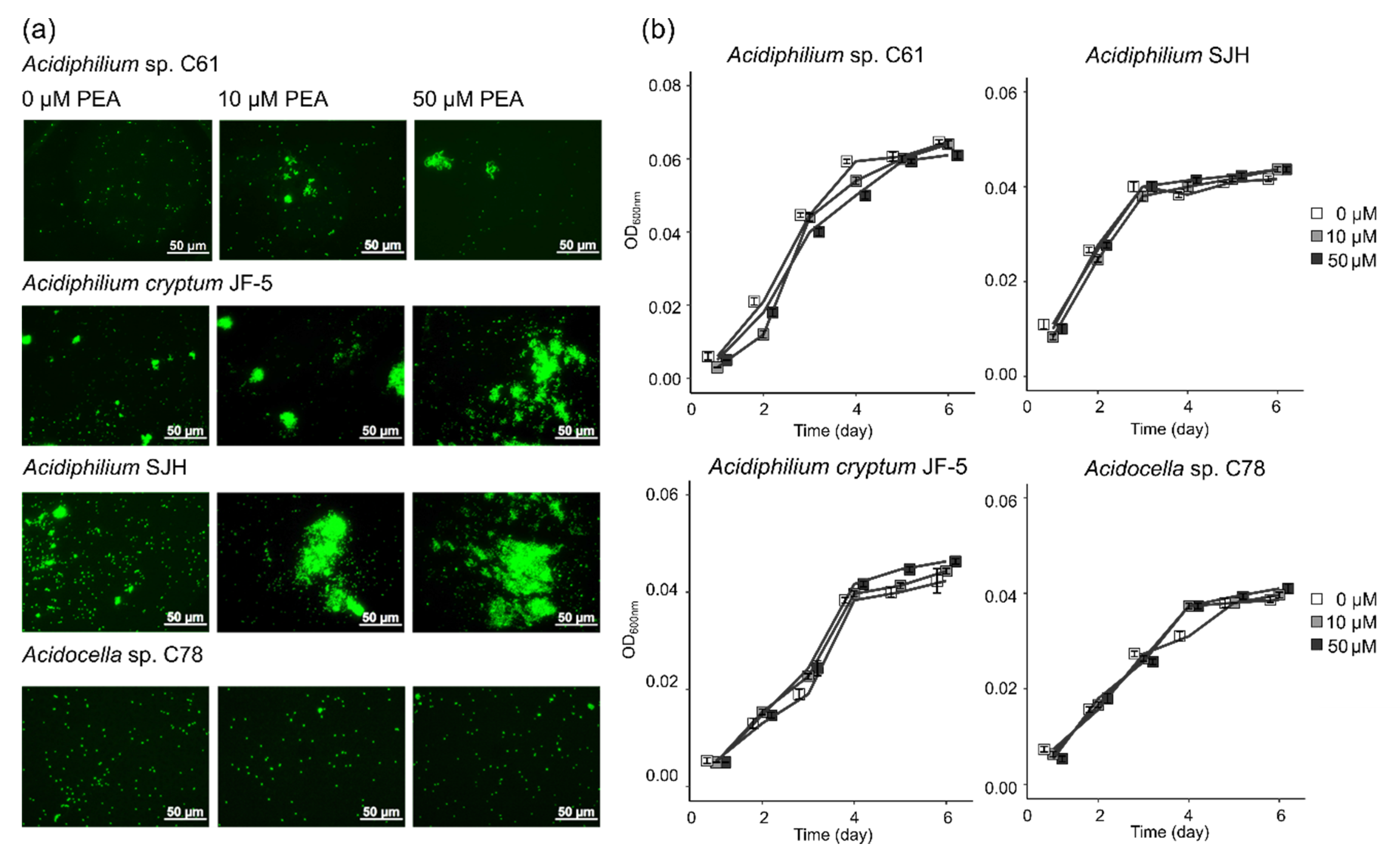
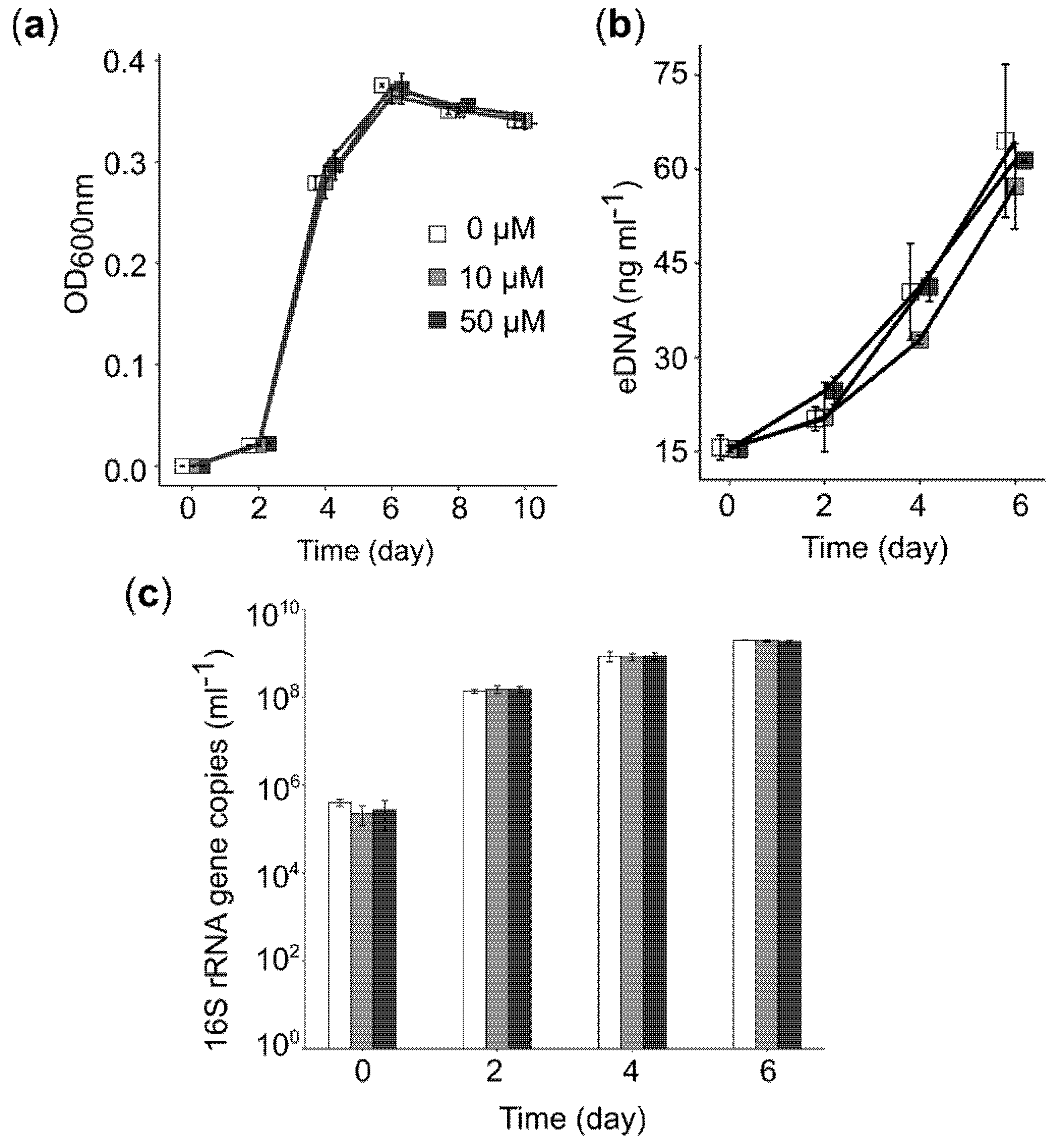
3.1. Effect of PEA on Phenotype and Growth of Iron Snow Key Players
3.2. Genome Sequencing, Assembly and Annotation of Acidiphilium sp. C61
3.3. Potential Mechanisms of Aggregate Formation in Acidiphilium sp. C61
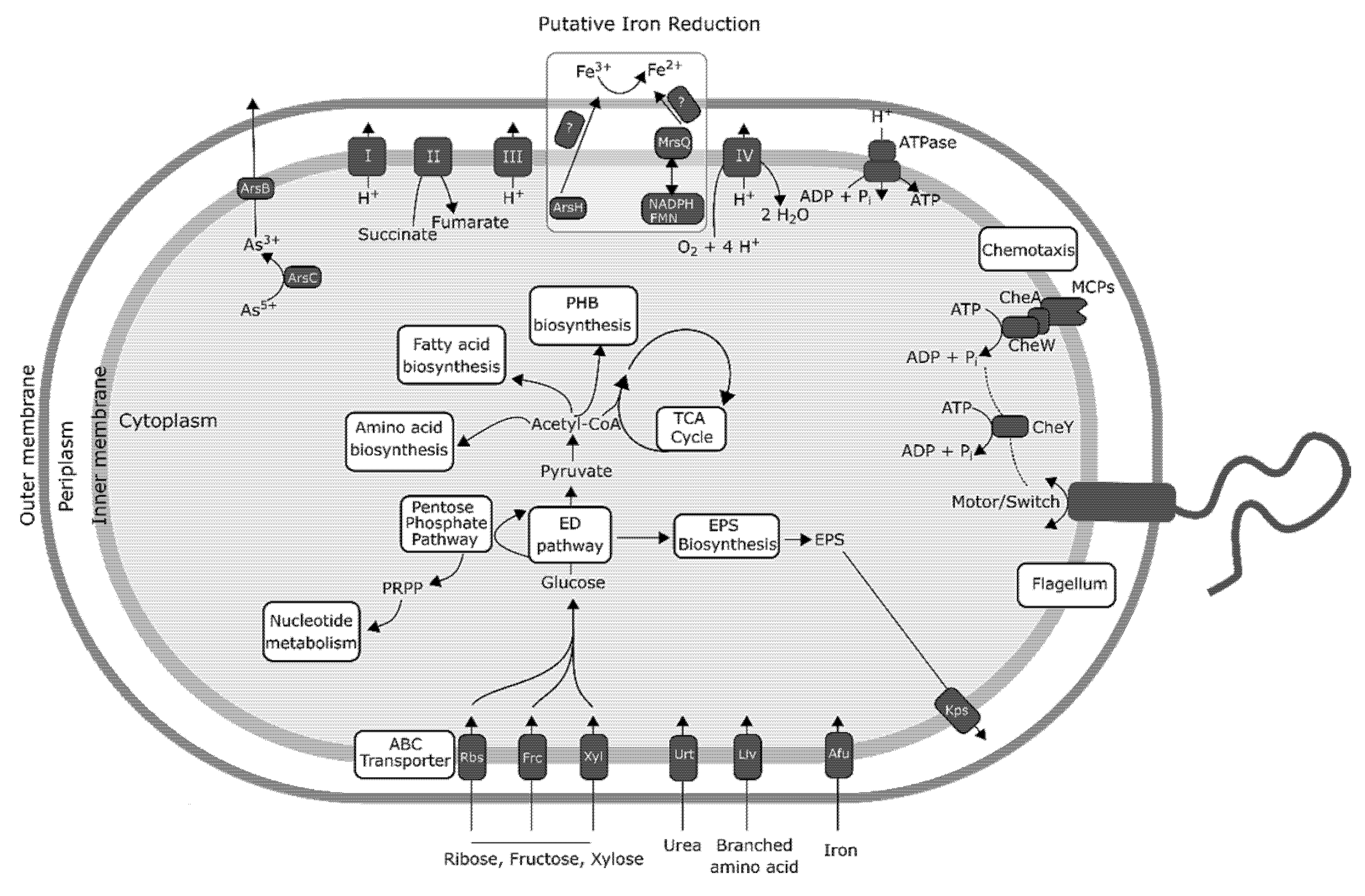
3.4. Central Metabolism and Iron Reduction Machinery in Acidiphilium sp. C61
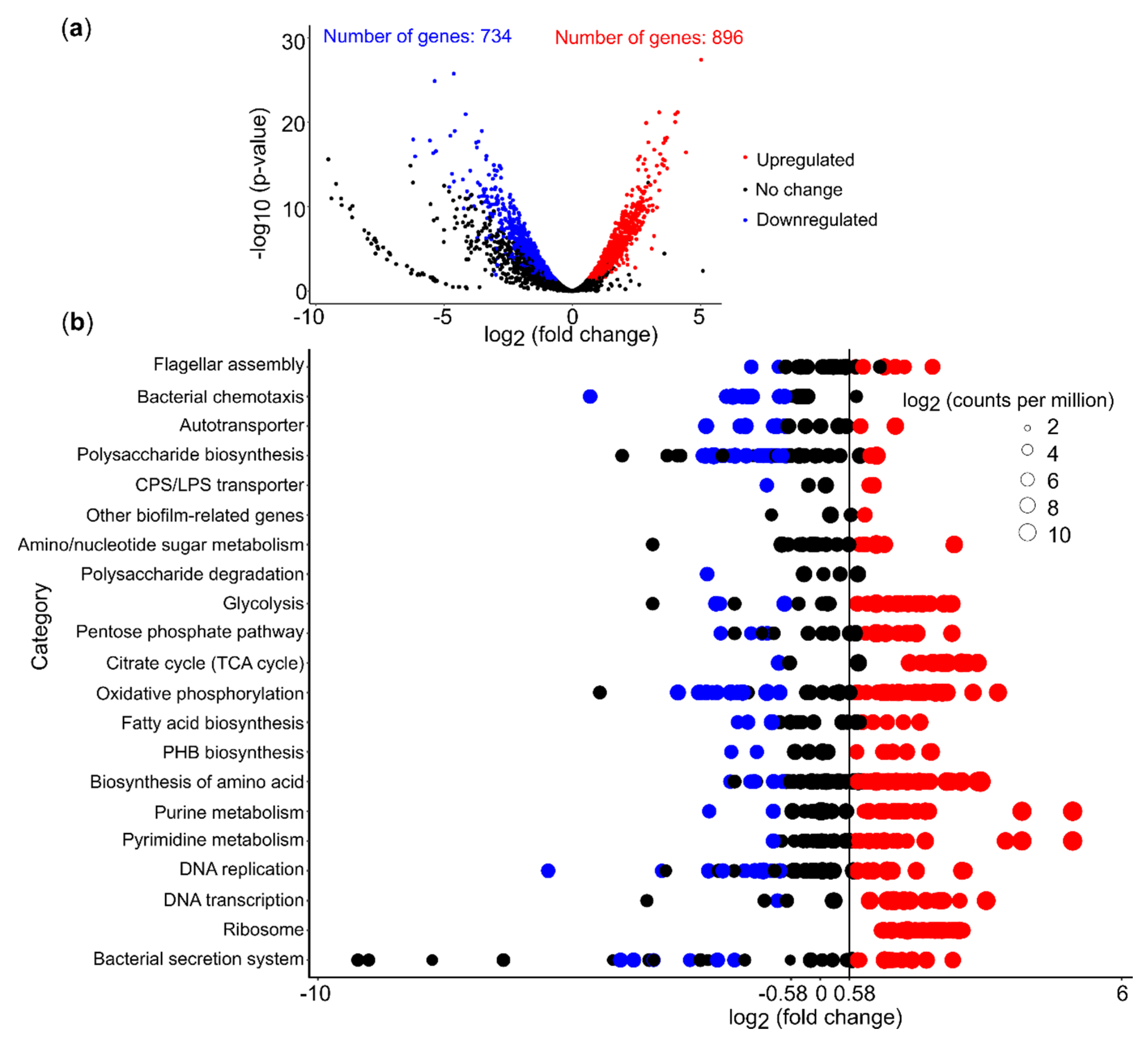
3.5. Differential Gene Expression Analysis
3.6. PEA Induced Upregulation of Central Cellular Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alldredge, A.L.; Silver, M.W. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 1988, 20, 41–82. [Google Scholar] [CrossRef]
- Simon, M.; Grossart, H.-P.; Schweitzer, B.; Ploug, H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 2002, 28, 175–211. [Google Scholar] [CrossRef]
- Thornton, D. Diatom aggregation in the sea: Mechanisms and ecological implications. Eur. J. Phycol. 2002, 37, 149–161. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Simon, M. Limnetic macroscopic organic aggregates (lake snow): Occurrence, characteristics, and microbial dynamics in Lake Constance. Limnol. Oceanogr. 1993, 38, 532–546. [Google Scholar] [CrossRef]
- Reiche, M.; Lu, S.; Ciobotǎ, V.; Neu, T.R.; Nietzsche, S.; Rösch, P.; Popp, J.; Küsel, K. Pelagic boundary conditions affect the biological formation of iron-rich particles (iron snow) and their microbial communities. Limnol. Oceanogr. 2011, 56, 1386–1398. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Grossart, H.-P.; Schlingloff, A.; Kiørboe, T. Possible quorum sensing in marine snow bacteria: Production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 2002, 68, 4111–4116. [Google Scholar] [CrossRef]
- Blodau, C. A review of acidity generation and consumption in acidic coal mine lakes and their watersheds. Sci. Total Environ. 2006, 369, 307–332. [Google Scholar] [CrossRef]
- Küsel, K. Microbial cycling of iron and sulfur in acidic coal mining lake sediments. Water Air Soil Pollut. Focus 2003, 3, 67–90. [Google Scholar]
- Bigham, J.M.; Schwertmann, U.; Traina, S.J.; Winland, R.L.; Wolf, M. Schwertmannite and the chemical modeling of iron in acid sulfate waters. Geochim. Cosmochim. Acta 1996, 60, 2111–2121. [Google Scholar] [CrossRef]
- Wang, H.; Bigham, J.M.; Tuovinen, O.H. Formation of schwertmannite and its transformation to jarosite in the presence of acidophilic iron-oxidizing microorganisms. Mater. Sci. Eng. C 2006, 26, 588–592. [Google Scholar] [CrossRef]
- Ciobotǎ, V.; Lu, S.; Tarcea, N.; Rösch, P.; Küsel, K.; Popp, J. Quantification of the inorganic phase of the pelagic aggregates from an iron contaminated lake by means of Raman spectroscopy. Vib. Spectrosc. 2013, 68, 212–219. [Google Scholar] [CrossRef]
- Mori, J.F.; Lu, S.; Händel, M.; Totsche, K.U.; Neu, T.R.; Iancu, V.V.; Tarcea, N.; Popp, J.; Küsel, K. Schwertmannite formation at cell junctions by a new filament-forming Fe(II)-oxidizing isolate affiliated with the novel genus Acidithrix. Microbiology 2016, 162, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Küsel, K.; Dorsch, T.; Acker, G.; Stackebrandt, E. Microbial reduction of Fe(III) in acidic sediments: Isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. Appl. Environ. Microbiol. 1999, 65, 3633–3640. [Google Scholar] [CrossRef]
- Lu, S.; Chourey, K.; Reiche, M.; Nietzsche, S.; Shah, M.B.; Neu, T.R.; Hettich, R.L.; Küsel, K. Insights into the structure and metabolic function of microbes that shape pelagic iron-rich aggregates (“Iron snow”). Appl. Environ. Microbiol. 2013, 79, 4272–4281. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Gischkat, S.; Reiche, M.; Akob, D.M.; Hallberg, K.B.; Küsel, K. Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl. Environ. Microbiol. 2010, 76, 8174–8183. [Google Scholar] [CrossRef]
- Mori, J.F.; Ueberschaar, N.; Lu, S.; Cooper, R.E.; Pohnert, G.; Küsel, K. Sticking together: Inter-species aggregation of bacteria isolated from iron snow is controlled by chemical signaling. ISME J. 2017, 11, 1075–1086. [Google Scholar] [CrossRef]
- Irsfeld, M.; Spadafore, M.; Prüß, B. β-Phenylethylamine, a small molecule with a large impact. Webmedcentral 2013, 4, 1–15. [Google Scholar]
- Paterson, I.A.; Juorio, A.V.; Boulton, A.A. 2-Phenylethylamine: A modulator of catecholamine transmission in the mammalian central nervous system? J. Neurochem. 1990, 55, 1827–1837. [Google Scholar] [CrossRef]
- Rothman, R.B.; Baumann, M.H. Balance between dopamine and serotonin release modulates behavioral effects of amphetamine-type drugs. Ann. N. Y. Acad. Sci. 2006, 1074, 245–260. [Google Scholar] [CrossRef]
- Marcobal, A.; De las Rivas, B.; Landete, J.M.; Tabera, L.; Muñoz, R. Tyramine and phenylethylamine biosynthesis by food bacteria. Crit. Rev. Food Sci. Nutr. 2012, 52, 448–467. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.G.; Rather, P.N. A novel gene involved in regulating the flagellar gene cascade in Proteus mirabilis. J. Bacteriol. 2006, 188, 7830–7839. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.G.; Szostek, B.A.; Clemmer, K.M.; Rather, P.N. Expression of the DisA amino acid decarboxylase from Proteus mirabilis inhibits motility and class 2 flagellar gene expression in Escherichia coli. Res. Microbiol. 2013, 164, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Belas, R. Surface sensing, swarmer cell differentiation, and biofilm development. Methods Enzymol. 2001, 336, 29–40. [Google Scholar] [PubMed]
- Belas, R.; Suvanasuthi, R. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 2005, 187, 6789–6803. [Google Scholar] [CrossRef] [PubMed]
- Sturgill, G.; Rather, P.N. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 2004, 51, 437–446. [Google Scholar] [CrossRef]
- Bridge, T.A.M.; Johnson, D.B. Reductive dissolution of ferric iron minerals by Acidiphilium SJH. Geomicrobiol. J. 2000, 17, 193–206. [Google Scholar]
- Tischler, J.S.; Jwair, R.J.; Gelhaar, N.; Drechsel, A.; Skirl, A.M.; Wiacek, C.; Janneck, E.; Schlömann, M. New cultivation medium for “Ferrovum” and Gallionella-related strains. J. Microbiol. Methods 2013, 95, 138–144. [Google Scholar] [CrossRef]
- Tamura, H.; Goto, K.; Yotsuyanagi, T.; Nagayama, M. Spectrophotometric determination of iron(II) with 1,10-phenanthroline in the presence of large amounts of iron(III). Talanta 1974, 21, 314–318. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Delmont, T.O.; Eren, E.M. Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ 2018, 2018, 1–23. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010, 11. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankavaram, U.T.; Rao, B.S.; Kiryutin, B.; Galperin, M.Y.; Fedorova, N.D.; Koonin, E.V. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2002, 29, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Benedict, M.N.; Henriksen, J.R.; Metcalf, W.W.; Whitaker, R.J.; Price, N.D. ITEP: An integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 2014, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; Dongen, S.V.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017, 46, D335–D342. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A fast, accurate, splice-aware aligner. In Proceedings of the Conference: 9th Annual Genomics of Energy & Environment Meeting, Walnut Creek, CA, USA, 19 March 2014. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Brühl, A.; Amann, R.; Schleifer, K.-H.; Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef]
- Loy, A.; Lehner, A.; Lee, N.; Adamczyk, J.; Meier, H.; Ernst, J.; Schleifer, K.-H.; Wagner, M. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 2002, 68, 5064–5081. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Hädrich, A.; Küsel, K. Predominance of thaumarchaeal ammonia oxidizer abundance and transcriptional activity in an acidic fen. Environ. Microbiol. 2012, 14, 3013–3025. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Schramm, A.; Neu, T.R.; Revsbech, N.P.; Meyer, R.L. Extracellular DNA in adhesion and biofilm formation of four environmental isolates: A quantitative study. FEMS Microbiol. Ecol. 2013, 86, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.R.; Poehlein, A.; Voget, S.; Hoppert, M.; Daniel, R.; Leimbach, A.; Tischler, J.S.; Schlömann, M.; Mühling, M. Permanent draft genome sequence of Acidiphilium sp. JA12-A1. Stand. Genomic Sci. 2015, 10, 1–10. [Google Scholar] [CrossRef]
- Ciobotǎ, V.; Burkhardt, E.M.; Schumacher, W.; Rösch, P.; Küsel, K.; Popp, J. The influence of intracellular storage material on bacterial identification by means of Raman spectroscopy. Anal. Bioanal. Chem. 2010, 397, 2929–2937. [Google Scholar] [CrossRef]
- Bird, L.J.; Bonnefoy, V.; Newman, D.K. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 2011, 19, 330–340. [Google Scholar] [CrossRef]
- Magnuson, T.S.; Swenson, M.W.; Paszczynski, A.J.; Deobald, L.A.; Kerk, D.; Cummings, D.E. Proteogenomic and functional analysis of chromate reduction in Acidiphilium cryptum JF-5, an Fe(III)-respiring acidophile. BioMetals 2010, 23, 1129–1138. [Google Scholar] [CrossRef]
- Mo, H.; Chen, Q.; Du, J.; Tang, L.; Qin, F.; Miao, B.; Wu, X.; Zeng, J. Ferric reductase activity of the ArsH protein from Acidithiobacillus ferrooxidans. J. Microbiol. Biotechnol. 2011, 21, 464–469. [Google Scholar] [CrossRef]
- Johnson, D.B.; Hallberg, K.B. The microbiology of acidic mine waters. Res. Microbiol. 2003, 154, 466–473. [Google Scholar] [CrossRef]
- Harrison, A.P.; Jarvis, B.W.; Johnson, J.L. Heterotrophic bacteria from cultures of autotrophic Thiobacillus ferrooxidans: Relationships as studied by means of deoxyribonucleic acid homology. J. Bacteriol. 1980, 143, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.P. The acidophilic Thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 1984, 38, 265–292. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B. Geomicrobiology of extremely acidic subsurface environments. FEMS Microbiol. Ecol. 2012, 81, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, K.B.; Johnson, D.B. Biodiversity of acidophilic prokaryotes. Adv. Appl. Microbiol. 2001, 49, 37–84. [Google Scholar] [PubMed]
- Lu, S.; Peiffer, S.; Lazar, C.S.; Oldham, C.; Neu, T.R.; Ciobota, V.; Näb, O.; Lillicrap, A.; Rösch, P.; Popp, J.; et al. Extremophile microbiomes in acidic and hypersaline river sediments of Western Australia. Environ. Microbiol. Rep. 2016, 8, 58–67. [Google Scholar] [CrossRef]
- Miot, J.; Jézéquel, D.; Benzerara, K.; Cordier, L.; Rivas-Lamelo, S.; Skouri-Panet, F.; Férard, C.; Poinsot, M.; Duprat, E. Mineralogical diversity in Lake Pavin: Connections with water column chemistry and biomineralization processes. Minerals 2016, 6, 24. [Google Scholar] [CrossRef]
- Peine, A.; Tritschler, A.; Küsel, K.; Peiffer, S. Electron flow in an iron-rich acidic sediment—Evidence for an acidity-driven iron cycle. Limnol. Oceanogr. 2000, 45, 1077–1087. [Google Scholar] [CrossRef]
- Rivas, M.; Seeger, M.; Holmes, D.S.; Jedlicki, E. A Lux-like quorum sensing system in the extreme acidophile Acidithiobacillus ferrooxidans. Biol. Res. 2005, 38, 283–297. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Zafra, O.; Lamprecht-Grandío, M.; de Figueras, C.G.; González-Pastor, J.E. Extracellular DNA release by undomesticated Bacillus subtilis is regulated by early competence. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Tapia, J.M.; Muñoz, J.; González, F.; Blázquez, M.L.; Malki, M.; Ballester, A. Extraction of extracellular polymeric substances from the acidophilic bacterium Acidiphilium 3.2Sup(5). Water Sci. Technol. 2009, 59, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Kermer, R.; Hedrich, S.; Taubert, M.; Baumann, S.; Schlömann, M.; Johnson, D.B.; Seifert, J. Elucidation of carbon transfer in a mixed culture of Acidiphilium cryptum and Acidithiobacillus ferrooxidans using protein-based stable isotope probing. J. Integr. OMICS 2012, 2, 37–45. [Google Scholar]
- Ullrich, S.R.; Poehlein, A.; Tischler, J.S.; González, C.; Ossandon, F.J.; Daniel, R.; Holmes, D.S.; Schlömann, M.; Mühling, M. Genome analysis of the biotechnologically relevant acidophilic iron oxidising strain JA12 indicates phylogenetic and metabolic diversity within the novel genus “Ferrovum”. PLoS ONE 2016, 11, e0146832. [Google Scholar] [CrossRef] [PubMed]
- Nitschke, W.; Bonnefoy, V. Energy acquisition in low pH environments. In Acidophiles: Life in Extremely Acidic Environments; Quatrini, R., Johnson, D.B., Eds.; Caister Academic Press: Poole, UK, 2016; pp. 19–48. [Google Scholar]
- Küsel, K.; Roth, U.; Drake, H.L. Microbial reduction of Fe(III) in the presence of oxygen under low pH conditions. Environ. Microbiol. 2002, 4, 414–421. [Google Scholar] [CrossRef]
- Coupland, K.; Johnson, D.B. Evidence that the potential for dissimilatory ferric iron reduction is widespread among acidophilic heterotrophic bacteria. FEMS Microbiol. Lett. 2008, 279, 30–35. [Google Scholar] [CrossRef]
- Johnson, D.B.; Kanao, T.; Hedrich, S. Redox transformations of iron at extremely low pH: Fundamental and applied aspects. Front. Microbiol. 2012, 3, 1–13. [Google Scholar] [CrossRef]
- Juillan-Binard, C.; Picciocchi, A.; Andrieu, J.P.; Dupuy, J.; Petit-Hartlein, I.; Caux-Thang, C.; Vivès, C.; Nivière, V.; Fieschi, F. A two-component NADPH oxidase (NOX)-like system in bacteria is involved in the electron transfer chain to the methionine sulfoxide reductase MsrP. J. Biol. Chem. 2017, 292, 2485–2494. [Google Scholar] [CrossRef]
- Osorio, H.; Mangold, S.; Denis, Y.; Ñancucheo, I.; Esparza, M.; Johnson, D.B.; Bonnefoy, V.; Dopson, M.; Holmesa, D.S. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2013, 79, 2172–2181. [Google Scholar] [CrossRef]
- Sugio, T.; Taha, T.M.; Takeuchi, F. Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grown Acidithiobacillus ferrooxidans ATCC 23270 cells. Biosci. Biotechnol. Biochem. 2009, 73, 1381–1386. [Google Scholar] [CrossRef][Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Cooper, R.E.; Wegner, C.-E.; Küsel, K. Molecular Mechanisms Underpinning Aggregation in Acidiphilium sp. C61 Isolated from Iron-Rich Pelagic Aggregates. Microorganisms 2020, 8, 314. https://doi.org/10.3390/microorganisms8030314
Li Q, Cooper RE, Wegner C-E, Küsel K. Molecular Mechanisms Underpinning Aggregation in Acidiphilium sp. C61 Isolated from Iron-Rich Pelagic Aggregates. Microorganisms. 2020; 8(3):314. https://doi.org/10.3390/microorganisms8030314
Chicago/Turabian StyleLi, Qianqian, Rebecca E. Cooper, Carl-Eric Wegner, and Kirsten Küsel. 2020. "Molecular Mechanisms Underpinning Aggregation in Acidiphilium sp. C61 Isolated from Iron-Rich Pelagic Aggregates" Microorganisms 8, no. 3: 314. https://doi.org/10.3390/microorganisms8030314
APA StyleLi, Q., Cooper, R. E., Wegner, C.-E., & Küsel, K. (2020). Molecular Mechanisms Underpinning Aggregation in Acidiphilium sp. C61 Isolated from Iron-Rich Pelagic Aggregates. Microorganisms, 8(3), 314. https://doi.org/10.3390/microorganisms8030314




