Diverse Microbial Community Profiles of Propionate-Degrading Cultures Derived from Different Sludge Sources of Anaerobic Wastewater Treatment Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Enrichment Process
2.2. Sample Collection and Molecular Analysis
2.3. Microbiome Analysis Based on the 16S rRNA Gene Sequences
3. Results
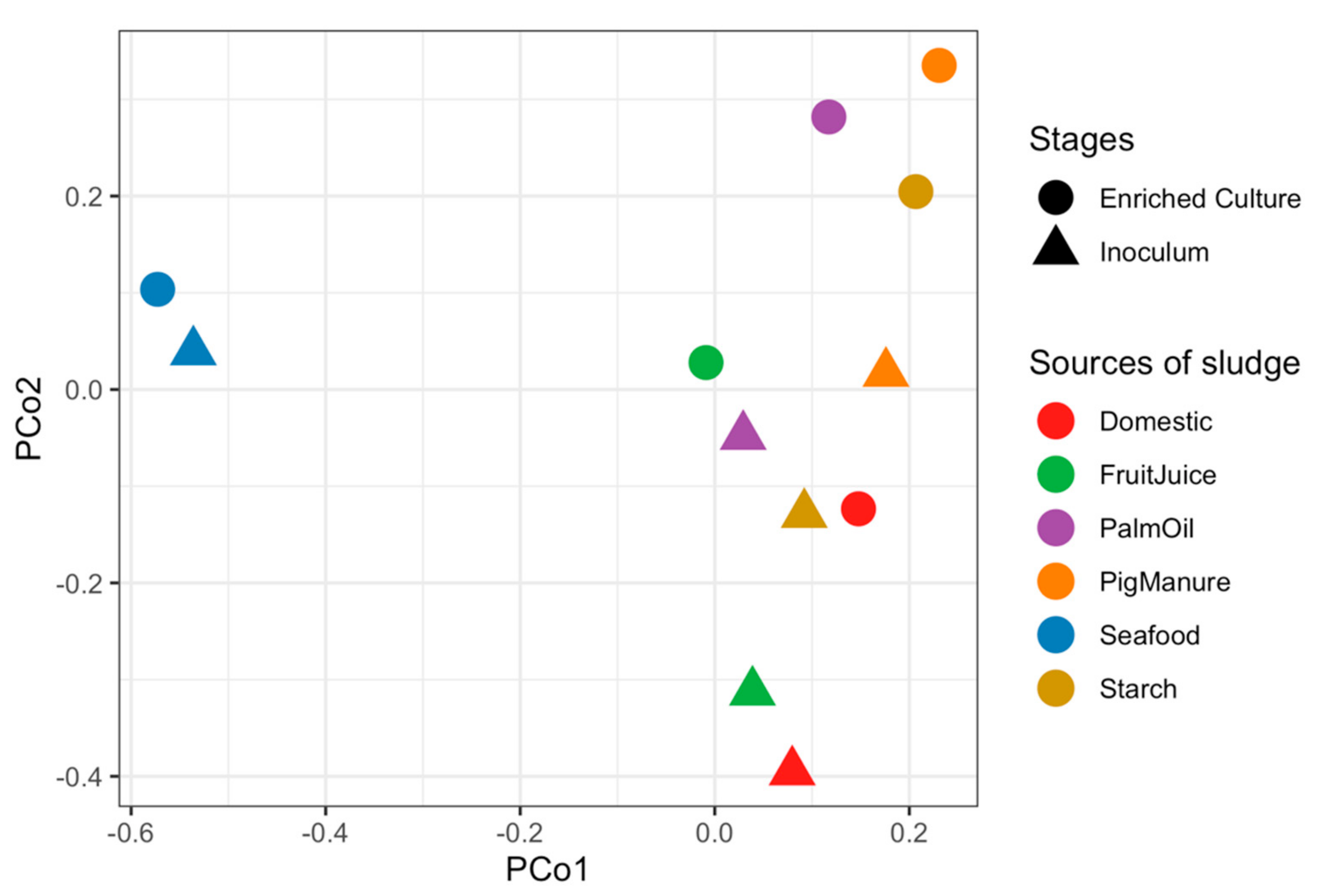
3.1. A Shift of Microbiome Profiles from Inoculums to Enriched Propionate-Degrading Cultures
3.2. Microbiome Profiles of Propionate-Degrading Cultures Enriched from Different Inoculum Sources
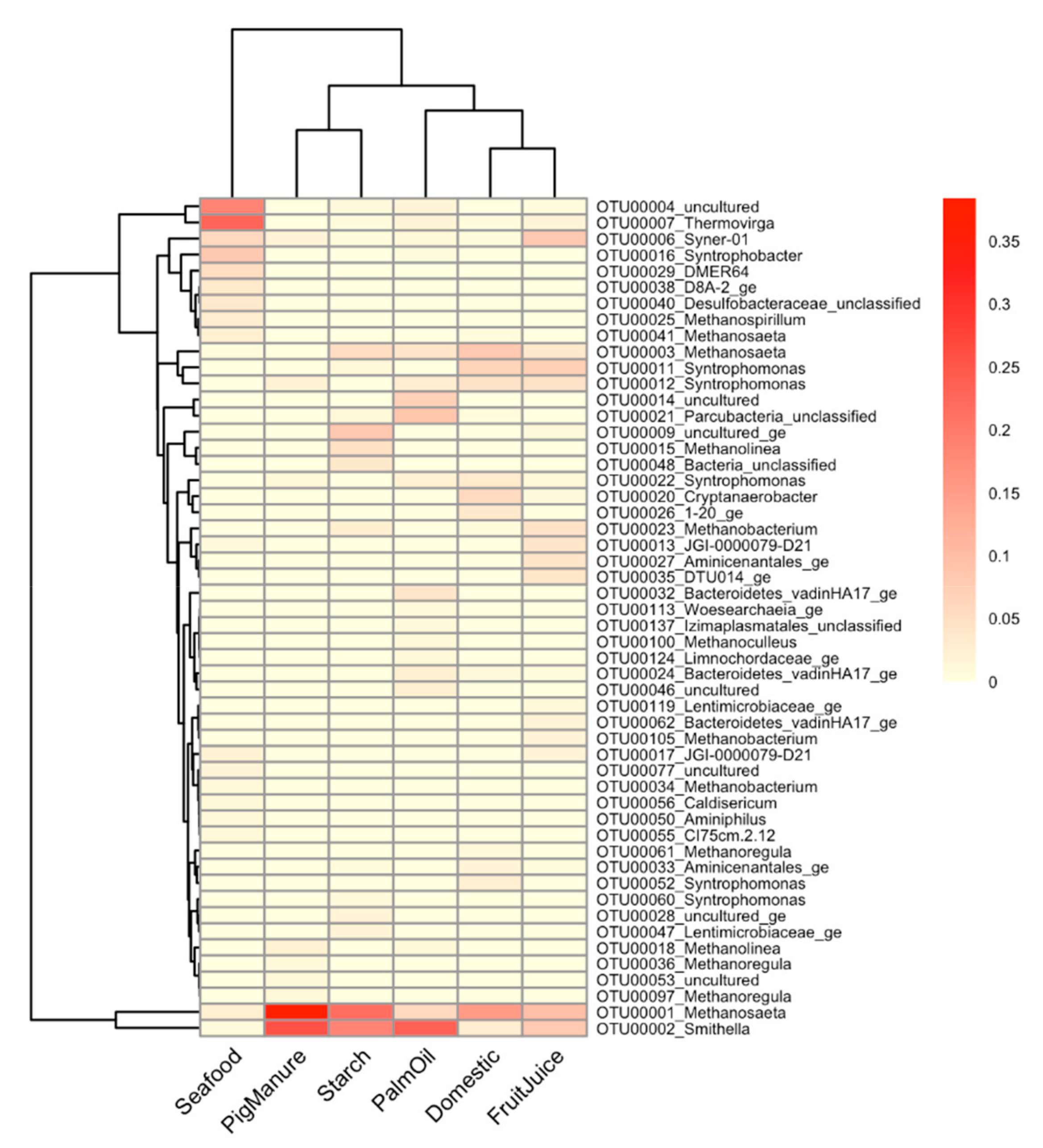
3.3. Common and Unique Microorganisms in Propionate-Degrading Cultures Enriched from Different Inoculum Sources
3.4. Several Uncultured Microbes Found in the Propionate-Degrading Cultures Using the Culture-Independent Amplicon-Based Sequencing Approach
4. Discussion
4.1. The Schematic Propionate-Degrading Pathway in the Enriched Cultures for Methane Production
4.2. Different Taxa of Hydrogenotrophic Methanogens Found Specifically to Different Sludge Sources
4.3. Unique Microbial Community in the Propionate-Degrading Culture Enriched from Seafood Sludge
4.4. Overall Microbial Profiles of Propionate-Degrading Cultures and Unculturable Microbes Revealed Through Amplicon-Based Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bajpai, P. Basics of Anaerobic Digestion Process. In Anaerobic Technology in Pulp and Paper Industry; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Abdelgadir, A.; Chen, X.; Liu, J.; Xie, X.; Zhang, J.; Zhang, K.; Wang, H.; Liu, N. Characteristics, process parameters, and inner components of anaerobic bioreactors. Biomed. Res. Int. 2014, 2014, 841573. [Google Scholar] [CrossRef] [PubMed]
- Amha, Y.M.; Anwar, M.Z.; Brower, A.; Jacobsen, C.S.; Stadler, L.B.; Webster, T.M.; Smith, A.L. Inhibition of anaerobic digestion processes: Applications of molecular tools. Bioresour. Technol. 2018, 247, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Grzelak, J.; Ślęzak, R.; Krzystek, L.; Ledakowicz, S. Effect of pH on the production of volatile fatty acids in dark fermentation process of organic waste. Ecol. Chem. Eng. S 2018, 25, 295–306. [Google Scholar] [CrossRef]
- Mir, M.A.; Hussain, A.; Verma, C. Design considerations and operational performance of anaerobic digester: A review. Cogent Eng. 2016, 3, 1181696. [Google Scholar] [CrossRef]
- Ward, A.J.; Hobbs, P.J.; Holliman, P.J.; Jones, D.L. Optimisation of the anaerobic digestion of agricultural resources. Bioresour. Technol. 2008, 99, 7928–7940. [Google Scholar] [CrossRef]
- Wrońska, I.; Cybulska, K. Quantity and quality of biogas produced from the poultry sludge optimized by filamentous fungi. Ecol. Chem. Eng. S 2018, 25, 395–404. [Google Scholar] [CrossRef]
- Wacławek, S.; Grübel, K.; Silvestri, D.; Padil, V.V.; Wacławek, M.; Černík, M.; Varma, R.S. Disintegration of wastewater activated sludge (WAS) for improved biogas production. Energies 2019, 12, 21. [Google Scholar] [CrossRef]
- Meyer, T.; Edwards, E.A. Anaerobic digestion of pulp and paper mill wastewater and sludge. Water Res. 2014, 65, 321–349. [Google Scholar] [CrossRef]
- Mao, C.; Feng, Y.; Wang, X.; Ren, G. Review on research achievements of biogas from anaerobic digestion. Renew. Sustain. Energy Rev. 2015, 45, 540–555. [Google Scholar] [CrossRef]
- Hu, D.; Su, H.; Chen, Z.; Cui, Y.; Ran, C.; Xu, J.; Xiao, T.; Li, X.; Wang, H.; Tian, Y. Performance evaluation and microbial community dynamics in a novel AnMBR for treating antibiotic solvent wastewater. Bioresour. Technol. 2017, 243, 218–227. [Google Scholar] [CrossRef]
- Fukuzaki, S.; Nishio, N.; Shobayashi, M.; Nagai, S. Inhibition of the fermentation of propionate to methane by hydrogen, acetate, and propionate. Appl. Env. Microbiol. 1990, 56, 719–723. [Google Scholar] [CrossRef]
- Lins, P.; Malin, C.; Wagner, A.O.; Illmer, P. Reduction of accumulated volatile fatty acids by an acetate-degrading enrichment culture. FEMS Microbiol. Ecol. 2010, 71, 469–478. [Google Scholar] [CrossRef]
- Amani, T.; Nosrati, M.; Mousavi, S.; Kermanshahi, R. Study of syntrophic anaerobic digestion of volatile fatty acids using enriched cultures at mesophilic conditions. Int. J. Env. Sci. Technol. 2011, 8, 83–96. [Google Scholar] [CrossRef]
- Tale, V.P.; Maki, J.S.; Struble, C.A.; Zitomer, D.H. Methanogen community structure-activity relationship and bioaugmentation of overloaded anaerobic digesters. Water Res. 2011, 45, 5249–5256. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Sun, Y.; Wu, S.; Kong, X.; Yuan, Z.; Dong, R. The performance efficiency of bioaugmentation to prevent anaerobic digestion failure from ammonia and propionate inhibition. Bioresour. Technol. 2017, 231, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Houwen, F.P.; Plokker, J.; Stams, A.J.; Zehnder, A.J. Enzymatic evidence for involvement of the methylmalonyl-CoA pathway in propionate oxidation by Syntrophobacter wolinii. Arch. Microbiol. 1990, 155, 52–55. [Google Scholar] [CrossRef]
- Kosaka, T.; Uchiyama, T.; Ishii, S.-I.; Enoki, M.; Imachi, H.; Kamagata, Y.; Ohashi, A.; Harada, H.; Ikenaga, H.; Watanabe, K. Reconstruction and regulation of the central catabolic pathway in the thermophilic propionate-oxidizing syntroph Pelotomaculum thermopropionicum. J. Bacteriol. 2006, 188, 202–210. [Google Scholar] [CrossRef]
- Ferry, J.G. Fundamentals of methanogenic pathways that are key to the biomethanation of complex biomass. Curr. Opin. Biotechnol. 2011, 22, 351–357. [Google Scholar] [CrossRef]
- Harmsen, H.J.; Van Kuijk, B.L.; Plugge, C.M.; Akkermans, A.D.; De Vos, W.M.; Stams, A.J. Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 1998, 48, 1383–1387. [Google Scholar] [CrossRef]
- Wallrabenstein, C.; Hauschild, E.; Schink, B. Syntrophobacter pfennigii sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Arch. Microbiol. 1995, 164, 346–352. [Google Scholar] [CrossRef]
- Boone, D.R.; Bryant, M.P. Propionate-degrading bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from methanogenic ecosystems. Appl. Env. Microbiol. 1980, 40, 626–632. [Google Scholar] [CrossRef]
- Li, J.; Ban, Q.; Zhang, L.; Jha, A.K. Syntrophic propionate degradation in anaerobic digestion: A review. Int. J. Agric. Biol. 2012, 14, 843–850. [Google Scholar]
- De Bok, F.A.; Stams, A.J.; Dijkema, C.; Boone, D.R. Pathway of propionate oxidation by a syntrophic culture of Smithella propionica and Methanospirillum hungatei. Appl. Env. Microbiol. 2001, 67, 1800–1804. [Google Scholar] [CrossRef]
- Dolfing, J. Syntrophic propionate oxidation via butyrate: A novel window of opportunity under methanogenic conditions. Appl. Env. Microbiol. 2013, 79, 4515–4516. [Google Scholar] [CrossRef]
- Liu, Y.; Balkwill, D.L.; Aldrich, H.C.; Drake, G.R.; Boone, D.R. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 1999, 49, 545–556. [Google Scholar] [CrossRef]
- Wawrik, B.; Marks, C.R.; Davidova, I.A.; McInerney, M.J.; Pruitt, S.; Duncan, K.E.; Suflita, J.M.; Callaghan, A.V. Methanogenic paraffin degradation proceeds via alkane addition to fumarate by ‘Smithella’spp. mediated by a syntrophic coupling with hydrogenotrophic methanogens. Environ. Microbiol. 2016, 18, 2604–2619. [Google Scholar] [CrossRef]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Vera-Ponce de Leon, A.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef]
- Wirth, R.; Kovacs, E.; Maroti, G.; Bagi, Z.; Rakhely, G.; Kovacs, K.L. Characterization of a biogas-producing microbial community by short-read next generation DNA sequencing. Biotechnol. Biofuels 2012, 5, 41. [Google Scholar] [CrossRef]
- Schluter, A.; Bekel, T.; Diaz, N.N.; Dondrup, M.; Eichenlaub, R.; Gartemann, K.H.; Krahn, I.; Krause, L.; Kromeke, H.; Kruse, O.; et al. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J. Biotechnol. 2008, 136, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wilkins, D.; Chen, J.; Ng, S.-K.; Lu, H.; Jia, Y.; Lee, P.K. Metagenomic reconstruction of key anaerobic digestion pathways in municipal sludge and industrial wastewater biogas-producing systems. Front. Microbiol. 2016, 7, 778. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Peng, Y.; Ni, B.J.; Han, X.; Fan, L.; Yuan, Z. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb. Cell Fact. 2015, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, Y.; Yang, G.; Hu, K.; Lv, P.; Li, L. Vertical distribution of microbial community and metabolic pathway in a methanogenic propionate degradation bioreactor. Bioresour. Technol. 2017, 245, 1022–1029. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Li, L.; Yuan, Z. Acclimation of acid-tolerant methanogenic propionate-utilizing culture and microbial community dissecting. Bioresour. Technol. 2018, 250, 117–123. [Google Scholar] [CrossRef]
- Scholten, J.C.; Conrad, R. Energetics of syntrophic propionate oxidation in defined batch and chemostat cocultures. Appl. Env. Microbiol. 2000, 66, 2934–2942. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Env. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. R package version 1.0.12. 2019. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 18 December 2019).
- Clarke, K.; Green, R. Statistical design and analysis for a‘biological effects’ study. Mar. Ecol. Prog. Ser. 1988, 213–226. [Google Scholar] [CrossRef]
- Koch, M.; Dolfing, J.; Wuhrmann, K.; Zehnder, A.J. Pathways of propionate degradation by enriched methanogenic cultures. Appl. Env. Microbiol. 1983, 45, 1411–1414. [Google Scholar] [CrossRef]
- Shigematsu, T.; Era, S.; Mizuno, Y.; Ninomiya, K.; Kamegawa, Y.; Morimura, S.; Kida, K. Microbial community of a mesophilic propionate-degrading methanogenic consortium in chemostat cultivation analyzed based on 16S rRNA and acetate kinase genes. Appl. Microbiol. Biotechnol. 2006, 72, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Chaiprasert, P.; Hudayah, N.; Auphimai, C. Efficacies of Various Anaerobic Starter Seeds for Biogas Production from Different Types of Wastewater. Biomed. Res. Int. 2017, 2017, 2782850. [Google Scholar] [CrossRef] [PubMed]
- McInerney, M.J.; Bryant, M.P.; Hespell, R.B.; Costerton, J.W. Syntrophomonas wolfei gen. nov. sp. nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium. Appl. Env. Microbiol. 1981, 41, 1029–1039. [Google Scholar] [CrossRef]
- Kitamura, K.; Fujita, T.; Akada, S.; Tonouchi, A. Methanobacterium kanagiense sp. nov., a hydrogenotrophic methanogen, isolated from rice-field soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1246–1252. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, Y.; Sakai, S.; Ehara, M.; Miyazaki, M.; Yamaguchi, T.; Imachi, H. Methanoregula formicica sp. nov., a methane-producing archaeon isolated from methanogenic sludge. Int. J. Syst. Evol. Microbiol. 2011, 61, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Kougias, P.G.; Campanaro, S.; Treu, L.; Zhu, X.; Angelidaki, I. A novel archaeal species belonging to Methanoculleus genus identified via de-novo assembly and metagenomic binning process in biogas reactors. Anaerobe 2017, 46, 23–32. [Google Scholar] [CrossRef]
- Imachi, H.; Sakai, S.; Sekiguchi, Y.; Hanada, S.; Kamagata, Y.; Ohashi, A.; Harada, H. Methanolinea tarda gen. nov., sp. nov., a methane-producing archaeon isolated from a methanogenic digester sludge. Int. J. Syst. Evol. Microbiol. 2008, 58, 294–301. [Google Scholar] [CrossRef]
- Dahle, H.; Birkeland, N.K. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int. J. Syst. Evol. Microbiol. 2006, 56, 1539–1545. [Google Scholar] [CrossRef]
- Yamada, T.; Sekiguchi, Y.; Imachi, H.; Kamagata, Y.; Ohashi, A.; Harada, H. Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl. Env. Microbiol. 2005, 71, 7493–7503. [Google Scholar] [CrossRef]
- Wang, S.; Hou, X.; Su, H. Exploration of the relationship between biogas production and microbial community under high salinity conditions. Sci. Rep. 2017, 7, 1149. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Wang, L.Y.; Mbadinga, S.M.; Liu, J.F.; Yang, S.Z.; Gu, J.D.; Mu, B.Z. Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 2015, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, S.J.; Kirkegaard, R.H.; Dueholm, M.S.; Fernando, E.; Karst, S.M.; Albertsen, M.; Nielsen, P.H. Culture-independent analyses reveal novel anaerolineaceae as abundant primary fermenters in anaerobic digesters treating waste activated sludge. Front. Microbiol. 2017, 8, 1134. [Google Scholar] [CrossRef] [PubMed]
- Imachi, H.; Sekiguchi, Y.; Kamagata, Y.; Ohashi, A.; Harada, H. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl. Env. Microbiol. 2000, 66, 3608–3615. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Anaerobic Sludge from Various Anaerobic Wastewater Treatment Plants | Reactor Performance | Specific Methanogenic Activity (SMA) (g COD/g VSS/d) | ||||
|---|---|---|---|---|---|---|
| pH | TVA/Alkalinity | COD Reduction (%) | Biogas Composition (%) | |||
| %CH4 | %CO2 | |||||
| Domestic | 7.50 | 0.30 | 86.5 | 60.0 | 35.5 | 0.22 ± 0.016 |
| FruitJuice | 7.50 | 0.30 | 85.0 | 62.5 | 34.0 | 0.17 ± 0.011 |
| PalmOil | 7.49 | 0.30 | 86.5 | 75.0 | 22.0 | 0.20 ± 0.009 |
| Starch | 7.56 | 0.25 | 90.0 | 73.5 | 23.5 | 0.22 ± 0.007 |
| PigManure | 7.57 | 0.27 | 89.0 | 75.5 | 21.0 | 0.28 ± 0.003 |
| Seafood | 7.52 | 0.35 | 80.0 | 80.0 | 17.5 | 0.14 ± 0.015 |
| OTUs | Taxonomic Lineage | Propionate-Enriched Culture | ||||
|---|---|---|---|---|---|---|
| Domestic | FruitJuice | PalmOil | PigManure | Starch | ||
| OTU00001 | Archaea; Euryarchaeota; Methanomicrobia; Methanosarcinales; Methanosaetaceae; Methanosaeta | ✓ | ✓ | ✓ | ✓ | ✓ |
| OTU00002 | Bacteria; Proteobacteria; Deltaproteobacteria; Syntrophobacterales; Syntrophaceae; Smithella | ✓ | ✓ | ✓ | ✓ | ✓ |
| OTU00003 | Archaea; Euryarchaeota; Methanomicrobia; Methanosarcinales; Methanosaeta; Methanosaeta | ✓ | ✓ | ✓ | ✓ | |
| OTU00006 | Bacteria; Synergistetes; Synergistia; Synergistales; Synergistaceae; Syner-01 | ✓ | ✓ | ✓ | ||
| OTU00011 | Bacteria; Firmicutes; Clostridia; Clostridiales; Syntrophomonadaceae; Syntrophomonas | ✓ | ✓ | ✓ | ||
| OTU00012 | Bacteria; Firmicutes; Cloastridia; Clostridiales; Syntrophomonadaceae; Syntrophomonas | ✓ | ✓ | ✓ | ✓ | |
| OTU00022 | Bacteria; Firmicutes; Clostridia; Clostridiales; Syntrophomonadaceae; Syntrophomonas | ✓ | ✓ | ✓ | ✓ | |
| OTU | Hydrogenotrophic Methanogen | Observed Sample | |
|---|---|---|---|
| Family | Genus | ||
| OTU00061 | Methanoregulaceae | Methanoregula | Domestic |
| OTU00023 | Methanobacteriaceae | Methanobacterium | FruitJuice |
| OTU00105 | Methanobacteriaceae | Methanobacterium | FruitJuice |
| OTU00100 | Methanomicrobiaceae | Methanoculleus | PalmOil |
| OTU00036 | Methanoregulaceae | Metanoregula | PigManure |
| OTU00097 | Methanoregulaceae | Metanoregula | PigManure |
| OTU00015 | Methanoregulaceae | Methanolinea | Starch |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puengrang, P.; Suraraksa, B.; Prommeenate, P.; Boonapatcharoen, N.; Cheevadhanarak, S.; Tanticharoen, M.; Kusonmano, K. Diverse Microbial Community Profiles of Propionate-Degrading Cultures Derived from Different Sludge Sources of Anaerobic Wastewater Treatment Plants. Microorganisms 2020, 8, 277. https://doi.org/10.3390/microorganisms8020277
Puengrang P, Suraraksa B, Prommeenate P, Boonapatcharoen N, Cheevadhanarak S, Tanticharoen M, Kusonmano K. Diverse Microbial Community Profiles of Propionate-Degrading Cultures Derived from Different Sludge Sources of Anaerobic Wastewater Treatment Plants. Microorganisms. 2020; 8(2):277. https://doi.org/10.3390/microorganisms8020277
Chicago/Turabian StylePuengrang, Pantakan, Benjaphon Suraraksa, Peerada Prommeenate, Nimaradee Boonapatcharoen, Supapon Cheevadhanarak, Morakot Tanticharoen, and Kanthida Kusonmano. 2020. "Diverse Microbial Community Profiles of Propionate-Degrading Cultures Derived from Different Sludge Sources of Anaerobic Wastewater Treatment Plants" Microorganisms 8, no. 2: 277. https://doi.org/10.3390/microorganisms8020277
APA StylePuengrang, P., Suraraksa, B., Prommeenate, P., Boonapatcharoen, N., Cheevadhanarak, S., Tanticharoen, M., & Kusonmano, K. (2020). Diverse Microbial Community Profiles of Propionate-Degrading Cultures Derived from Different Sludge Sources of Anaerobic Wastewater Treatment Plants. Microorganisms, 8(2), 277. https://doi.org/10.3390/microorganisms8020277




