Regulatory Patterns of Crp on Monensin Biosynthesis in Streptomyces cinnamonensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Chemicals and Growth Conditions
2.2. Plasmid Constructions for Gene Overexpression and Silencing
2.3. Construction of Engineered S. cinnamonensis Strains
2.4. RNA-Seq Library Preparation, Clustering and Sequencing
2.5. Quantification and Differential Analysis of Gene Expression
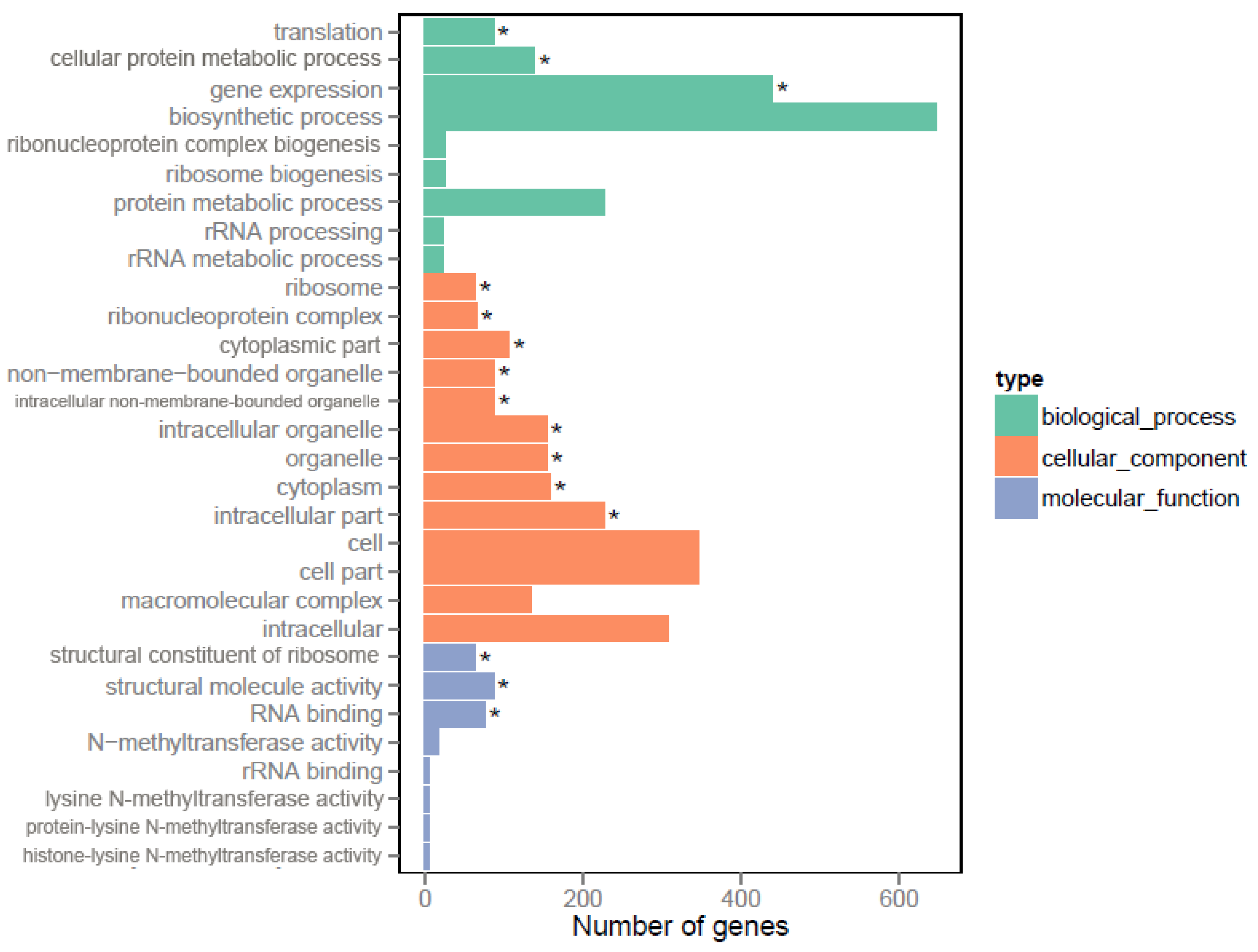
2.6. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
2.7. Semi-Quantitative RT-PCR
2.8. Extraction and Analysis of Monensin
3. Results
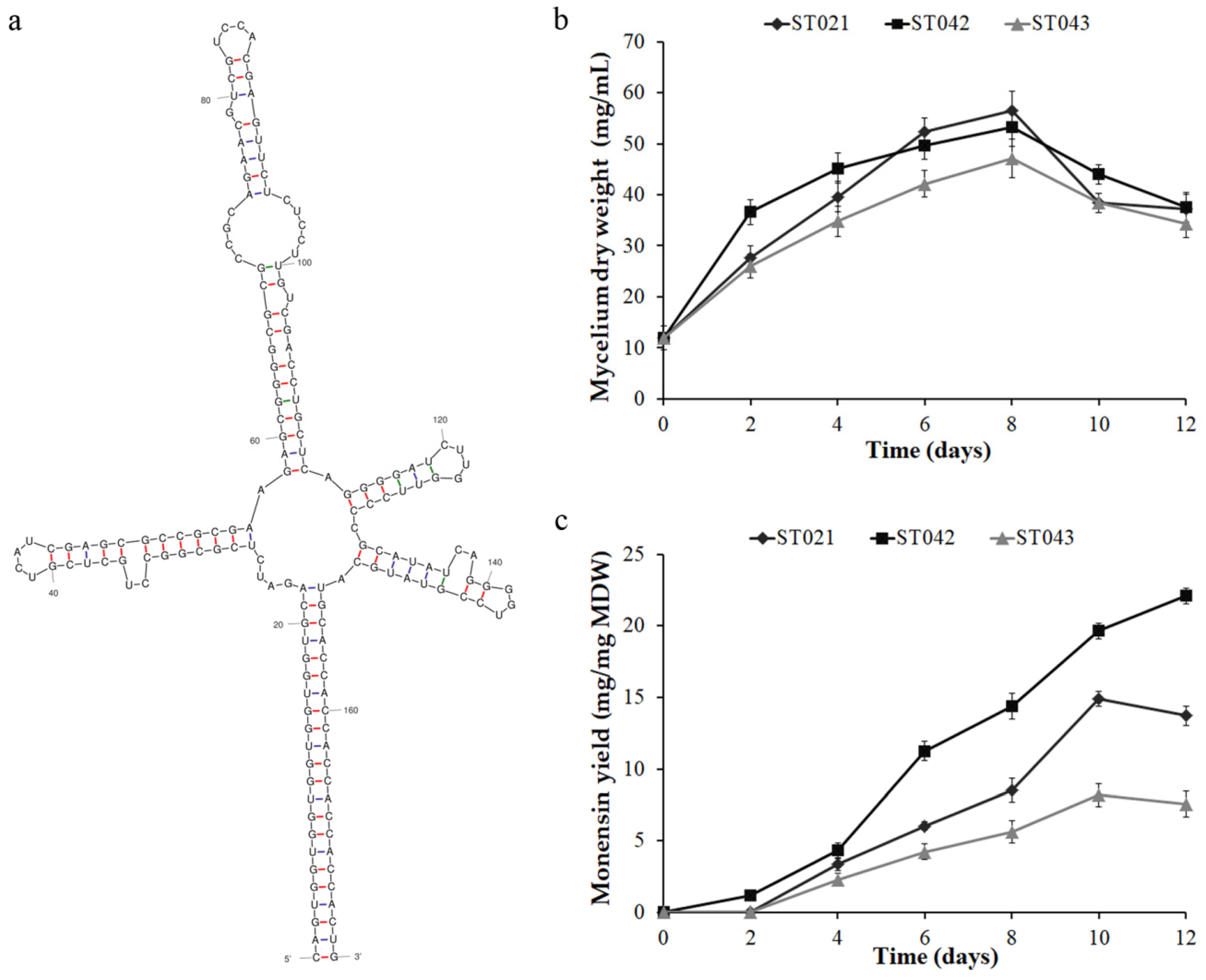
3.1. Crp Positively Regulates Monensin Biosynthesis
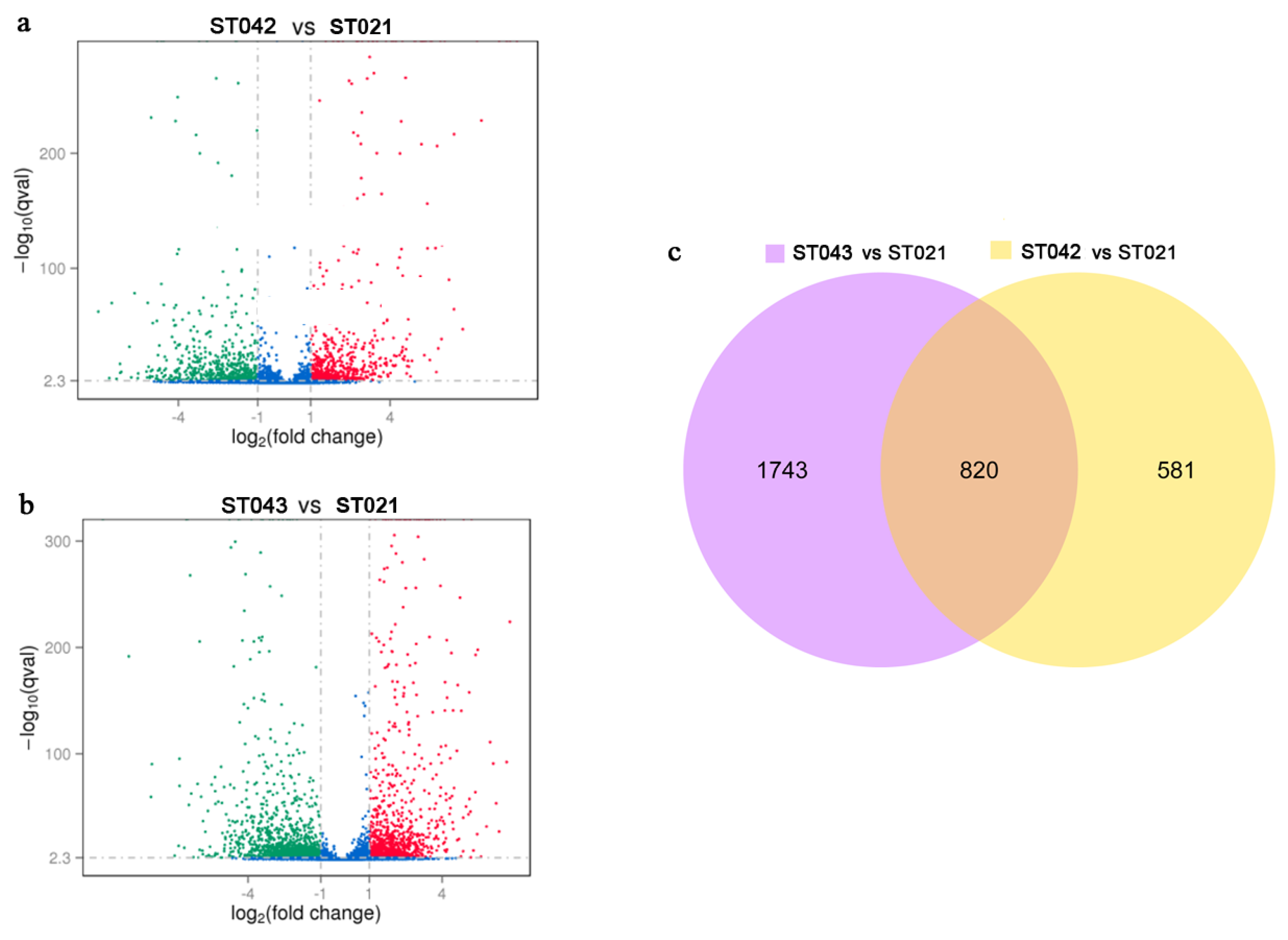
3.2. RNA-Seq Shows Global Regulatory Roles of Crp
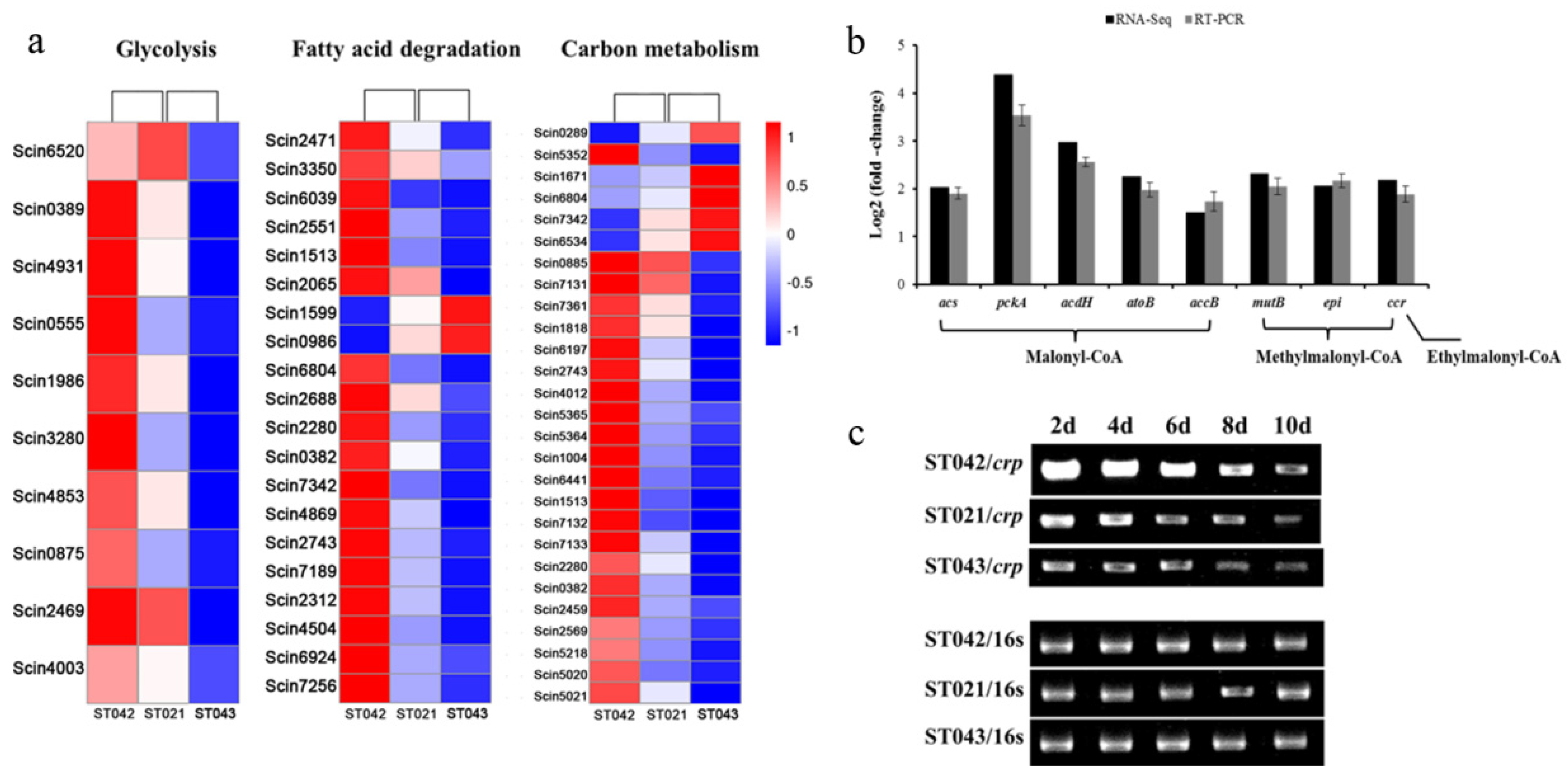
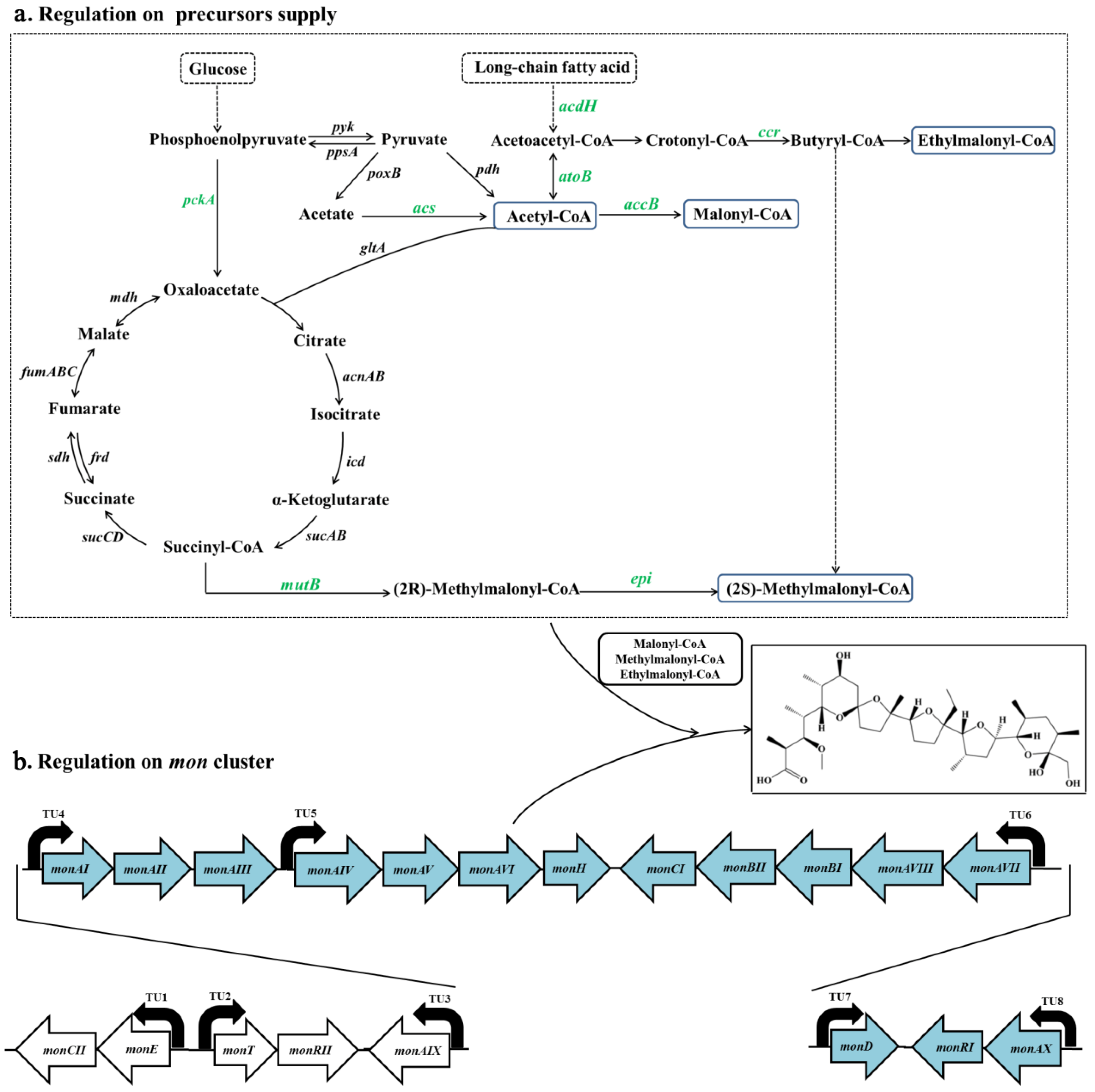
3.3. Crp Upregulates Biosynthetic Pathways of Monensin Precursors
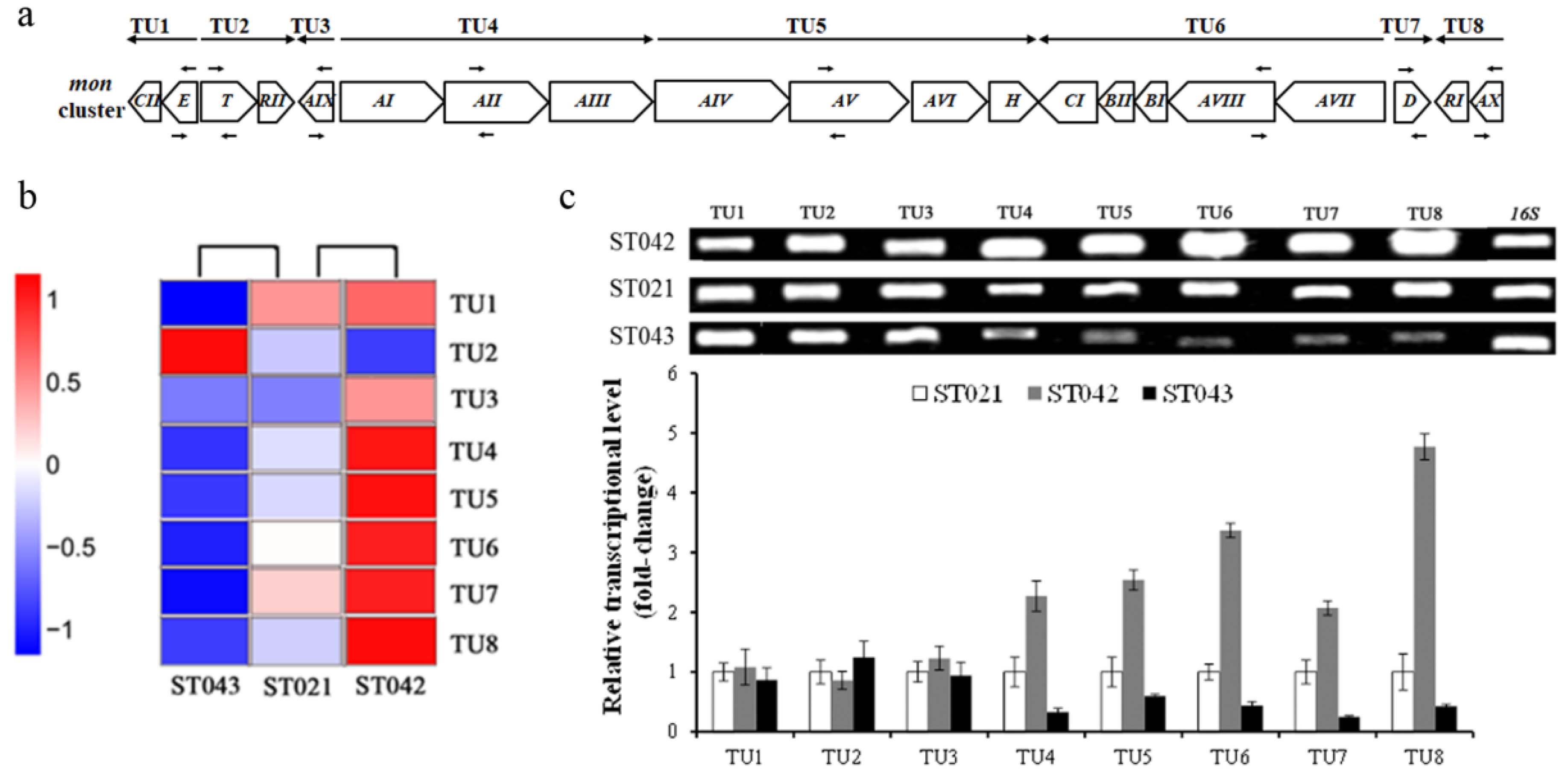
3.4. Crp Upregulates Transcription of mon Cluster
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Baltz, R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 2016, 43, 343–370. [Google Scholar] [CrossRef]
- Urem, M.; Świątek-Połatyńska, M.A.; Rigali, S.; van Wezel, G.P. Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol. Microbiol. 2016, 102, 183–195. [Google Scholar] [CrossRef]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Lett. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Nosho, K.; Fukushima, H.; Asai, T.; Nishio, M.; Takamaru, R.; Kobayashi-Kirschvink, K.J.; Ogawa, T.; Hidaka, M.; Masaki, H. cAMP-CRP acts as a key regulator for the viable but non-culturable state in Escherichia coli. Microbiology 2018, 164, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Grasso, L.L.; Martino, D.C.; Alduina, R. Production of antibacterial compounds from Actinomycetes. In Actinobacteria-Basics and Biotechnological Applications; IntechOpen: London, UK, 2016; pp. 177–198. [Google Scholar]
- Derouaux, A.; Dehareng, D.; Lecocq, E.; Halici, S.; Nothaft, H.; Giannotta, F.; Moutzourelis, G.; Dusart, J.; Devreese, B.; Titgemeyer, F. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem. Biophys. Res. Commun. 2004, 325, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Derouaux, A.; Halici, S.; Nothaft, H.; Neutelings, T.; Moutzourelis, G.; Dusart, J.; Titgemeyer, F.; Rigali, S. Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. J. Bacteriol. 2004, 186, 1893–1897. [Google Scholar] [CrossRef] [PubMed]
- Piette, A.; Derouaux, A.; Gerkens, P.; Noens, E.E.; Mazzucchelli, G.; Vion, S.; Koerten, H.K.; Titgemeyer, F.; De Pauw, E.; Leprince, P. From dormant to germinating spores of Streptomyces coelicolor A3 (2): New perspectives from the crp null mutant. J. Proteome Res. 2005, 4, 1699–1708. [Google Scholar] [CrossRef]
- Gao, C.; Hindra; Mulder, D.; Yin, C.; Elliot, M.A. Crp is a global regulator of antibiotic production in streptomyces. mBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.H.; Yao, L.l.; Ye, B.C. Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators GlnR, DasR, and CRP. Mol. Microbiol. 2014, 94, 1065–1084. [Google Scholar] [CrossRef]
- Chapman, H.D.; Jeffers, T.K.; Williams, R.B. Forty years of monensin for the control of coccidiosis in poultry. Poult. Sci. 2010, 89, 1788–1801. [Google Scholar] [CrossRef]
- Ketola, K.; Vainio, P.; Fey, V.; Kallioniemi, O.; Iljin, K. Monensin is a potent inducer of oxidative stress and inhibitor of androgen signaling leading to apoptosis in prostate cancer cells. Mol. Cancer Ther. 2010, 9, 3175–3185. [Google Scholar] [CrossRef] [PubMed]
- Tumova, L.; Pombinho, A.R.; Vojtechova, M.; Stancikova, J.; Gradl, D.; Krausova, M.; Sloncova, E.; Horazna, M.; Kriz, V.; Machonova, O. Monensin inhibits canonical Wnt signaling in human colorectal cancer cells and suppresses tumor growth in multiple intestinal neoplasia mice. Mol. Cancer Ther. 2014, 13, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhang, J.; Wang, Z.; Yan, Z.; Qiao, M.; Ye, J.; Wei, Q.; Wang, J.; Wang, X.; Zhao, L.; et al. Antibiotic monensin synergizes with EGFR inhibitors and oxaliplatin to suppress the proliferation of human ovarian cancer cells. Sci. Rep. 2015, 5, 17523. [Google Scholar] [CrossRef]
- Leadlay, P.; Staunton, J.; Oliynyk, M.; Bisang, C.; Cortes, J.; Frost, E.; Hughes-Thomas, Z.; Jones, M.; Kendrew, S.; Lester, J. Engineering of complex polyketide biosynthesis—insights from sequencing of the monensin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 2001, 27, 360–367. [Google Scholar] [CrossRef]
- Oliynyk, M.; Stark, C.B.; Bhatt, A.; Jones, M.A.; Hughes-Thomas, Z.A.; Wilkinson, C.; Oliynyk, Z.; Demydchuk, Y.; Staunton, J.; Leadlay, P.F. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol. Microbiol. 2003, 49, 1179–1190. [Google Scholar] [CrossRef]
- Risdian, C.; Mozef, T.; Wink, J. Biosynthesis of polyketides in Streptomyces. Microorganisms. 2019, 7, 124. [Google Scholar] [CrossRef]
- Gallimore, A.R.; Stark, C.B.; Bhatt, A.; Harvey, B.M.; Demydchuk, Y.; Bolanos-Garcia, V.; Fowler, D.J.; Staunton, J.; Leadlay, P.F.; Spencer, J.B. Evidence for the role of the monB genes in polyether ring formation during monensin biosynthesis. Chem. Biol. 2006, 13, 453–460. [Google Scholar] [CrossRef]
- Sato, K.; Minami, A.; Ose, T.; Oguri, H.; Oikawa, H. Remarkable synergistic effect between MonBI and MonBII on epoxide opening reaction in ionophore polyether monensin biosynthesis. Tetrahedron Lett. 2011, 52, 5277–5280. [Google Scholar] [CrossRef]
- Minami, A.; Ose, T.; Sato, K.; Oikawa, A.; Kuroki, K.; Maenaka, K.; Oguri, H.; Oikawa, H. Allosteric regulation of epoxide opening cascades by a pair of epoxide hydrolases in monensin biosynthesis. ACS Synth. Biol. 2013, 9, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Hüttel, W.; Spencer, J.B.; Leadlay, P.F. Intermediates in monensin biosynthesis: A late step in biosynthesis of the polyether ionophore monensin is crucial for the integrity of cation binding. Beilstein. J. Org. Chem. 2014, 10, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Harvey, B.M.; Hong, H.; Jones, M.A.; Hughes-Thomas, Z.A.; Goss, R.M.; Heathcote, M.L.; Bolanos-Garcia, V.M.; Kroutil, W.; Staunton, J.; Leadlay, P.F. Evidence that a novel thioesterase is responsible for polyketide chain release during biosynthesis of the polyether ionophore monensin. ChemBioChem. 2006, 7, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.-K.; Li, X.-M.; Pang, A.-P.; Lin, C.-Y.; Zhang, Y.; Zhang, J.; Qiao, J.; Zhao, G.-R. Characterization of three pathway-specific regulators for high production of monensin in Streptomyces cinnamonensis. Appl. Microbiol. Biotechnol. 2017, 101, 6083–6097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, C.-Y.; Li, X.-M.; Tang, Z.-K.; Qiao, J.; Zhao, G.-R. DasR positively controls monensin production at two-level regulation in Streptomyces cinnamonensis. J. Ind. Microbiol. Biotechnol. 2016, 43, 1681–1692. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical streptomyces genetics; John Innes Foundation Norwich: Norwich, UK, 2000. [Google Scholar]
- Uguru, G.C.; Mondhe, M.; Goh, S.; Hesketh, A.; Bibb, M.J.; Good, L.; Stach, J.E. Synthetic RNA silencing of actinorhodin biosynthesis in Streptomyces coelicolor A3 (2). PLoS ONE 2013, 8, e67509. [Google Scholar] [CrossRef][Green Version]
- Pang, A.-P.; Du, L.; Lin, C.-Y.; Qiao, J.; Zhao, G.-R. Co-overexpression of lmbW and metK led to increased lincomycin A production and decreased byproduct lincomycin B content in an industrial strain of Streptomyces lincolnensis. J. Appl. Microbiol. 2015, 119, 1064–1074. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009, 26, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome. Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, O.; Luzhetskyy, A. Metabolic engineering of natural product biosynthesis in actinobacteria. Curr. Opin. Biotechnol. 2016, 42, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Haydock, S.F.; Aparicio, J.F.; Molnár, I.; Schwecke, T.; Khaw, L.E.; König, A.; Marsden, A.F.; Galloway, I.S.; Staunton, J.; Leadlay, P.F. Divergent sequence motifs correlated with the substrate specificity of (methyl) malonyl-CoA: Acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 1995, 374, 246–248. [Google Scholar] [CrossRef]
- Zhang, W.; Reynolds, K.A. MeaA, a putative coenzyme B12-dependent mutase, provides methylmalonyl coenzyme A for monensin biosynthesis in Streptomyces cinnamonensis. J. Bacteriol. 2001, 183, 2071–2080. [Google Scholar] [CrossRef]
- Li, C.; Florova, G.; Akopiants, K.; Reynolds, K.A. Crotonyl-coenzyme A reductase provides methylmalonyl-CoA precursors for monensin biosynthesis by Streptomyces cinnamonensis in an oil-based extended fermentation. Microbiology. 2004, 150, 3463–3472. [Google Scholar] [CrossRef]
- Shen, B.; Hutchinson, C.R. Enzymatic synthesis of a bacterial polyketide from acetyl and malonyl coenzyme A. Science 1993, 262, 1535–1540. [Google Scholar] [CrossRef] [PubMed]
- Cummings, M.; Breitling, R.; Takano, E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol. Lett. 2014, 351, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.-G.; Butler, M.J.; Chater, K.F.; Lee, K.J. Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl. Environ. Microbiol. 2006, 72, 7132–7139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zabala, D.; Braña, A.F.; Flórez, A.B.; Salas, J.A.; Méndez, C. Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab. Eng. 2013, 20, 187–197. [Google Scholar] [CrossRef]
- Dayem, L.C.; Carney, J.R.; Santi, D.V.; Pfeifer, B.A.; Khosla, C.; Kealey, J.T. Metabolic engineering of a methylmalonyl-CoA mutase-epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry. 2002, 41, 5193–5201. [Google Scholar] [CrossRef]
- Chan, Y.A.; Podevels, A.M.; Kevany, B.M.; Thomas, M.G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009, 26, 90–114. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
| Strains/Plasmids | Characteristics | Reference or Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | Recipient for cloning experiments | Invitrogen |
| ET12567/pUZ8002 | Strain for conjugal transfer, with pUZ8002 helper plasmids, ChlR, KanR | [28] |
| Streptomyces cinnamonensis | ||
| ST021 | The parental strain derived from Streptomyces cinnamonensis ATCC15413 | This work |
| ST042 | ST021 with pLCY009-crp integrative expression vector, HygR | This work |
| ST043 | ST021 with pLCY010-csRNA replicative anti-crp RNA vector, HygR | This work |
| Plasmids | ||
| pLCY009 | pIB139 derivative, ΦC31 attachment site, integrative, carrying PermE* promoter, HygR | [27] |
| pLCY010 | pUWL201 derivative, pIJ101 replicon, replicative, carrying PermE* promoter, AmpR, TsrR, HygR | [30] |
| pLCY009-crp | crp overexpression vector, ΦC31 attachment site, integrative, carrying PermE* promoter, HygR | This work |
| pLCY010-csRNA | Anti-crp RNA vector, pIJ101 replicon, replicative, carrying PermE* promoter, HygR | This work |
| Samples | ST021 | ST042 | ST043 |
|---|---|---|---|
| Raw reads | 16,537,074 | 21,162,804 | 23,277,776 |
| Clean reads | 15,709,834 | 20,599,352 | 22,714,616 |
| Clean bases (G) | 1.96 | 2.57 | 2.84 |
| GC content (%) | 65.57 | 63.56 | 66.01 |
| Q20 Base ratio (%) * | 93.94 | 97.27 | 97.26 |
| Q30 Base ratio (%) | 88.06 | 93.11 | 93.05 |
| Mapped reads | 15,597,581 | 17,869,047 | 20,707,208 |
| Mapped reads ratio (%) | 99.29 | 86.75 | 91.16 |
| Pathway ID | Pathway | Number of DEGs | Representative Genes * |
|---|---|---|---|
| sco00010 | Glycolysis | 10 | acs, pckA |
| sco00071 | Fatty acid degradation | 20 | acdH, atoB |
| sco01200 | Carbon metabolism | 27 | accB, mutB, epi, ccr |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Zhang, Y.; Wu, J.-H.; Xie, R.-H.; Qiao, J.; Zhao, G.-R. Regulatory Patterns of Crp on Monensin Biosynthesis in Streptomyces cinnamonensis. Microorganisms 2020, 8, 271. https://doi.org/10.3390/microorganisms8020271
Lin C-Y, Zhang Y, Wu J-H, Xie R-H, Qiao J, Zhao G-R. Regulatory Patterns of Crp on Monensin Biosynthesis in Streptomyces cinnamonensis. Microorganisms. 2020; 8(2):271. https://doi.org/10.3390/microorganisms8020271
Chicago/Turabian StyleLin, Chun-Yan, Yue Zhang, Ji-Hua Wu, Rong-Hui Xie, Jianjun Qiao, and Guang-Rong Zhao. 2020. "Regulatory Patterns of Crp on Monensin Biosynthesis in Streptomyces cinnamonensis" Microorganisms 8, no. 2: 271. https://doi.org/10.3390/microorganisms8020271
APA StyleLin, C.-Y., Zhang, Y., Wu, J.-H., Xie, R.-H., Qiao, J., & Zhao, G.-R. (2020). Regulatory Patterns of Crp on Monensin Biosynthesis in Streptomyces cinnamonensis. Microorganisms, 8(2), 271. https://doi.org/10.3390/microorganisms8020271





