Processing of Metals and Metalloids by Actinobacteria: Cell Resistance Mechanisms and Synthesis of Metal(loid)-Based Nanostructures
Abstract
1. Introduction
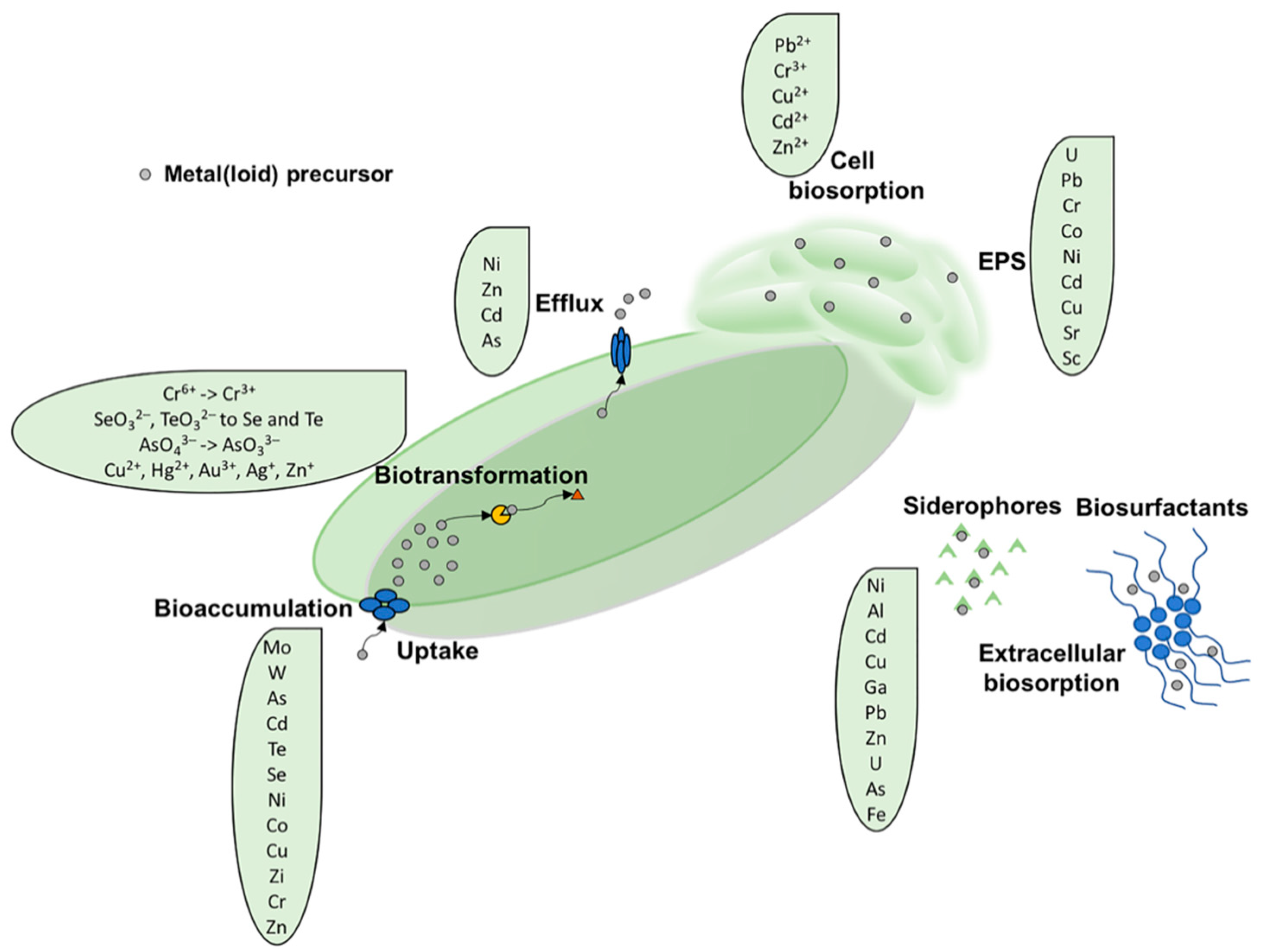
2. Mechanism(s) of Metal Tolerance and Resistance in Actinobacteria
2.1. Biosorption
2.1.1. Extracellular Sequestration by Siderophores
2.1.2. Extracellular Sequestration Mediated by Extracellular Polymeric Substance (EPS)
2.2. Bioaccumulation
2.3. Biotic Metal(loid) Reduction Reaction
2.4. Metal Efflux Systems
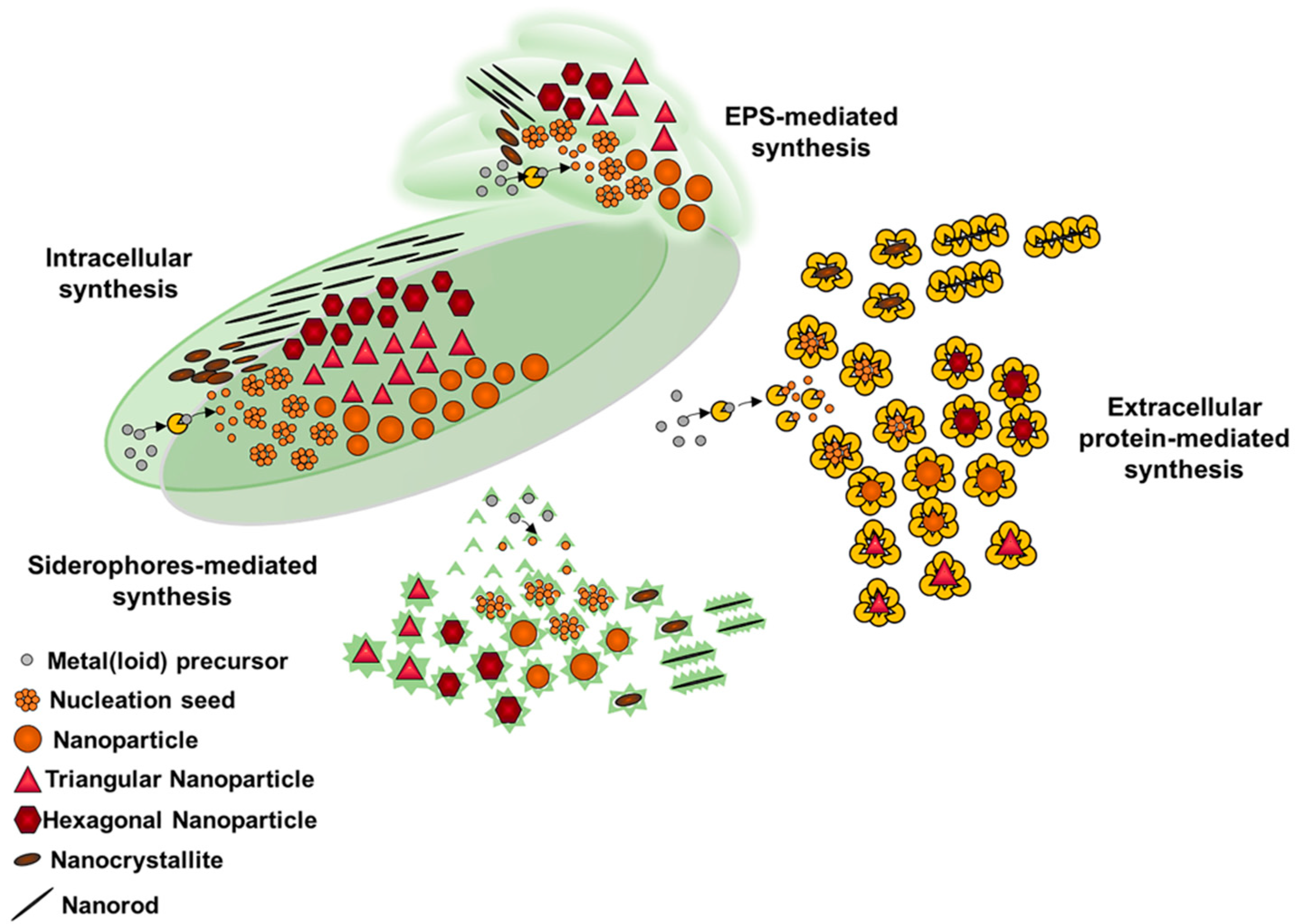
3. Metal(loid) Nanomaterial Biosynthesis by Actinobacteria
Properties and Applications of Metal(loid) NMs Produced by Actinobacteria
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kabata-Pendias, A.; Pendias, H. Trace Elements in Soils and Plants; CRC Press, Inc.: Boca Raton, FL, USA, 1984. [Google Scholar]
- Maret, W. The metals in the biological periodic system of the elements: Concepts and conjectures. Int. J. Mol. Sci. 2016, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Ledin, M. Accumulation of metals by microorganisms-processes and importance for soil systems. Earth Sci. Rev. 2000, 51, 1–31. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy Metal Toxicity and the Environment. In Molecular, Clinical and Environmental Toxicology; Luch, A., Ed.; Springer: Basel, Switzerland, 2012; Volume 3, pp. 133–164. [Google Scholar] [CrossRef]
- Barbosa, F., Jr. Toxicology of metals and metalloids: Promising issues for future studies in environmental health and toxicology. J. Toxicol. Environ. Health A 2017, 80, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Hamelink, J.; Landrum, P.F.; Bergman, H.; Benson, W.H. Bioavailability: Physical, Chemical and Biological Interactions; CRC Press: Boca Raton, FL, USA, 1994; ISBN 9781566700863. [Google Scholar]
- Presentato, A.; Cappelletti, M.; Sansone, A.; Ferreri, C.; Piacenza, E.; Demeter, M.A.; Crognale, S.; Petruccioli, M.; Milazzo, G.; Fedi, S.; et al. Aerobic growth of Rhodococcus aetherivorans BCP1 using selected naphthenic acids as the sole carbon and energy sources. Front. Microbiol. 2018, 9, 672. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, I.; Sathe, S.M.; Khuman, C.N.; Ghangrekar, M.M. Bioelectrochemically powered remediation of xenobiotic compounds and heavy metal toxicity using microbial fuel cell and microbial electrolysis cell. Mater. Sci. Technol. 2020, 3, 104–115. [Google Scholar] [CrossRef]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 2014, 5, e01918-14. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef]
- Alduina, R.; Gambino, D.; Presentato, A.; Gentile, A.; Sucato, A.; Savoca, D.; Filippello, S.; Visconti, G.; Caracappa, G.; Vicari, D.; et al. Is Caretta caretta a carrier of antibiotic resistance in the Mediterranean sea? Antibiotics 2020, 9, 116. [Google Scholar] [CrossRef]
- Ali, H.; Khanb, E.; Sajad, A.M. Phytoremediation of heavy metals-Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef]
- Abdel-Ghani, N.; El-Chagbaby, G. Biosorption for metal ions removal from aqueous solutions: A review of recent studies. Int. J. Latest Res. Sci. Technol. 2014, 3, 24–42. [Google Scholar]
- Alvarez, A.; Saez, J.M.; Davila Costa, J.S.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M. Microbial Control of Heavy Metal Pollution. In Microbial Control of Heavy Metal Pollution; Fry, J., Gadd, G.M., Herbert, R.A., Jones, C.W., Watson-Craik, I.A., Eds.; Cambridge University Press: Cambridge, UK, 1992; pp. 59–88. [Google Scholar]
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannoni, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nesvera, J.; Kren, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef]
- Goodfellow, M.; Kämpfer, P.; Busse, H.-J.; Trujillo, M.E.; Suzuki, K.-I.; Ludwig, W.; Whitman, W.B. Bergey’s Manual of Systematic Bacteriology: The Actinobacteria; Springer: New York, NY, USA, 2012; Volume 5. [Google Scholar]
- Cappelletti, M.; Zampolli, J.; Di Gennaro, P.; Zannoni, D. Chapter 2: Genomics of Rhodococcus. In Biology of Rhodococcus, 2nd ed.; Alvarez, H.M., Ed.; Springer International Publishing: Basel, Switzerland, 2019; pp. 23–60. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lampis, S.; Vallini, G.; Turner, R.J. Selenium and Tellurium nanomaterials. Phys. Sci. Rev. 2018, 3, 20170100. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Turner, R.J. Stability of biogenic metal(loid) nanomaterials related to the colloidal stabilization theory of chemical nanostructures. Crit. Rev. Biotechnol. 2018, 38, 1137–1156. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, B.S.; Chon, C.M. Characterization of iron and manganese minerals and their associated microbiota in different mine sites to reveal the potential interactions of microbiota with mineral formation. Chemosphere 2018, 191, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Solecka, J.; Zajko, J.; Postek, M.; Rajnisz, A. Biologically active secondary metabolites from Actinomycetes. Open Life Sci. 2012, 7, 373–390. [Google Scholar] [CrossRef]
- Liao, L.; Chen, R.; Jiang, M.; Tian, X.; Liu, H.; Yu, Y.; Fan, C.; Chen, B. Bioprospecting potential of halogenases from Arctic marine actinomycetes. BMC Microbiol. 2016, 16, 34. [Google Scholar] [CrossRef]
- Shivlata, L.; Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1014. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Volesky, B. Biosorption and Biosorbents. In Biosorption of Heavy Metals; Volesky, B., Ed.; CRC Press: Boca Raton, FL, USA, 1990; pp. 3–6. ISBN 9780849349171. [Google Scholar]
- Wilde, E.W.; Benemann, J.R. Bioremoval of heavy metals by the use of microalgae. Biotechnol. Adv. 1993, 4, 781–812. [Google Scholar] [CrossRef]
- Bankar, A.; Kumar, A.; Zinjarde, S. Removal of chromium (VI) ions from aqueous solution by adsorption onto two marine isolates of Yarrowia lipolytica. J. Hazard. Mater. 2009, 170, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Bankar, A.V.; Zinjarde, S.S.; Kapadnis, B.P. Management of Heavy Metal Pollution by Using Yeast Biomass. In Microorganisms in Environmental Management; Satyanarayana, T., Johri, B., Prakash, A., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 335–363. [Google Scholar] [CrossRef]
- Gadd, G.M.; White, C. Microbial treatment of metal pollution-a working biotechnology? Trends Biotechnol. 1993, 11, 353–359. [Google Scholar] [CrossRef]
- Goksungur, Y.; Üren, S.; Guvenc, U. Biosorption of cadmium and lead ions by ethanol treated waste baker’s yeast biomass. Bioresour. Technol. 2005, 96, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Plette, A.C.C.; van Riemsdijk, W.H.; Benedetti, M.F.; van der Wal, A. pH dependent charging behavior of isolated cell walls of a gram-positive soil bacterium. J. Colloid Interface Sci. 1995, 173, 354–363. [Google Scholar] [CrossRef]
- Stratton, H.; Brooks, P.; Griffiths, P.; Seviour, R. Cell surface hydrophobicity and mycolic acid composition of Rhodococcus strains isolated from activated sludge foam. J. Ind. Microbiol. Biotechnol. 2002, 28, 264–267. [Google Scholar] [CrossRef]
- Botero, A.E.C.; Torem, M.L.; de Mesquita, L.M.S. Fundamental studies of Rhodococcus opacus as a biocollector of calcite and magnesite. Miner. Eng. 2007, 20, 1026–1032. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, X.; Wang, B.; Mohamad, O.; Wei, G. Bioaccumulation characterization of zinc and cadmium by Streptomyces zinciresistens, a novel actinomycete. Ecotoxicol. Environ. Saf. 2012, 77, 717. [Google Scholar] [CrossRef]
- Sheng, P.X.; Tan, L.H.; Chen, J.P.; Ting, Y.P. Biosorption performance of two brown marine algae for removal of chromium and cadmium. J. Disper. Sci. Technol. 2004, 25, 679–686. [Google Scholar] [CrossRef]
- Vasquez, T.G.P.; Botero, A.E.C.; de Mesquita, L.M.S.; Torem, M.L. Biosorptive removal of Cd and Zn from liquid streams with a Rhodococcus opacus strain. Miner. Eng. 2007, 20, 939–944. [Google Scholar] [CrossRef]
- Bueno, B.Y.M.; Torem, M.L.; Molina, F.; de Mesquita, L.M.S. Biosorption of lead (II), chromium (III) and copper (II) by R. opacus: Equilibrium and kinetic studies. Miner. Eng. 2008, 21, 65–75. [Google Scholar] [CrossRef]
- Cayllahua, J.E.B.; Torem, M.L. Biosorption of aluminum ions onto Rhodococcus opacus from wastewaters. Chem. Eng. J. 2010, 161, 1–8. [Google Scholar] [CrossRef]
- Kumar, S.; Krishnan, K. Biosorption of Cd (II) and Pb (II) ions by aqueous solutions of novel alkalophillic Streptomyces VITSVK5 spp. biomass. J. Ocean Univ. China 2011, 10, 61–66. [Google Scholar] [CrossRef]
- Kumar, S.; Krishnan, K. Biosorption of Cr (III) and Cr (VI) by Streptomyces VITSVK9 spp. Ann. Microbiol. 2011, 61, 833–841. [Google Scholar] [CrossRef]
- Latha, S.; Vinothini, G.; Dhanasekaran, D. Chromium [Cr (VI)] biosorption property of the newly isolated actinobacterial probiont Streptomyces werraensis LD22. 3 Biotech 2015, 5, 423432. [Google Scholar] [CrossRef]
- Kirova, G.; Velkova, Z.; Gochev, V. Copper (II) removal by heat inactivated Streptomyces fradiae biomass: Surface chemistry characterization of the biosorbent. J. BioSci. Biotechnol. 2012, 2012, 77–82. [Google Scholar]
- Doyle, R.J.; Matthews, T.H.; Streips, U.N. Chemical basis for the selectivity of metal ions by the Bacillus subtilis wall. J Bacteriol. 1980, 143, 471–480. [Google Scholar] [CrossRef]
- Xue, H.B.; Stumm, W.; Sigg, L. The binding of heavy metals to algal surfaces. Water Res. 1988, 22, 917–926. [Google Scholar] [CrossRef]
- Flemming, C.A.; Ferris, F.G.; Beveridge, T.J.; Bailey, G.W. Remobilization of toxic heavy metals absorbed to wall-clay composites. Appl. Environ. Microbiol. 1990, 56, 3191–3209. [Google Scholar] [CrossRef]
- Plette, A.C.C.; Benedetti, M.F.; Vanriemsdijk, W.H. Competitive binding of protons, calcium, cadmium, and zinc to isolated cell walls of a Gram-positive soil bacterium. Environ. Sci. Technol. 1996, 30, 1902–1910. [Google Scholar] [CrossRef]
- Koch, A.L. Growth and form of the bacterial cell wall. Am. Sci. 1990, 78, 327–341. [Google Scholar]
- Kraemer, S.M. Iron oxide dissolution and solubility in the presence of siderophores. Aquat. Sci. 2004, 66, 3–18. [Google Scholar] [CrossRef]
- Timkova, I.; Sedlakova-Kadukova, J.; Pristas, P. Biosorption and bioaccumulation abilities of Actinomycetes/Streptomyces isolated from metal contaminated sites. Separations 2018, 5, 54. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Hopwood, D.A. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 2003, 100, 14555–14561. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Matzanke, B.F. Structures, coordination chemistry and functions of microbial iron chelates. In Handbook of Microbial Iron Chelates; Matzanke, B.F., Ed.; CRC Press: Boca Raton, FL, USA, 1991; pp. 23–72. [Google Scholar]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Schalk, I.J.; Hannauer, M.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef]
- John, S.G.; Ruggiero, C.E.; Hersman, L.E.; Tung, C.S.; Neu, M.P. Siderophore mediated Plutonium accumulation by Microbacterium flavescens (JG-9). Environ. Sci. Technol. 2001, 35, 2942–2948. [Google Scholar] [CrossRef]
- Barzanti, R.; Ozino, F.; Bozzicalupo, M.; Gabbrielli, R.; Galardi, F.; Gonnelli, C.; Mengoni, A. Isolation and characterization of endophytic bacteria from Nickel hyperaccumulator plant Alyssum bertolonii. Microb. Ecol. 2007, 53, 306–316. [Google Scholar] [CrossRef]
- Kuffner, M.; Puschenreiter, M.; Wieshammer, G.; Gorfer, M.; Sessitsch, A. Rhizosphere bacteria affect growth and metal uptake of heavy metal accumulating willows. Plant Soil 2008, 304, 35–44. [Google Scholar] [CrossRef]
- Sheng, X.F.; Xia, J.J.; Jiang, C.Y.; He, L.Y.; Qian, M. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 2008, 156, 1164–1170. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Svatos, A.; Merten, D.; Buchel, G.; Kothe, E. Hydroxamate siderophores produced by Streptomyces acidiscabies E13 bind nickel and promote growth in cowpea (Vigna unguiculate L.) under nickel stress. Can. J. Microbiol. 2008, 54, 163–172. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Svatos, A.; Dabrowska, P.; Schmidt, A.; Boland, W.; Kothe, E. Involvement of siderophores in the reduction of metal-induced inhibition of auxin synthesis in Streptomyces spp. Chemosphere 2008, 74, 19–25. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Merten, D.; Svatos, A.; Buchel, G.; Kothe, E. Siderophores mediate reduced and increased uptake of cadmium by Streptomyces tendae F4 and sunflower (Helianthus annuus), respectively. J. Appl. Microbiol. 2009, 107, 1687–1696. [Google Scholar] [CrossRef]
- Kuffner, M.; De Maria, S.; Puschenreiter, M.; Fallmann, K.; Wieshammer, G.; Gorfer, M.; Strauss, J.; Rivelli, A.R.; Sessitsch, A. Culturable bacteria from Zn- and Cd-accumulating Salix caprea with differential effects on plant growth and heavy metal availability. J. Appl. Microbiol. 2010, 108, 1471–1484. [Google Scholar] [CrossRef]
- Sun, L.N.; Zhang, Y.F.; He, L.Y.; Chen, Z.J.; Qang, Q.Y.; Qian, M.; Sheng, X.F. Genetic diversity and characterization of heavy metal-resistant-endophytic bacteria from two copper-tolerant species on copper mine wasteland. Biores. Technol. 2010, 101, 501–509. [Google Scholar] [CrossRef]
- Visioli, G.; D’Egidio, S.; Vamerali, T.; Mattarozzi, M.; Sanangelantoni, A.M. Culturable endophytica bacteria enhance Ni translocation in the hyperaccumulator Noccaea caerulescens. Chemosphere 2014, 117, 538–544. [Google Scholar] [CrossRef]
- Schutze, E.; Ahmed, E.; Voit, A.; Klose, M.; Greyer, M.; Svatos, A.; Merten, D.; Roth, M.; Holmstrom, S.J.M.; Kothe, E. Siderophores production by Streptomycetes—Stability and alteration of ferrihydroxamates in heavy metal-contaminated soil. Environ. Sci. Pollut. Res. 2015, 22, 19376–19383. [Google Scholar] [CrossRef]
- El Baz, S.; Baz, M.; Barakate, M.; Hassani, L.; El Gharmali, A.; Imziln, B. Resistance to and accumulation of heavy metals by Actinobacteria isolated from abandoned mining areas. Sci. World J. 2015, 2015, 761834. [Google Scholar] [CrossRef]
- Zloch, M.; Thiem, D.; Gadzala-Kopciuch, R.; Hrynkiewicz, K. Synthesis of siderophores by plant-associated metallotolerant bacteria under exposure to Cd2+. Chemosphere 2016, 156, 312–325. [Google Scholar] [CrossRef]
- Retamal-Morales, G.; Menhert, M.; Schwabe, R.; Tischer, D.; Zapata, C.; Chavez, R. Detection of arsenic-binding siderophores in arsenic-tolerating Actinobacteria by a modified CAS assay. Ecotoxicol. Environ. Saf. 2018, 157, 176–181. [Google Scholar] [CrossRef]
- Uranga, C.; Arroyo, P.J.; Duggan, B.M.; Gerwick, W.H.; Edlund, A. Commensal oral Rothia mucilaginosa produces enterobactin—A metal chelating siderophore. mSystems 2020, 5, e001621-20. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, Q.; Liu, X.; Chen, P.; Guo, X.; Ma, L.Z.; Dong, H.; Huang, Y. Iron reduction by diverse actinobacteria under oxic and pH-neutral conditions and the formation of secondary minerals. Chem. Geol. 2019, 525, 390–399. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Freitas, H. Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 2009, 77, 153–160. [Google Scholar] [CrossRef]
- Van der Lelie, D.; Corbisier, P.; Diels, L.; Gilis, A.; Lodewyckx, C.; Mergeay, M.; Taghavi, S.; Spelmans, N.; Vangronsveld, J. The role of bacteria in the phytoremediation of heavy metals. In Phytoremediation of Contaminated Soil and Water; Terry, N., Banuelos, G.S., Eds.; CRC Press: Boca Raton, FL, USA, 2000; pp. 265–281. [Google Scholar]
- Djuric, A.; Gojgic-Cvijovic, G.; Jakovljevic, D.; Kekez, B.; Stefanovic Kojic, J.; Mattinen, M.L.; Haru, I.E.; Vrvic, M.M.; Beskoski, V. Brachybacterium sp. CH-KOV3 isolated from an oil-polluted environment—A new producer of levan. Int. J. Biol. Macromol. 2017, 104, 311–321. [Google Scholar] [CrossRef]
- Presentato, A.; Piacenza, E.; Cappelletti, M.; Turner, R.J. Interaction of Rhodococcus with metals and biotechnological applications. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; pp. 333–357. [Google Scholar]
- Harrison, J.J.; Turner, R.J.; Marques, L.L.R.; Ceri, H. Biofilms: A new understanding of these microbial communities is driving a revolution that may transform the science of microbiology. Amer. Sci. 2005, 93, 508–515. [Google Scholar] [CrossRef]
- Shuhong, Y.; Meiping, Z.; Hong, Y.; Han, W.; Shan, X. Biosorption of Cu2+, Pb2+ and Cr6+ by a novel exopolysaccharide from Artrhobacter ps-5. Carbohydr. Polym. 2014, 101, 50–56. [Google Scholar] [CrossRef]
- Bankar, A.; Nagaraja, G. Recent trends in biosorption of heavy metals by Actinobacteria. In New and Future Developments in Microbial Biotechnology and Bioengineering—Actinobacteria: Diversity and Biotechnological Applications; Singh, B.P., Gupta, K., Passari, A.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 257–275. [Google Scholar]
- Xiong, Y.W.; Ju, X.Y.; Li, X.W.; Gong, Y.; Xu, M.J.; Zhang, C.M.; Yuan, B.; Lv, Z.P.; Qin, S. Fermentation conditions optimization, purification, and antioxidant activity of exopolysaccharides obtained from the plant growth-promoting endophytic actinobacterium Glutamicibacter halophytocola KLBMP 5180. Int. J. Biol. Macromol. 2019, 153, 1176–1185. [Google Scholar] [CrossRef]
- Neal, A.L.; Dublin, S.N.; Taylor, J.; Bates, D.J.; Burns, J.L.; Apkarian, R.; DiChristina, T.J. Terminal electron acceptors influence the quantity and chemical composition of capsular exopolymers produced by anaerobically growing Shewanella spp. Biomacromolecules 2007, 8, 166–174. [Google Scholar] [CrossRef]
- Solis, M.; Solis, A.; Perez, H.I.; Manjarrez, N.; Flores, M. Microbial decolouration of azo dyes: A review. Process Biochem. 2012, 47, 1723–1748. [Google Scholar] [CrossRef]
- Dobrowolski, R.; Szczes, A.; Czemierska, M.; Jarosz-Wikolazka, A. Studies of cadmium(II), lead(II), nickel(II), cobalt(II) and chromium(IV) sorption on extracellular polymeric substances produced by Rhodococcus opacus and Rhodococcus rhodochrous. Biores. Technol. 2017, 225, 113–120. [Google Scholar] [CrossRef]
- Sivaperumal, P.; Kamala, K.; Rajaram, R. Adsorption of cesium ion by marine actinobacterium Nocardiopsis sp. 13H and their extracellular polymeric substances (EPS) role in bioremediation. Environ. Sci. Pollut. Res. 2018, 25, 4254–4267. [Google Scholar] [CrossRef]
- Kamala, K.; Sivaperumal, P.; Thilagaraj, R.; Natarajan, E. Bioremediation of Sr2+ ion radionuclide by using marine Streptomyces sp. CuOff24 extracellular polymeric substances. J. Chem. Technol. Biotechnol. 2020, 95, 893–903. [Google Scholar] [CrossRef]
- Liu, H.; Fang, H.H. Characterization of electrostatic binding sites of extracellular polymers by linear programming analysis of titration data. Biotechnol. Bioeng. 2002, 30, 806–811. [Google Scholar] [CrossRef]
- Wei, X.; Fang, L.; Cai, P.; Huang, Q.; Chen, H.; Liang, W.; Rong, X. Influence of extracellular polymeric substances (EPS) on Cd adsorption by bacteria. Environ. Pollut. 2011, 159, 1369–1374. [Google Scholar] [CrossRef]
- Roane, T.M.; Josephson, K.L.; Pepper, I.L. Dual-Bioaugmentation Strategy to Enhance Remediation of Cocontaminated Soil. Appl. Environ. Microbiol. 2001, 67, 3208–3215. [Google Scholar] [CrossRef]
- Albarracin, V.H.; Winik, B.; Kothe, E.; Amoroso, M.J.; Abate, C.M. Copper bioaccumulation by the actinobacterium Amycolatopsis sp. AB0. J. Basic Microbiol. 2008, 48, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, J.; Diestra, E.; Huang, L.; Domenech, A.M.; Villagrasa, E.; Puyen, Z.M.; Duran, R.; Esteve, I.; Solé, A. Isolation and identification of a bacterium with high tolerance to lead and copper from a marine microbial mat in Spain. Ann. Microbiol. 2010, 60, 113–120. [Google Scholar] [CrossRef]
- Puyen, Z.M.; Villagrasa, E.; Maldonado, J.; Diestra, E.; Esteve, I.; Sole, A. Biosorption of lead and copper by heavy-metal tolerant Micrococcus luteus DE2008. Biores. Technol. 2012, 126, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Quintelas, C.; da Silva, V.B.; Silva, B.; Figueiredo, H.; Tavares, T. Optimization of production of extracellular polymeric substances by Arthrobacter viscosus and their interaction with a 13X zeoilite for the biosorption of Cr(IV). Environ. Technol. 2011, 32, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- More, T.T.; Yan, S.; John, R.P.; Tyagi, R.D.; Surampalli, R.Y. Biochemical diversity of the bacterial strains and their biopolymer producing capabilities in wastewater sludge. Biores. Technol. 2012, 121, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Vijay, V.; Vandana, K.B.; Kumar, R.M.; Prabagaran, S.R. Genetic analysis of arsenic metabolism in Micrococcus luteus BPB1, isolated from the Bengal basin. Ann. Microbiol. 2017, 67, 79–89. [Google Scholar] [CrossRef]
- Furnholm, T.; Rehan, M.; Wishart, J.; Tisa, L.S. Pb2+ tolerance by Frankia sp. strain EAN1pec involves surface-binding. Microbiology 2017, 163, 472–487. [Google Scholar] [CrossRef]
- Mulik, A.; Bhadekar, R. Extracellular polymeric substance (EPS) from Kocuria sp. BRI 36: A key component in heavy metal resistance. Int. J. Pharm. Pharm. Sci. 2018, 10, 50–54. [Google Scholar] [CrossRef][Green Version]
- Sivaperumal, P.; Kamala, K.; Rajaram, R. Biosorption of long half-life radionuclide of Strontium ion (Sr+) by marine actinobacterium Nocardiopsis sp. 13H. Geomicrobiol. J. 2018, 35, 300–310. [Google Scholar] [CrossRef]
- Pal, A.; Paul, A.K. Microbial extracellular polymeric substances central elements in heavy metal bioremediation. Indian J. Microbiol. 2008, 48, 49–64. [Google Scholar] [CrossRef]
- Han, R.P.; Li, H.K.; Li, Y.H.; Zhang, J.H.; Xiao, H.J.; Shi, J. Biosorption of copper and lead ions by waste beer yeast. J. Hazard. Mater. 2006, 137, 1569–1576. [Google Scholar] [CrossRef]
- Lamelas, C.; Benedetti, M.; Wilkinson, K.J.; Slaveykova, V.I. Characterization of H+ and Cd(II) binding properties of the bacterial exopolysaccharides. Chemosphere 2006, 65, 1362–1370. [Google Scholar] [CrossRef]
- Veglio, F.; Beolchini, F. Removal of metals by biosorption: A review. Hydrometallurgy 1997, 44, 301–306. [Google Scholar] [CrossRef]
- Avery, S.V. Microbial interactions with caesium—Implications for biotechnology. J. Chem. Technol. Biotechnol. 1995, 62, 3–16. [Google Scholar] [CrossRef]
- Brooks, A.N.; Turkarslan, S.; Beer, K.D.; Lo, F.Y.; Baliga, N.S. Adaptation of cells to new environments. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 544–561. [Google Scholar] [CrossRef] [PubMed]
- Mowll, J.L.; Gadd, G.M. Cadmium uptake by Aureobasidium pullulans. J. Gen. Microbiol. 1984, 130, 279–284. [Google Scholar] [CrossRef][Green Version]
- Konings, W.N.; Albers, S.; Koning, S.; Driessen, A.J.M. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Van Leeuwenhoek 2002, 81, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, H.L. Microbes and metals. Appl. Microbiol. Biotechnol. 1997, 48, 687–692. [Google Scholar] [CrossRef]
- Valls, M.; de Lorenzo, V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 2002, 4, 327–338. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Pau, R.N.; Klipp, W.; Leimkühler, S. Molybdenum transport, processing and gene regulation. In Transition Metals in Microbial Metabolism; Winkelmann, G., Carrano, C.J., Eds.; Harwood: Amsterdam, The Netherlands, 1997; pp. 217–234. [Google Scholar]
- Taylor, D.E. Bacterial tellurite resistance. Trends Microbiol. 1999, 7, 111–115. [Google Scholar] [CrossRef]
- Natasha, S.M.; Niazi, N.K.; Khalid, S.; Murtaza, B.; Bibi, I.; Rashid, I.M. A critical review of selenium biogeochemical behavior in soil-plant system with an inference to human health. Environ. Pollut. 2018, 234, 915–934. [Google Scholar] [CrossRef]
- Presentato, A.; Turner, R.J.; Vásquez, C.C.; Yurkov, V.; Zannoni, D. Tellurite-dependent blackening of bacteria emerges from the dark ages. Environ. Chem. 2019, 16, 266–288. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lemire, J.A.; Demeter, M.; Vallini, G.; Turner, R.J.; Lampis, S. Antimicrobial activity of biogenically produced spherical Se nanomaterials embedded in organic material against Pseudomonas aeruginosa and Staphylococcus aureus strains on hydroxyapatite-coated surfaces. Microb. Biotechnol. 2017, 10, 804–818. [Google Scholar] [CrossRef]
- Springer, S.E.; Huber, R.E. Sulfate and selenate uptake and transport in wild and in two selenate-tolerant strains of Escherichia coli K12. Arch. Biochem. Biophys. 1973, 156, 595–603. [Google Scholar] [CrossRef]
- Brown, T.A.; Shrift, A. Assimilation of selenate and selenite by Salmonella thyphimurium. Can. J. Microbiol. 1980, 26, 671–675. [Google Scholar] [CrossRef]
- Bryant, R.D.; Laishley, E.J. Evidence for proton motive force dependent transport of selenite by Clostridium paesteurianum. Can. J. Microbiol. 1989, 35, 481–486. [Google Scholar] [CrossRef]
- Bebien, M.; Chauvin, J.P.; Adriano, J.M.; Grosse, S.; Vermeglio, A. Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2001, 67, 4440–4447. [Google Scholar] [CrossRef]
- Tomas, J.M.; Kay, W.W. Tellurite susceptibility and non-plasmid mediated resistance in Escherichia coli. Antimicrob. Agents Chemother. 1986, 30, 127–131. [Google Scholar] [CrossRef]
- Van Veen, H.W. Phosphate transporter in prokaryotes: Molecules, mediators and mechanisms. Antoine Van Leeuwenhoek 1997, 72, 299–315. [Google Scholar] [CrossRef]
- Harris, R.M.; Webb, D.C.; Howitt, S.M.; Cox, G.B. Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 2001, 183, 5008–5014. [Google Scholar] [CrossRef]
- Turner, M.S.; Tan, Y.P.; Giffard, P.M. Inactivation of an iron transporter in Lactococcus lactis results in resistance to tellurite and oxidative stress. Appl. Environ. Microbiol. 2007, 73, 6144–6149. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zannoni, D.; Borsetti, F.; Harrison, J.J.; Turner, R.J. The bacterial response to the chalcogen metalloids Se and Te. Adv. Microb. Physiol. 2008, 53, 1–71. [Google Scholar] [CrossRef] [PubMed]
- Borghese, R.; Zannoni, D. Acetate permease (ActP) is responsible for tellurite (TeO32−) uptake and resistance in cells of the facultative phototroph Rhodobacter capsulatus. Appl. Environ. Microbiol. 2010, 76, 942–944. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Edwards, C. Effects of metals on a range of Streptomyces species. Appl. Environ. Microbiol. 1990, 55, 2030–2035. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.J.; Castro, G.R.; Carlino, F.J.; Romero, N.C.; Hill, R.T.; Oliver, G. Screening of heavy metal-tolerant actinomycetes isolated from the Salí River. J. Gen. Appl. Microbiol. 1998, 44, 129–132. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, R.L.; Maguire, M.E. Microbial magnesium transport: Unusual transporters searching for identity. Mol. Microbiol. 1998, 28, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Javedan, S.; Moshiri, F.; Maier, R.J. Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc. Natl. Acad. Sci. USA 1994, 91, 5099–5103. [Google Scholar] [CrossRef]
- Eitinger, T.; Friedrich, B. Microbial nickel transport and incorporation into hydrogenases. In Transition Metals in Microbial Metabolism; Winkelmann, G., Carrano, C.J., Eds.; Harwood: Amsterdam, The Netherlands, 1997; pp. 235–256. [Google Scholar]
- Fulkerson, J.F., Jr.; Garner, R.M.; Mobley, H.L.T. Conserved motifs and residues in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J. Biol. Chem. 1998, 273, 235–241. [Google Scholar] [CrossRef]
- Cole, S.T.; Broch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeir, K.; Gas, S.; Barry, C.E., III; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Kim, E.J.; Chung, H.J.; Suh, B.; Hah, Y.C.; Roe, J.H. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Müller. Mol. Microbiol. 1998, 27, 187–195. [Google Scholar] [CrossRef]
- Ahn, B.E.; Cha, J.; Lee, E.J.; Han, A.R.; Thompson, C.J.; Roe, J.H. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 2006, 59, 1848–1858. [Google Scholar] [CrossRef]
- Lu, M.; Jiang, Y.L.; Wang, S.; Jin, H.; Zhang, R.G.; Virolle, M.J.; Chen, Y.; Zhou, C.Z. Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein. PLoS ONE 2014, 9, e109660. [CrossRef]
- Komeda, H.; Kobayashi, M.; Shimizu, S. A novel transporter involved in cobalt uptake. Proc. Natl. Acad. Sci. USA 1997, 94, 36–41. [Google Scholar] [CrossRef]
- Pogorelova, T.E.; Ryabchenko, L.E.; Sunzow, N.I.; Yanenko, A.S. Cobalt-dependent transcription of nitrile hydratase gene in Rhodococcus rhodochrous M8. FEMS Microbiol. Lett. 1996, 144, 191–195. [Google Scholar] [CrossRef]
- Wolfram, L.; Friedrich, B.; Eitinger, T. The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J. Bacteriol. 1995, 177, 1840–1843. [Google Scholar] [CrossRef]
- Degen, O.; Kobayashi, M.; Shimizu, S.; Eitinger, T. Selective transport of divalent cations by transition metal permeases: The Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch. Microbiol. 1999, 171, 139–145. [Google Scholar] [CrossRef]
- Amoroso, M.J.; Schubert, D.; Mitscherlich, P.; Schumann, P.; Kothe, E. Evidence for high affinity nickel transporter genes in heavy metal resistant Streptomyces spec. J. Basic Microbiol. 2000, 40, 295–301. [Google Scholar] [CrossRef]
- Hendricks, J.K.; Mobley, H.L. Helicobacter pylori ABC transporter: Effect of allelic exchange mutagenesis on urease activity. J. Bacteriol. 1997, 179, 5892–5902. [Google Scholar] [CrossRef][Green Version]
- Locatelli, F.M.; Goo, K.S.; Ulanova, D. Effects of trace metal ions on secondary metabolism and the morphological development of Streptomycetes. Metallomics 2016, 8, 469–480. [Google Scholar] [CrossRef]
- Aagaard, A.; Brzezinski, P. Zinc ions inhibit oxidation of cytochrome c oxidase by oxygen. FEBS Lett. 2001, 494, 157–160. [Google Scholar] [CrossRef]
- Mills, D.A.; Schmidt, B.; Hiser, C.; Westley, E.; Ferguson-Miller, S. Membrane potential-controlled inhibition of cytochrome c oxidase by zinc. J. Biol. Chem. 2002, 277, 14894–14901. [Google Scholar] [CrossRef]
- Mangold, S.; Potrykus, J.; Björn, E.; Lövgren, L.; Dopson, M. Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 2013, 17, 75–85. [Google Scholar] [CrossRef]
- Shin, J.H.; Oh, A.Y.; Kim, A.J.; Roe, J.H. The Zinc-Responsive Regulator Zur Controls a Zinc Uptake System and Some Ribosomal Proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 2007, 189, 4070–4077. [Google Scholar] [CrossRef]
- Choi, S.H.; Lee, K.L.; Shin, J.H.; Cho, Y.B.; Cha, S.S.; Roe, J.H. Zinc-dependent regulation of zinc import and export genes by Zur. Nat. Commun. 2017, 8, 15812. [Google Scholar] [CrossRef]
- Sedlakova-Kadukova, J.; Kopcakova, A.; Gresakova, L.; Godany, A.; Pristas, P. Bioaccumulation and biosorption of zinc by a novel Streptomyces K11 strain isolated from highly alkaline aluminium brown mud disposal site. Ecotoxicol. Environ. Saf. 2019, 167, 204–211. [Google Scholar] [CrossRef]
- Gadd, G.M. Microbial influence on metal mobility and application for bioremediation. Geoderma 2004, 122, 109–119. [Google Scholar] [CrossRef]
- Malik, A. Metal bioremediation through growing cells. Environ. Int. 2004, 30, 261–278. [Google Scholar] [CrossRef]
- Polti, M.A.; Amoroso, M.J.; Abate, C.M. Chromium(VI) resistance and removal by actinomycete strains isolated from sediments. Chemosphere 2007, 67, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Polti, M.A.; Amoroso, M.J.; Abaste, C.M. Intracellular chromium accumulation by Streptomyces sp. MC1. Water Air Soil Pollut. 2011, 214, 49–57. [Google Scholar] [CrossRef]
- Tomioka, N.; Uchiyama, H.; Yagi, O. Cesium accumulation and growth characteristics of Rhodococcus erythropolis CS98 and Rhodococcus sp. strain CS402. Appl. Environ. Microbiol. 1994, 60, 2227–2231. [Google Scholar] [CrossRef]
- Avery, S.V.; Codd, G.A.; Gadd, G.M. Caesium accumulation and interactions with other monovalent cations in the cyanobacterium Synechocystis PCC 6803. J. Gen. Microbiol. 1991, 137, 405–413. [Google Scholar] [CrossRef][Green Version]
- Avery, S.V.; Codd, G.A.; Gadd, G.M. Interactions of cyanobacteria and microalgae with caesium. In Impact of Heavy Metals on the Environment; Vernet, J.P., Ed.; Elsevier: Amsterdam, The Netherlands, 1992; pp. 133–182. [Google Scholar]
- Woolfolk, C.A.; Whiteley, H.R. Reduction of inorganic compounds with molecular hydrogen by Micrococcus lactilyticus. I. Stoichiometry with compounds of arsenic, selenium, tellurium, transition and other elements. J. Bacteriol. 1962, 84, 647–658. [Google Scholar] [CrossRef]
- Nakahara, H.; Schottel, J.L.; Yamada, T.; Miyakawa, Y.; Asakawa, M.; Harville, J.; Silver, S. Mercuric Reductase Enzymes from Streptomyces Species and Group B Streptococcus. J. Gen. Microbiol. 1985, 131, 1053–1059. [Google Scholar] [CrossRef]
- Das, S.; Chandra, A.L. Chromate reduction in Streptomyces. Experientia 1990, 46, 731–733. [Google Scholar] [CrossRef]
- Pattanapipitpaisal, P.; Brown, N.L.; Macaskie, L.E. Chromate reduction by Microbacterium liquefaciens immobilised in polyvinyl alcohol. Biotechnol. Lett. 2001, 23, 61–62. [Google Scholar] [CrossRef]
- Laxman, R.S.; More, S. Reduction of hexavalent chromium by Streptomyces griseus. Miner. Eng. 2002, 15, 831–837. [Google Scholar] [CrossRef]
- Ahmad, A.; Senapati, S.; Islam Khan, M.; Kumar, R.; Sastry, M. Extracellular Biosynthesis of Monodisperse Gold Nanoparticles by a Novel Extremophilic Actinomycete, Thermomonospora sp. Langmuir 2003, 19, 3550–3553. [Google Scholar] [CrossRef]
- Ahmad, A.; Senapati, S.; Islam Khan, M.; Kumar, R.; Ramani, R.; Srinivas, V.; Sastry, M. Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete, Rhodococcus species. Nanotechnology 2003, 14, 824–828. [Google Scholar] [CrossRef]
- Desjardin, V.; Bayard, R.; Lejeune, P.; Gourdon, R. Utilisation of supernatants of pure cultures of Streptomyces thermocarboxidus NH50 to reduce Chromium toxicity and mobility in contaminated soils. Water Air Soil Pollut. 2003, 3, 153–160. [Google Scholar] [CrossRef]
- Camargo, F.; Bento, F.; Okeke, B.; Frankenberger, W. Hexavalent chromium reduction by an actinomycete, Arthrobacter crystallopoietes ES 32. Biol. Trace Elem. Res. 2004, 97, 183–194. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Q.; Lu, Y.; Sun, D.; Lin, X.; Deng, X.; He, N.; Zheng, S. Biosorption and bioreduction of diamine silver complex by Corynebacterium. J. Chem. Technol. Biotechnol. 2005, 80, 285–290. [Google Scholar] [CrossRef]
- Bharde, A.; Wani, A.; Shouche, Y.; Joy, P.A.; Prasad, B.L.V.; Sastry, M. Bacterial Aerobic Synthesis of Nanocrystalline Magnetite. J. Am. Chem. Soc. 2005, 127, 9326–9327. [Google Scholar] [CrossRef]
- Mateos, L.M.; Ordonez, E.; Letek, M.; Gil, J.A. Corynebacterium glutamicum as a model bacterium for the bioremediation of arsenic. Int. Microbiol. 2006, 9, 207–215. [Google Scholar]
- Bharde, A.; Kulkarni, A.; Rao, M.; Prabhune, A.; Sastry, M. Bacterial Enzyme Mediated Biosynthesis of Gold Nanoparticles. J. Nanosci. Nanotechnol. 2007, 7, 4369–4377. [Google Scholar] [CrossRef]
- Bharde, A.; Parikh, R.Y.; Baidakova, M.; Jouen, S.; Hannoyer, B.; Enoki, T.; Prasad, B.L.V.; Shouche, Y.S.; Ogale, S.; Sastry, M. Bacteria-Mediated Precursor-Dependent Biosynthesis of Superparamagnetic Iron Oxide and Iron Sulfide Nanoparticles. Langmuir 2008, 24, 5787–5794. [Google Scholar] [CrossRef]
- Mabrouk, M.E.M. Statistical optimization of medium components for chromate reduction by halophilic Streptomyces sp. MS-2. Afr. J. Microbiol. Res. 2008, 2, 103–110. [Google Scholar]
- Kumar, U.; Shete, A.; Harle, A.S.; Kasyutich, O.; Schwarzacher, W.; Pundle, A.; Poddar, P. Extracellular Bacterial Synthesis of Protein-Functionalized Ferromagnetic Co3O4 Nanocrystals and Imaging of Self-Organization of Bacterial Cells under Stress after Exposure to Metal Ions. Chem. Mater. 2008, 20, 1484–1491. [Google Scholar] [CrossRef]
- Polti, M.A.; Garcia, R.O.; Amoroso, M.J.; Abate, C.M. Bioremediation of chromium(VI) contaminated soil by Streptomyces sp. MC1. J. Basic Microbiol. 2009, 49, 285–292. [Google Scholar] [CrossRef]
- Polti, M.A.; Amoroso, M.J.; Abate, C.M. Chromate reductase activity in Streptomyces sp. MC1. J. Gen. Appl. Microbiol. 2010, 56, 11–18. [Google Scholar] [CrossRef]
- Elangovan, R.; Philip, L.; Chandraraj, K. Hexavalent chromium reduction by free and immobilized cell-free extract of Arthrobacter rhombi-RE. Appl. Biochem. Biotechnol. 2010, 160, 81. [Google Scholar] [CrossRef]
- Sadhasivam, S.; Shanmugam, P.; Yun, K.S. Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms. Colloids Surf. B Biointerfaces 2010, 81, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Sneha, K.; Sathishkumar, M.; Mao, J.; Kwak, I.S.; Yun, Y.S. Corynebacterium glutamicum—Mediated crystallization of silver ions through sorption and reduction processes. Chem. Eng. J. 2010, 162, 989–996. [Google Scholar] [CrossRef]
- Bafana, A.; Krishnamurthi, K.; Patil, M.; Chakrabarti, T. Heavy metal resistance in Arthrobacter ramosus strain G2 isolated from mercuric salt-contaminated soil. J. Hazard. Mater. 2010, 177, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Patra, R.C.; Malik, S.; Beer, M.; Megharaj, M.; Naidu, R. Molecular characterization of chromium (VI) reducing potential in gram positive bacteria isolated from contaminated sites. Soil Biol. Biochem. 2010, 42, 1857–1863. [Google Scholar] [CrossRef]
- Sugiyama, T.; Sugito, H.; Mamiya, K.; Suzuki, Y.; Ando, K.; Ohnuki, T. Hexavalent chromium reduction by an actinobacterium Flexivirga alba ST13 in the family Dermacoccaceae. J. Biosci. Bioeng. 2012, 113, 367–371. [Google Scholar] [CrossRef]
- Alani, F.; Moo-Young, M.; Anderson, W. Biosynthesis of silver nanoparticles by a new strain of Streptomyces sp. compared with Aspergillus fumigatus. World J. Microbiol. Biotechnol. 2012, 28, 1081–1086. [Google Scholar] [CrossRef]
- Oza, G.; Pandey, S.; Gupta, A.; Kesarkar, R.; Sharon, M. Biosynthetic Reduction of Gold Ions to Gold Nanoparticles by Nocardia farcinica. J. Microbiol. Biotech. Res. 2012, 2, 511–515. [Google Scholar]
- Dey, S.; Paul, A.K. Optimization of cultural conditions for growth associated chromate reduction by Arthrobacter sp. SUK 1201 isolated from chromite mine overburden. J. Hazard. Mater. 2012, 213–214, 200–206. [Google Scholar] [CrossRef]
- Dey, S.; Paul, A.K. In vitro bioreduction of hexavalent Chromium by viable whole cells of Arthrobacter sp. SUK 1201. J. Microbiol. Biotech. Food Sci. 2014, 4, 19–23. [Google Scholar] [CrossRef][Green Version]
- Dey, S.; Paul, A.K. Optimization of Chromate Reduction by Whole Cells of Arthrobacter sp. SUK 1205 Isolated from Metalliferous Chromite Mine Environment. Geomaterials 2012, 2, 73–81. [Google Scholar] [CrossRef][Green Version]
- Javaid, M.; Sultan, S. Plant growth promotion traits and Cr (VI) reduction potentials of Cr (VI) resistant Streptomyces strains. J. Basic Microbiol. 2013, 53, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Otari, S.V.; Patil, R.M.; Nadaf, N.H.; Gosh, S.J.; Pawar, S.H. Green biosynthesis of silver nanoparticles from an actinobacteria Rhodococcus sp. Mat. Lett. 2012, 72, 92–94. [Google Scholar] [CrossRef]
- Otari, S.V.; Patil, R.M.; Gosh, S.J.; Thorat, N.D.; Pawar, S.H. Intracellular synthesis of silver nanoparticle by actinobacteria and its antimicrobial activity. Spectrochim. Acta A Mol. Biomol. Spectr. 2015, 136, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Kumar, A.; Abraham, J. A Biological Approach to the Synthesis of Silver Nanoparticles with Streptomyces sp JAR1 and its Antimicrobial Activity. Sci. Pharm. 2013, 81, 607–621. [Google Scholar] [CrossRef]
- Arunkumar, P.; Thanalakshmi, M.; Kumar, P.; Premkumar, K. Micrococcus luteus mediated dual mode synthesis of gold nanoparticles: Involvement of extracellular α-amylase and cell wall teichuronic acid. Colloid Surf. B Biointerfaces 2013, 103, 517–522. [Google Scholar] [CrossRef]
- Gopal, J.V.; Thenmozhi, M.; Kannabiran, K.; Rajakumar, G.; Velayutham, K.; Rahuman, A.A. Actinobacteria mediated synthesis of gold nanoparticles using Streptomyces sp. VITDDK3 and its antifungal activity. Mat. Lett. 2013, 93, 360–362. [Google Scholar] [CrossRef]
- Gren, T.; Ostash, B.; Hrubskyy, Y.; Tistechok, S.; Fedorenko, V. Influence of transition metals on Streptomyces coelicolor and S. sioyaensis and generation of chromate-reducing mutants. Folia Microbiologica 2014, 59, 147–153. [Google Scholar] [CrossRef]
- Kundu, D.; Hazra, C.; Chatterjee, A.; Chaudhari, A.; Mishra, S. Extracellular biosynthesis of zinc oxide nanoparticles using Rhodococcus pyridinivorans NT2: Multifunctional textile finishing, biosafety evaluation and in vitro drug delivery in colon carcinoma. J. Photochem. Photobiol. B 2014, 140, 194–204. [Google Scholar] [CrossRef]
- Polti, M.A.; Aparicio, J.D.; Benimeli, C.S.; Amoroso, M.J. Simultaneous bioremediation of Cr(VI) and lindane in soil by actinobacteria. Int. Biodet. Biodegr. 2014, 88, 48–55. [Google Scholar] [CrossRef]
- Kaur, H.; Dolma, K.; Kaur, N.; Malhotra, A.; Kumar, N.; Dixit, P.; Sharma, D.; Mayilraj, S.; Choudhury, A.R. Marine Microbe as Nano-factories for Copper Biomineralization. Biotechnol. Bioproc. Eng. 2015, 20, 51–57. [Google Scholar] [CrossRef]
- Ramya, S.; Shanmugasundaram, T.; Balagurunathan, R. Biomedical potential of actinobacterially synthesized selenium nanoparticles with special reference to anti-biofilm, anti-oxidant, wound healing, cytotoxic and anti-viral activities. J. Trace Elem. Med. Biol. 2015, 32, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.M.; Johnson, D.B. Acidithrix ferrooxidans gen. nov., sp. nov.; a filamentous and obligately heterotrophic, acidophilic member of the Actinobacteria that catalyzes dissimilatory oxido-reduction of iron. Res. Microbiol. 2015, 166, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.S.; Yasser, M.S.; Sholkamy, E.N.; Ali, M.A.; Mehanni, M.M. Anticancer activity of biostabilized selenium nanorods synthesized by Streptomyces bikiniensis strain Ess_amA-1. Int. J. Nanomed. 2015, 10, 3389–3401. [Google Scholar] [CrossRef]
- Liu, H.; Huang, J.; Zhang, S.; Xu, B.; Wang, G. Chromate interaction with chromate reducing actinobacterium Intrasporangium chromatireducens Q5-1. Geomicrobiol. J. 2015, 32, 616–623. [Google Scholar] [CrossRef]
- Abd-Elnaby, H.M.; Abo-Elala, G.M.; Abdel-Raouf, U.M.; Hamed, M.Z. Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egypt. J. Aquat. Res. 2016, 42, 301–312. [Google Scholar] [CrossRef]
- Dey, S.; Paul, A.K. Assessment of heavy metal tolerance and hexavalent chromium reducing potential of Corynebacterium paurometabolum SKPD 1204 isolated from chromite mine seepage. AIMS Bioeng. 2016, 3, 337–351. [Google Scholar] [CrossRef]
- Bennur, T.; Khan, Z.; Kshirsagar, R.; Javdekar, V.; Zinjarde, S. Biogenic gold nanoparticles from the Actinomycete Gordonia amarae: Application in rapid sensing of copper ions. Sens. Actuat. B 2016, 233, 684–690. [Google Scholar] [CrossRef]
- Tan, Y.; Yao, R.; Wang, R.; Wang, D.; Wang, G.; Zheng, S. Reduction of selenite to Se(0) nanoparticles by filamentous bacterium Streptomyces sp. ES2-5 isolated from a selenium mining soil. Microb. Cell. Fact. 2016, 15, 157. [Google Scholar] [CrossRef]
- Presentato, A.; Piacenza, E.; Anikovskiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Rhodococcus aetherivorans BCP1 as cell factory for the production of intracellular tellurium nanorods under aerobic conditions. Microb. Cell. Fact. 2016, 15, 204. [Google Scholar] [CrossRef]
- Składanowski, M.; Wypij, M.; Laskowski, D.; Golinksa, P.; Dahm, H.; Rai, M. Silver and gold nanoparticles synthesized from Streptomyces sp. isolated from acid forest soil with special reference to its antibacterial activity against pathogens. J. Clust. Sci. 2017, 28, 59–79. [Google Scholar] [CrossRef]
- Wypij, M.; Golinska, P.; Dahm, H.; Rai, M. Actinobacterial-mediated synthesis of silver nanoparticles and their activity against pathogenic bacteria. IET Nanobiotechnol. 2017, 11, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Silva-Vinhote, N.M.; Duran Caballero, N.E.; de Amorim Silva, T.; Veras Quelemes, P.; de Araujo, A.R.; de Moraes, A.C.M.; dos Santos Camara, A.L.; Longo, J.P.F.O.; Azevedo, R.B.; da Silva, D.A.; et al. Extracellular biogenic synthesis of silver nanoparticles by Actinomycetes from amazonic biome and its antimicrobial efficiency. Afr. J. Biotechnol. 2017, 16, 2072–2082. [Google Scholar] [CrossRef]
- Presentato, A.; Piacenza, E.; Anikovskiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. New Biotechnol. 2018, 41, 1–8. [Google Scholar] [CrossRef]
- Presentato, A.; Piacenza, E.; Darbandi, A.; Anikovskiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Assembly, growth and conductive properties of tellurium nanorods produced by Rhodococcus aetherivorans BCP1. Sci. Rep. 2018, 8, 3923. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, J.D.; Saez, J.M.; Raimondo, E.E.; Benimeli, C.S.; Polti, M.A. Comparative study of single and mixed cultures of actinobacteria for the bioremediation of co-contaminated matrices. J. Environ. Chem. Eng. 2018, 6, 2310–2318. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Ghilan, A.K.M.; Arasu, M.V.; Duraipandiyan, V. Green biosynthesis of silver nanoparticles produced from marine Streptomyces sp. Al-Dhabi-89 and their potential applications against wound infection and drug resistant clinical pathogens. J. Photochem. Photobiol. B 2018, 189, 176–184. [Google Scholar] [CrossRef]
- Wypij, M.; Czarnecka, J.; Świecimska, M.; Dahm, H.; Rai, M.; Golinska, P. Synthesis, characterization and evaluation of antimicrobial and cytotoxic activities of biogenic silver nanoparticles synthesized from Streptomyces xinghaiensis OF1 strain. World J. Microbiol. Biotechnol. 2018, 34, 23. [Google Scholar] [CrossRef]
- Hassan, S.E.D.; Salem, S.S.; Fouda, A.; Awad, M.A.; El-Gamal, M.S.; Abdo, A.M. New approach for antimicrobial activity and bio-control of various pathogens by biosynthesized copper nanoparticles using endophytic actinomycetes. J. Radiat. Res. Appl. Sci. 2018, 11, 262–270. [Google Scholar] [CrossRef]
- Katyal, P.; Kaur, G. Reduction of Cr(VI) by Micrococcus luteus isolate from common effluent treatment plants (CETPs). Int. J. Curr. Microbiol. App. Sci. 2018, 7, 693–710. [Google Scholar] [CrossRef]
- Firrincieli, A.; Presentato, A.; Favoino, G.; Marabottini, R.; Allevato, E.; Stazi, S.R.; Scarascia Mugnozza, G.; Harfouche, A.; Petruccioli, M.; Turner, R.J.; et al. Identification of Resistance Genes and Response to Arsenic in Rhodococcus aetherivorans BCP1. Front. Microbiol. 2019, 10, 888. [Google Scholar] [CrossRef]
- Hassan, S.E.D.; Fouda, A.; Radwan, A.A.; Salem, S.S.; Barghoth, M.G.; Awad, M.A.; Abdo, A.M.; El-Gamal, M.S. Endophytic actinomycetes Streptomyces spp mediated biosynthesis of copper oxide nanoparticles as a promising tool for biotechnological applications. J. Biol. Inorg. Chem. 2019, 24, 377–393. [Google Scholar] [CrossRef]
- Rehan, M.; Alsohim, A.S.; El-Fadly, G.; Tisa, L.S. Detoxification and reduction of selenite to elemental red selenium by Frankia. Antonie Van Leeuwenhoek 2019, 112, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Maas, D.; Valerio, A.; Lourenco, L.A.; de Oliveira, D.; Hotza, D. Biosynthesis of iron oxide nanoparticles from mineral coal tailings in a stirred tank reactor. Hydrometallurgy 2019, 184, 199–205. [Google Scholar] [CrossRef]
- Maas, D.; de Medeiros, M.M.; Cesa Rovaris, B.; Bernardin, A.M.; de Oliveira, D.; Hotza, D. Biomining of iron-containing nanoparticles from coal tailings. Appl. Microbiol. Biotechnol. 2019, 103, 7231–7240. [Google Scholar] [CrossRef] [PubMed]
- Devagi, P.; Suresh, T.C.; Sandhiya, R.V.; Sairandhry, M.; Bharathi, S.; Velmurugan, P.; Radhakrishnan, M.; Sathiamoorthi, T.; Suresh, G. Actinobacterial-Mediated Fabrication of Silver Nanoparticles and Their Broad Spectrum Antibacterial Activity Against Clinical Pathogens. J. Nanosci. Nanotechnol. 2020, 20, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Dhanaraj, S.; Thirunavukkarasu, S.; John, H.A.; Pandian, S.; Salmen, S.H.; Chinnathambi, A.; Alharbi, S.A. Novel marine Nocardiopsis dassonvillei-DS013 mediated silver nanoparticles characterization and its bactericidal potential against clinical isolates. Saudi J. Biol. Sci. 2020, 27, 991–995. [Google Scholar] [CrossRef]
- Ramya, S.; Shanmugasundaram, T.; Balagurunathan, R. Actinobacterial enzyme mediated synthesis of selenium nanoparticles for antibacterial, mosquito larvicidal and anthelminthic applications. Part. Sci. Technol. 2020, 38, 63–72. [Google Scholar] [CrossRef]
- Desjardin, V.; Bayard, R.; Huck, N.; Manceau, A.; Gourdon, R. Effect of microbial activity on the mobility of chromium in soils. Waste Manag. 2002, 22, 195–200. [Google Scholar] [CrossRef]
- Bansal, V.; Bharde, A.; Ramanathan, R.; Bargava, S.K. Inorganic materials using ‘unusual’ microorganisms. Adv. Colloid Interf. Sci. 2012, 179–182, 150–168. [Google Scholar] [CrossRef]
- Newton, G.L.; Ta, P.; Fahey, R.C. A mycothiol synthase mutant of Mycobacterium smegmatis produces novel thiols and has an altered thiol redox status. J. Bacteriol. 2005, 187, 7309–7316. [Google Scholar] [CrossRef][Green Version]
- Painter, E.P. The chemistry and toxicity of selenium compounds with special reference to the selenium problem. Chem. Rev. 1941, 28, 179–213. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, M.; Fedi, S.; Zampolli, J.; Di Canito, A.; D’Ursi, P.; Orro, A.; Viti, C.; Milanesi, L.; Zannoni, D.; Di Gennaro, P. Phenotype microarray analysis may unravel genetic determinants of the stress response by Rhodococcus aetherivorans BCP1 and Rhodococcus opacus R7. Res. Microbiol. 2016, 167, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Villadangos, A.F.; Van Belle, K.; Whani, K.; Tamu Dufe, V.; Freitas, S.; Nur, H.; De Galan, S.; Gil, G.A.; Collet, J.F.; Mateos, L.M.; et al. Corynebacterium glutamicum survives arsenic stress with arsenate reductases coupled to two distinct redox mechanisms. Mol. Microbiol. 2011, 82, 998–1014. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Y.; Zhang, X.; Xum, H.; Lescop, E.; Xia, B.; Jin, C. Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis. J. Biol. Chem. 2007, 282, 11078–11083. [Google Scholar] [CrossRef]
- Ordóñez, E.; Van Belle, K.; Roos, G.; De Galan, S.; Letek, M.; Gil, J.A.; Wyns, L.; Mateos, L.M.; Messens, J. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 2009, 284, 15107–15116. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Srikumar, R. Multidrug efflux in Pseudomonas aeruginosa: Components, mechanisms and clinical significance. Curr. Top. Med. Chem. 2001, 1, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Yarnall, B.; Doyle, D.A. Efflux drug transporters at the forefront of antimicrobial resistance. Eur. Biophys. J. 2017, 46, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Nakaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef]
- Kuroda, T.; Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 2009, 1794, 763–768. [Google Scholar] [CrossRef]
- Locher, K.P. Review. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Bay, D.C.; Rommens, K.L.; Turner, R.J. Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim. Biophys. Acta 2008, 1778, 1814–1838. [Google Scholar] [CrossRef] [PubMed]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef] [PubMed]
- Getsin, I.; Nalbandian, G.H.; Yee, D.C.; Vastermark, A.; Paparoditis, P.C.G.; Reddy, V.S.; Saier, M.H. Comparative genomics of transport proteins in developmental bacteria: Myxococcus xanthus and Streptomyces coelicolor. BMC Microbiol. 2013, 13, 279. [Google Scholar] [CrossRef]
- Schmidt, A.; Haferburg, M.; Sineriz, M.; Merten, D.; Büchel, G.; Kothe, E. Heavy metal resistance mechanisms in actinobacteria for survival in AMD contaminated soils. Geochemistry 2005, 65, 131–144. [Google Scholar] [CrossRef]
- Raytapadar, S.; Datta, R.; Paul, A.K. Effects of some heavy metals on growth, pigment and antibiotic production by Streptomyces albus. Acta Microbiol. Acta Microbiol. Immunol. Hung. 1995, 42, 171–177. [Google Scholar]
- Mirimanoff, N.; Wilkinson, K.J. Regulation of Zn accumulation by a freshwater Gram-positive bacterium (Rhodococcus opacus). Environ. Sci. Technol. 2000, 34, 616–622. [Google Scholar] [CrossRef]
- Yang, H.C.; Fu, H.L.; Lin, Y.F.; Rosen, B.P. Pathways of arsenic uptake and efflux. Curr. Top. Membr. 2012, 69, 325–358. [Google Scholar] [CrossRef]
- Fu, H.L.; Meng, Y.; Ordóñez, E.; Villadangons, V.A.; Bhattacharjee, H.; José, A.G.; Mateos, L.M.; Rosen, B.P. Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J. Biol. Chem. 2009, 284, 19887–19895. [Google Scholar] [CrossRef]
- Yumei, L.; Yamei, L.; Qiang, L.; Jie, B. Rapid Biosynthesis of Silver Nanoparticles Based on Flocculation and Reduction of an Exopolysaccharide from Arthrobacter sp. B4: Its Antimicrobial Activity and Phytotoxicity. J. Nanomater. 2017, 2017, 9703614. [Google Scholar] [CrossRef]
- Balagurunathan, R.; Radhakrishnan, M.; Babu Rajendran, R.; Velmurugan, D. Biosynthesis of gold nanoparticles by actinomycete Streptomyces viridogens strain HM10. Indian J. Biochem. Biophys. 2011, 48, 331–335. [Google Scholar]
- Kalabegishvili, T.L.; Kirkesali, E.I.; Rcheulishvili, A.N.; Ginturi, E.N.; Murusidze, I.G.; Pataraya, D.T.; Gurielidze, M.A.; Tsertsvadze, G.I.; Gabunia, V.N.; Lomidze, L.G.; et al. Synthesis of Gold Nanoparticles by Some Strains of Arthrobacter Genera. J. Mat. Sci. Eng. A 2012, 2, 164–173. [Google Scholar]
- Derakhshan, F.K.; Dehnad, A.; Salouti, M. Extracellular Biosynthesis of Gold Nanoparticles by Metal Resistance Bacteria: Streptomyces griseus. Synth. React. Inorg. Met. Org. Nano Met. Chem. 2012, 42, 868–871. [Google Scholar] [CrossRef]
- Faghri Zonooz, N.; Salouti, M.; Shapouri, R.; Nasseryan, J. Biosynthesis of Gold Nanoparticles by Streptomyces sp. ERI-3 Supernatant and Process Optimization for Enhanced Production. J. Clust. Sci. 2012, 23, 375–382. [Google Scholar] [CrossRef]
- Verma, V.C.; Anand, S.; Ulrichs, C.; Singh, S.K. Biogenic gold nanotriangles from Saccharomonospora sp., an endophytic actinomycetes of Azadirachta indica A. Juss. Intern. Nano Lett. 2013, 3, 21. [Google Scholar] [CrossRef]
- Waghmare, S.S.; Deshmukh, A.M.; Sadowski, Z. Biosynthesis, optimization, purification and characterization of gold nanoparticles. Afr. J. Microbiol. Res. 2014, 8, 138–146. [Google Scholar] [CrossRef]
- Dehnad, A.; Hamedi, J.; Derakhshan-Khadivi, F.; Abuşov, R. Green Synthesis of Gold Nanoparticles by a Metal Resistant Arthrobacter nitroguajacolicus Isolated from Gold Mine. IEEE Trans. NanoBiosci. 2015, 14, 393–396. [Google Scholar] [CrossRef]
- Sowani, H.; Mohite, P.; Munot, H.; Shouche, Y.; Bapat, T.; Kumar, A.R.; Kulkarni, M.; Zinjarde, S. Green synthesis of gold and silver nanoparticles by an actinomycete Gordonia amicalis HS-11: Mechanistic aspects and biological application. Proc. Biochem. 2016, 51, 374–383. [Google Scholar] [CrossRef]
- Ranjitha, V.R.; Ray, V.R. Actinomycetes mediated synthesis of gold nanoparticles from the culture supernatant of Streptomyces griseoruber with special reference to catalytic activity. 3 Biotech 2017, 7, 299. [Google Scholar] [CrossRef]
- Tsibakhashvili, N.Y.; Kirkesali, E.I.; Pataraya, D.T.; Gurielidze, M.A.; Kalabegishvili, T.L.; Gvarjaladze, D.N.; Tsertsvadze, G.I.; Frontasyeva, M.V.; Zinicovscaia, I.I.; Wakstein, M.S.; et al. Microbial Synthesis of Silver Nanoparticles by Streptomyces glaucus and Spirulina platensis. Adv. Sci. Lett. 2011, 4, 3408–3417. [Google Scholar] [CrossRef][Green Version]
- Selvakumar, P.; Viveka, S.; Prakash, S.; Jasminebeaula, S.; Uloganathan, R. Antimicrobial activity of extracellularly synthesized silver nanoparticles from marine derived Streptomyces rochei. Int. J. Pharm. Biol. Sci. 2012, 3, 188–197. [Google Scholar]
- Otari, S.V.; Patil, R.M.; Nadaf, N.H.; Ghosh, S.J.; Pawar, S.H. Green synthesis of silver nanoparticles by microorganism using organic pollutant: Its antimicrobial and catalytic application. Environ. Sci. Pollut. Res. 2014, 21, 1503–1513. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Senthilkumar, K.; Sivakumar, K.; Kim, S.K. Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. BioMed. Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Priyaragini, S.; Sathishkumar, S.; Bhaskararao, K. Biosynthesis of silver nanoparticles using actinobacteria and evaluating its antimicrobial and cytotoxicity activity. Int. J. Pharm. Pharm. Sci. 2013, 5, 709–712. [Google Scholar]
- Babu Vimalanathan, A.; Ernest, V.; Arumugasamay, K.; Tyagi, M.G. Biosynthesis of silver nano-particles by the bacterium Micrococcus luteus. Int. J. Appl. Biol. Pharm. Technol. 2013, 4, 77–83. [Google Scholar]
- Shanmuga Priya, T.; Balasubramanian, V. Enzyme Mediated Synthesis of Silver Nanoparticles using Marine Actinomycetes and Their Characterization. Biosci. Biotechnol. Res. Asia 2014, 11, 159–165. [Google Scholar] [CrossRef]
- Prakash, N.K.U.; Bhuvaneswari, S.; Prabha, S.B.; Kavitha, K.; Sandhya, K.V.; Sathyabhuvaneshwari, P.; Bharathiraja, B. Green Synthesis of Silver Nanoparticles using Airborne Actinomycetes. Int. J. ChemTech. Res. 2014, 6, 4123–4127. [Google Scholar]
- Karthik, L.; Kumar, G.; Vishnu Kirthi, A.; Rahuman, A.A.; Bhaskara Rao, K.V. Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application. Bioprocess Biosyst. Eng. 2014, 37, 261–267. [Google Scholar] [CrossRef]
- Mohanta, Y.K.; Behera, S.K. Biosynthesis, characterization and antimicrobial activity of silver nanoparticles by Streptomyces sp. SS2. Bioprocess Biosyst. Eng. 2014, 37, 2263–2269. [Google Scholar] [CrossRef]
- Anasane, N.; Golinska, P.; Wypij, M.; Rathod, D.; Dahm, H.; Rai, M. Acidophilic actinobacteria synthesised silver nanoparticles showed remarkable activity against fungi-causing superficial mycoses in humans. Mycoses 2016, 59, 157–166. [Google Scholar] [CrossRef]
- Gowramma, B.; Keerthi, U.; Rafi, M.; Muralidhara Rao, D. Biogenic silver nanoparticles production and characterization from native stain of Corynebacterium species and its antimicrobial activity. 3 Biotech 2015, 5, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Kamel, Z.; Saleh, M.; El Namoury, N. Biosynthesis, Characterization, and Antimicrobial Activity of Silver Nanoparticles from Actinomycetes. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 119–127. [Google Scholar]
- Raja, M.M.M.; John, S.A. Biosynthesis of Silver Nanoparticles by Novel Isolate of Marine Micromonospora species (KU 867645) and its Antibacterial Activity against Multidrug Resistant Hospital-acquired Uropathogens in Reference with Standard Antibiotics. Indian J. Pharm. Sci. 2017, 79, 369–376. [Google Scholar] [CrossRef]
- Avilala, J.; Golla, N. Antibacterial and antiviral properties of silver nanoparticles synthesized by marine Actinomycetes. Int. J. Pharm. Sci. Res. 2019, 10, 1223–1228. [Google Scholar] [CrossRef]
- Hamed, A.A.; Kabary, H.; Khedr, M.; Emam, A.N. Antibiofilm, antimicrobial and cytotoxic activity of extracellular green-synthesized silver nanoparticles by two marine-derived actinomycete. RSC Adv. 2020, 10, 10361–10367. [Google Scholar] [CrossRef]
- Usha, R.; Prabu, E.; Palaniswamy, M.; Venil, C.K.; Rajendran, R. Synthesis of Metal Oxide Nano Particles by Streptomyces sp. for Development of Antimicrobial Textiles. Glob. J. Biotech. Biochem. 2010, 5, 153–160. [Google Scholar]
- Nabila, M.I.; Kannabiran, K. Biosynthesis, characterization and antibacterial activity of copper oxide nanoparticles (CuO NPs) from actinomycetes. Biocatal. Agric. Biotechnol. 2018, 15, 56–62. [Google Scholar] [CrossRef]
- Waghmare, S.; Deshmukh, A.; Kulkarni, S.W.; Oswaldo, L. Biosynthesis and characterization of manganese and zinc nanoparticles. Univ. J. Environ. Res. Technol. 2011, 1, 64–69. [Google Scholar]
- Rajamanickam, U.; Mylsamy, P.; Viswanathan, S.; Muthusamy, P. Biosynthesis of Zinc Nanoparticles Using Actinomycetes for Antibacterial Food Packaging. In Proceedings of the Internation Conference on Nutriotion and Food Science IPCBEE, Singapore, 23–24 July 2012; IACSIT Press: Singapore, 2012; Volume 39, pp. 195–199. [Google Scholar]
- Piacenza, E.; Presentato, A.; Amborsi, E.; Speghini, A.; Turner, R.J.; Vallini, G.; Lampis, S. Physical-Chemical Properties of Biogenic Selenium Nanostructures Produced by Stenotrophomonas maltophilia SeITE02 and Ochrobactrum sp. MPV1. Front. Microbiol. 2018, 9, 3178. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Heyne, B.; Turner, R.J. Tunable photoluminescence properties of selenium nanoparticles: Biogenic versus chemogenic synthesis. Nanophotonics 2020, 9, 3615–3628. [Google Scholar] [CrossRef]
- Rahman, M.M.; Khan, S.B.; Jamal, A.; Faisal, M.; Aisiri, A.M. Chapter 3: Iron Oxide Nanoparticles. In Nanomaterials; Rhaman, M.M., Ed.; IntechOpen: London, UK, 2011; pp. 43–66. [Google Scholar] [CrossRef]
- Turner, R.J. Metal-based antimicrobial strategies. Microb. Biotechnol. 2017, 10, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Bernardos, A.; Piacenza, E.; Sancenon, F.; Hamidi, M.; Maleki, A.; Turner, R.J.; Martinez-Manez, R. Mesoporous Silica-Based Materials with Bactericidal Properties. Small 2019, 15, 1900669. [Google Scholar] [CrossRef] [PubMed]
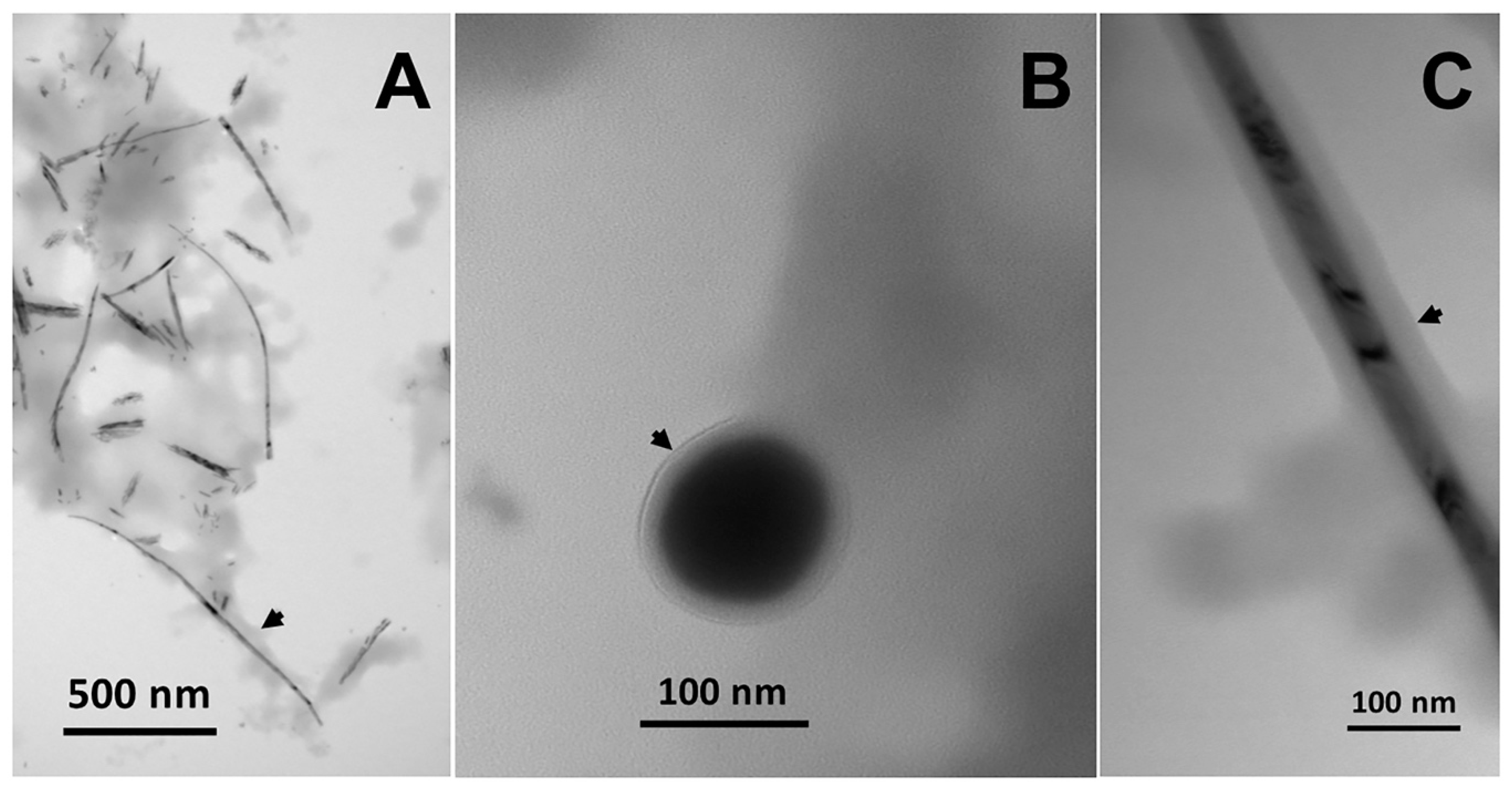
| Actinobacteria Strains | Siderophores | Metal Tolerance | References |
|---|---|---|---|
| Microbacterium flavescens JG-9 | Desferrioxamine-B | U and Pu | [59] |
| Arthrobacter spp., Microbacterium spp., Curtobacterium spp., Leifsonia spp. | N.D. 1 | Co, Cr, Cu, Ni, Zn | [60] |
| Streptomyces sp. AR16 and AR36 | N.D. 1 | Zn, Cd, Pb | [61] |
| Microbacterium G16 | N.D. 1 | Cu, Cd, Ni, Pb, Zn | [62] |
| Streptomyces acidiscabies E13 | Coelichelin and desferrioxamines | Ni | [63] |
| Streptomyces tendae F4 | Coelichelin and desferrioxamines | Ni, Cd | [64,65] |
| Methylobacterium and Frigobacterium spp. | N.D. 1 | Zn, Cd | [66] |
| Arthrobacter spp. and M. kitamiense YM31 | N.D. 1 | Cu | [67] |
| Microbacterium sp. NCr-8, Arthrobacter sp. NCr-1, Kokuria sp. NCr-3 | N.D. 1 | Ni | [68] |
| Streptomyces mirabilis P16B-1 | Linear and cyclic ferrioxamines | Zn, Al, Ni, Cu | [69] |
| Streptomyces spp. | Hydroxamates | Pb | [70] |
| Streptomyces spp. | Ferrioxamines, phenolates, and catecholates | Cd | [71] |
| Arthrobacter oxydans ATW2 and ATW3, Kocuria rosea ATW4, Rhodococcus erythropolis ATW1 and S43 | N.D. 1 | As | [72] |
| Rothia mucilaginosa ATCC 25,296 | Enterobactin | Zn, Mg | [73] |
| Actinobacteria Strains | EPS Features | Metal Tolerance | References |
|---|---|---|---|
| Arthrobacter D9 | N.D. 1 | Cd | [91] |
| Amycolatopsis sp. AB0 | Polysaccharides containing glucose | Cu | [92] |
| Kocuria rizophila DE2008 | Polysaccharides and uronic acid | Cu, Pb | [93,94] |
| Arthrobacter viscosus | N.D. 1 | Cr | [95] |
| Nocardia amarae | N.D. 1 | Na, Ca, Al, Fe | [96] |
| Arthrobacter ps-5 | Polysaccharides containing glucose and galactose | Pb, Cu, Cr | [81] |
| Rhododoccus opacus and Rhodococcus rhodochrous | N.D. 1 | Ni, Cd, Pb, Co, Cr | [86] |
| Kocuria rizophila BPB1 | N.D. 1 | As | [97] |
| Frankia sp. strain EAN1pec | Polysaccharides and proteins | Pb | [98] |
| Kokuria sp. BRI 36 | Polysaccharides and uronic acid | Pb, Cd, Cr | [99] |
| Nocardiopsis sp. 13H | Polysaccharides, proteins, and nucleic acids | Sr and Cs | [87,100] |
| Streptomyces sp. CuOff24 | Arabinose, galactose, mannose, glucose, and uronic acid | Sr | [88] |
| Glutamicibacter halophytocola KLBMP 5180 | Rhamnose, glucuronic acid, glucose, galactose, xylose, and arabinose | Mn, Cu, Fe, Ca, Mg | [83] |
| Actinobacteria Strains | Metal(loid) Reduction | Mediating Biomolecules | References |
|---|---|---|---|
| Micrococcus lactilyticus | Au3+, Ag+, SeO32−, TeO32−, TeO42−, AsO43−, bismuthate (BiO3−), vanadate (VO34−), molybdate (MoO42−), ruthenium (Ru3+) | N.D. 1 | [157] |
| Streptomyces lividans 1326 | Hg2+ | Hg-constitutive reductase | [158] |
| Streptomyces sp. 3 M | CrO42− | Constitutive reductase | [159] |
| Microbacterium sp. MP30 | CrO42− | N.D. 1 | [160] |
| Streptomyces griseus | CrO42− | N.D. 1 | [161] |
| Thermomonospora sp. | Au3+ | Four proteins and enzymes (80–100 kDa) | [162] |
| Rhodococcus sp. | Au3+ | Intracellular proteins | [163] |
| Streptomyces thermocarboxydus NH50 | CrO42− | Extracellular “nonenzymatic” substance | [164] |
| Arthrobacter crystallopoietes ES 32 | CrO42− | NADH-dependent chromate reductase | [165] |
| Corynebacterium strain SH09 | Ag+ | Aldehyde and ketone groups of biomolecules | [166] |
| Actinobacter sp. | K3Fe(CN)6/ K4Fe(CN)6 mixture | Extracellular hydrolases | [167] |
| Corynebacterium glutamicum | AsO43− | NAD(P)H-dependent intracellular ArsC proteins | [168] |
| Streptomyces spp., Amycolatopsis sp. | CrO42− | N.D. 1 | [152] |
| Actinobacter sp. | Au3+ | Cytochrome oxidase | [169] |
| Actinobacter sp. | FeCl3 or FeCl3/FeSO4 mixture | Ferrisiderophore reductase, phosphoadenosyl sulfate, and sulfite reductases | [170] |
| Streptomyces sp. MS-2 | CrO42− | N.D. 1 | [171] |
| Brevibacterium casei AP6 | Co2+ | Intra- and extracellular proteins | [172] |
| Streptomyces sp. MC1 | CrO42− | NAD(P)H-dependent constitutive chromate reductase | [153,173,174] |
| Arthroobacter rhombi-RE | CrO42− | Intracellular enzymes | [175] |
| Streptomyces hygroscopicus | Ag+ | Secondary metabolites | [176] |
| Corynebacterium glutamicum | Ag+ | Proteins | [177] |
| Arthrobacter ramosus | Hg2+, CrO42− | MerA enzyme, intracellular enzymes | [178] |
| Arthrobacter aurescens MM10 | CrO42− | Intracellular chromate reductase | [179] |
| Flexivirga alba ST13 | CrO42− | Intracellular proteins | [180] |
| Streptomyces sp. | Ag+ | Intracellular enzymes | [181] |
| Nocardia farcinica | Au3+ | extracellular nitrate reductase | [182] |
| Arthrobacter sp. SUK 1201 | CrO42− | N.D. 1 | [183,184] |
| Arthrobacter sp. SUK 1205 | CrO42− | N.D. 1 | [185] |
| Streptomyces sp. RSF17, CRF14, Streptomyces matansis BG5, Streptomyces vinaceus CRF2, Streptomyces pulcher CRF17, Streptomyces griseoincarnatus SCF18 | CrO42− | N.D. 1 | [186] |
| Rhodococcus NCIM 2891 | Ag+ | Intracellular NADH-dependent nitrate reductase; peptides, proteins, and carbohydrates | [187,188] |
| Streptomyces sp. JAR1 | Ag+ | Intracellular NADH-dependent nitrate reductase | [189] |
| Micrococcus luteus NCIM 2379 | Au3+ | Extracellular α-amylase and TUA | [190] |
| Streptomyces sp.VITDDK3 | Au3+ | (2S,5R,6R)-2-hydroxy-3,5,6-trimethyloctan-4-one | [191] |
| Streptomyces sioyaensis Lv81-138 | CrO42− | Intracellular reductases | [192] |
| Rhodococcus pyridinivorans NT2 | Zn2+ | NAD(P)H-dependent reductase and secreted reductase | [193] |
| Streptomyces spp. and Amycolatopsis tucumanensis | CrO42− and lindane | N.D. 1 | [194] |
| Kocuria flava M-7 | Cu2+ | Intracellular proteins | [195] |
| Streptomyces minutiscleroticus M10A6 | SeO32− | Intracellular proteins | [196] |
| Acidithrix ferrooxidans PY-F3 | Fe3+ | N.D. 1 | [197] |
| Streptomyces bikiniensis strain Ess_amA-I | SeO32− | Intracellular proteins and enzymes | [198] |
| Intraspongium chromatireducens Q5-1 | CrO42− | Extracellular constitutive enzyme | [199] |
| Streptomyces rochei MHM13 | Ag+ | Intracellular proteins | [200] |
| Corynebacterium paurometabolum SKPD 1204 | CrO42− | N.D. 1 | [201] |
| Gordonia amarae | Au3+ | Glycolipids | [202] |
| Streptomyces sp. ES2-5 | SeO32− | Mycothiols and thiol-containing molecules | [203] |
| Rhodococcus etherivorans BCP1 | TeO32− | Mycothiols and thiol-containing molecules | [204] |
| Streptomyces sp. NH21 | Au3+, Ag+ | Intra- and extracellular proteins | [205] |
| Streptomyces kasugaensis M338-M1T, Streptomyces celluloflavus NRRL B-2493T | Ag+ | Proteins | [206] |
| Streptomyces parvulus DPUA 1549, Streptomyces owasiensis DPUA 1748 | Ag+ | Proteins | [207] |
| Rhodococcus etherivorans BCP1 | SeO32− | Mycothiols and thiol-containing molecules | [208] |
| Rhodococcus etherivorans BCP1 | TeO32− | Mycothiols and thiol-containing molecules | [209] |
| Streptomyces spp. M7, A5, and MC1, Amycolatopsis tucumanensis | CrO42− | N.D. 1 | [210] |
| Streptomyces sp. Al-Dhabi 89 | Ag+ | Extracellular biomolecules | [211] |
| Streptomyces xinghaiensis OF1 | Ag+ | Organic compounds featuring amino bonds | [212] |
| Streptomyces capillispiralis Ca-1 | Cu2+ | Intracellular proteins | [213] |
| Micrococcus luteus HM 2 and HM 16 | CrO42− | Extracellular proteins | [214] |
| Rhodococcus etherivorans BCP1 | AsO43− | Mycothiols and NAD(P)H-dependent intracellular ArsC proteins | [215] |
| Streptomyces zaomyceticus Oc-5, Streptomyces pseudogriseolus Acv-11 | Cu2+ | Biomolecules containing peptide bonds, -NH, -OH, -CN, and C=C reactive groups | [216] |
| Frankia inefficax strain EuI1c | SeO32− | NADH-dependent reductase and dehydrogenase | [217] |
| Rhodococcus erythropolis ATCC 4277 | Fe3+ | N.D. 1 | [218,219] |
| Streptomyces spongiicola AS-3 (cell-free extracts) | Ag+ | Proteins, alcohols, and terpenoids | [220] |
| Nocardiopsis. dassonvillei-DS013 | Ag+ | Biomolecules containing -OH, -C+O, -CCl, -CBr, and -C≡C reactive groups | [221] |
| Streptomyces sp. M10A65 | SeO32− | Intracellular NAD(P)H-dependent reductase | [222] |
| Actinobacteria | Growth Conditions | Shape | Size (nm) | Crystal Structure | Stabilization | Properties | References |
|---|---|---|---|---|---|---|---|
| AuNMs | |||||||
| Thermomonospora sp. | growing cells | AuNPs 1 | ca. 10 | fcc 2 | extracellular secreted proteins | N.D. 3 | [162] |
| Rhodococcus sp. | resting cells | AuNPs | ca. 9 | fcc | N.D. | N.D. | [163] |
| Actinobacter sp. | BSA-exposed biomass | triangular hexagonal AuNPs | 50–500 | fcc | extracellular proteases | N.D. | [169] |
| BSA-exposed biomass (anoxic) | triangular AuNPs | 30–50 | fcc | ||||
| BSA-exposed biomass 25 C | AuNPs | N.D. | fcc | ||||
| Streptomyces viridogens HM10 | resting cells | AuNPs | 18–20 | fcc | N.D. | antibacterial | [246] |
| Arthrobacter sp. 61B; Arthrobacter globiformis 151B | resting cells | AuNPs | 8–40 | fcc | N.D. | N.D. | [247] |
| Streptomyces griseus | resting cells | AuNPs | 50 | fcc | N.D. | N.D. | [248] |
| Nocardia farcinica | cell-free extract | AuNPs | 15–20 | fcc | N.D. | N.D. | [182] |
| Streptomyces hygroscopicus | resting cells (neutral pH) | AuNPs | 2–10 | fcc | intracellular bioactive compounds | antibacterial, electrochemical | [176] |
| Streptomyces hygroscopicus | resting cells (acidic pH) | hexagonal, pentagonal AuNPs | 30–1500 | fcc | intracellular bioactive compounds | antibacterial, electrochemical | [176] |
| Streptomyces sp. ERI-3 | cell-free extract | AuNPs | ca. 9 | fcc | N.D. | N.D. | [249] |
| Micrococcus luteus NCIM 2379 | extracted α-amylase | AuNPs | ca. 6 | N.D. | extracted α-amylase | N.D. | [192] |
| extracted TUA | hexagonal AuNPs | ca. 50 | extracted TUA | ||||
| Streptomyces sp. VITDDK3 | cell-free extract | hexagonal, cubical AuNPs | ca. 90 | fcc | (2S,5R,6R)-2-hydroxy-3,5,6-trimethyloctan-4-one | antifungal | [191] |
| Sacchomonospora sp. | cell-free extract | triangular AuNPs | 40–80 | fcc | polypeptides | N.D. | [250] |
| Streptomyces hygroscopicus | resting cells | AuNPs | 10–20 | N.D. | N.D. | N.D. | [251] |
| Arthrobacter nitroguajacolicus | resting cells | AuNPs | ca.40 | fcc | N.D. | N.D. | [252] |
| Gordonia amarae | cell-free extract | AuNPs | 15–40 | fcc | N.D. | sensor for Cu detection | [202] |
| Gordonia amicalis HS-11 | cell-free extract | AuNPs | 5–25 | fcc | glycolipids | antioxidant | [253] |
| Streptomyces grisoruber | cell-free extract | triangular, hexagonal AuNPs | 5–50 | fcc | extracellular biomolecules | catalytic | [254] |
| Streptomyces sp. NH21 | cell-free extract | AuNPs | 10 | fcc | proteins | antibacterial | [205] |
| Nocardiopsis dassonvillei DS013 | growing cells | AuNPs | 30–80 | fcc | proteins | antibacterial | [221] |
| AgNMs | |||||||
| Corynebacterium strain SH09 | resting cells | AgNPs | 10–15 | fcc | proteins | N.D. | [166] |
| Streptomyces hygroscopicus | cell-free extract | AgNPs | 20–30 | fcc | extracellular biomolecules | antibacterial | [176] |
| Corynebacterium glutamicum | resting cells | irregular AgNPs | 5–50 | fcc | proteins | N.D. | [177] |
| Streptomyces glaucus 71MD | resting cells | AgNPs | 4–25 | fcc | N.D. | N.D. | [255] |
| Streptomyces sp. | cell-free extract | AgNPs | 15–25 | fcc | extracellular proteins | N.D. | [181] |
| Streptomyces rochei | cell-free extract | N.D. | N.D. | N.D. | N.D. | antibacterial | [256] |
| Rhodococcus NCIM 2891 | growing cells cell-free extract | AgNPs | ca.10 | fcc | N.D. | N.D. | [187] |
| 5–50 | proteins | antibacterial | [257] | ||||
| ca. 15 | mesophilic proteins | fluorescent, antimicrobial, catalytic | [188] | ||||
| Nocardiopsis sp. MBRC-1 | cell-free extract | AgNPs | 30–80 | fcc | proteins | antimicrobial, anticancer | [258] |
| Actinobacteria sp. PSBVIT-13 | cell-free extract | AgNPs | ca. 45 | fcc | proteins | antibacterial | [259] |
| Streptomyces sp. JAR1 | cell-free extract | N.D. | ca. 68 | fcc | extracellular proteins | antibacterial, antifungal | [189] |
| Micrococcus luteus | growing cells | AgNPs | <100 | fcc | N.D. | N.D. | [260] |
| Streptomyces sp. P-311 | crude enzyme extract | AgNPs | 100–200 | fcc | N.D. | antibacterial | [261] |
| Streptomyces sp., Streptoverticillium sp. | growing cells | AgNPs | ca. 8 < 70 | N.D. | N.D. | N.D. | [262] |
| Streptomyces sp. LK3 | cell-free extract | AgNPs | ca. 5 | fcc | N.D. | acaricidal | [263] |
| Streptomyces sp. SS2 | cell-free extract and resting cells | AgNPs | ca. 67 | N.D. | secondary metabolites | antibacterial | [264] |
| Pilimelia columellifera subsp. pallida SF23 and C9 | cell-free extract | AgNPs | 4–36 8–60 | N.D. | extracellular proteins | antifungal | [265] |
| Corynebacterium glutamicum | resting cells | AgNPs | 15 | N.D. | N.D. | antibacterial | [266] |
| Streptomyces rochei MHM13 | cell-free extract | AgNPs | 22–85 | N.D. | proteins | antibacterial, antibiofouling, anticancer | [200] |
| Gordonia amicalis HS-11 | cell-free extract | AgNPs | 5–25 | fcc | glycolipids | antioxidant | [253] |
| purified glycolipid | ca. 20 | ||||||
| Streptomyces sp. M-13 and M-24 | cell-free extract | AgNPs | 10–20 | N.D. | N.D. | antibacterial | [267] |
| Streptomyces parvulus DPUA 1549, Streptomyces owasiensis DPUA 1748 | cell-free extract | AgNPs | 1–40 | fcc | amino acids and peptides | antibacterial | [207] |
| Streptomyces sp. NH21 | cell-free extract | AgNPs | ca. 44 | fcc | proteins | antibacterial | [205] |
| resting cells | ca. 8 | ||||||
| Arthrobacter sp. B4 | EPS | AgNPs | 9–72 | fcc | EPS | antibacterial | [245] |
| Micromonospora KPMS10 | cell-free extract | AgNPs | ca.80 | N.D. | N.D. | antibacterial | [268] |
| Streptomyces sp. Al-Dhabi 89 | cell-free extract | cubic AgNPs | 4–11 | fcc | extracellular metabolites | antibacterial | [211] |
| Nocardiopsis alba | cell-free extract | AgNPs | 20–60 | fcc | N.D. | antibacterial, antiviral | [269] |
| Streptomyces spongiicola AS-3 | cell-free extract | AgNPs | ca.22 | fcc | proteins | antibacterial | [220] |
| Streptomyces sp. 192ANMG and 17ANMG | cell-free extract | AgNPs | ca. 9 ca. 35 | hcp 4 | proteins | antibacterial, antibiofilm | [270] |
| Fe-based NMs | |||||||
| Actinobacter strain EC5 | growing cells | mixture of Fe2O3 and Fe3O4 NPs | 10–40 | fcc | extracellular biomolecules | superpara magnetic | [167] |
| Actinobacter sp. | growing cells | Fe2O3 NPs | 5–7 | fcc | proteins | superpara magnetic | [170] |
| mixture of Fe3S4 and FeS2 NPs | ca. 20 | ||||||
| Rhodococcus erythropolis ATCC 4277 | growing cells, cell-free extract | Fe2O3 NPs | 50–100 | fcc | N.D. | N.D. | [218,219] |
| Cu-based NMs | |||||||
| Streptomyces sp. KUA106 | cell-free extract | CuO NPs | 100–150 | N.D. | reductases | antibacterial, antifungal | [271] |
| growing cells | |||||||
| Kocuria flava M-7 | cell-free extract | CuNPs | 5–30 | N.D. | proteins | N.D. | [195] |
| Streptomyces capillispiralis Ca-1 | cell-free extract | CuNPs | 5–59 | fcc | proteins | antimicrobial, larvicidal | [213] |
| Actinomycete sp. VITBN4 | cell-free extract | CuO NPs | ca. 60 | N.D. | proteins | antimicrobial | [272] |
| Streptomyces zaomyceticus Oc-5; Streptomyces pseudogriseolus Acv-11 | cell-free extract | CuO NPs | ca. 80 | fcc | N.D. | antimicrobial, antioxidant, larvicidal | [216] |
| Zn-based NMs | |||||||
| Streptomyces sp. KUA106 | growing cells, cell-free extract | ZnO NPs | 100–150 | N.D. | reductases | antimicrobial | [271] |
| Streptomyces sp. HBUM171191 | growing cells | ZnSO4 NPs | 10–20 | N.D. | N.D. | N.D. | [273] |
| Streptomyces sp. | growing cells | ZnO NPs | N.D. | N.D. | N.D. | antibacterial | [274] |
| Rhodococcus pyridinivorans NT2 | growing cells, cell-free extract | ZnO NPs | ca. 100 | N.D. | metabolites and proteins | UV-protective, photocatalytic, self-cleaning, antibacterial | [193] |
| SeNMs | |||||||
| Streptomyces bikiniensis strain Ess_amA-I | growing cells | SeNPs | 50–100 | N.D. | proteins and enzymes | N.D. | [198] |
| SeNRs 5 | 600 | anticancer | |||||
| Streptomyces minutiscleroticus M10A62 | resting cells | SeNPs | 100–250 | monoclinic | proteins | antibiofilm, antioxidant, wound healing, anticancer, antiviral | [198] |
| Streptomyces sp. ES2-5 | growing cells | SeNPs | 100–500 | N.D. | N.D. | N.D. | [203] |
| Rhodococcus etherivorans BCP1 | growing cells | SeNPs SeNRs | 50–600 | N.D. | amphiphilic biomolecules | N.D. | [208] |
| Frankia inefficax strain EuI1c | growing cells | SeNPs | N.D. | N.D. | N.D. | N.D. | [217] |
| Streptomyces sp. M10A65 | resting cells | SeNPs | 20–150 | monoclinic | proteins | antibacterial, larvicidal, anthelminthic | [222] |
| TeNMs | |||||||
| Rhodococcus etherivorans BCP1 | growing cells | TeNRs | 100–500 | N.D. | amphiphilic biomolecules | N.D. | [204] |
| resting cells | TeNPs TeNRs | 200–700 | trigonal | electrically conductive | [209] | ||
| Co-based NMs | |||||||
| Brevibacterium casei | cell-free extract | Co3O4 NPs | ca. 6 | crystals | proteins | magnetic | [172] |
| Mn-based NMs | |||||||
| Streptomyces sp. HBUM | growing cells | MnSO4 NPs | 10–20 | N.D. | N.D. | N.D. | [273] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Presentato, A.; Piacenza, E.; Turner, R.J.; Zannoni, D.; Cappelletti, M. Processing of Metals and Metalloids by Actinobacteria: Cell Resistance Mechanisms and Synthesis of Metal(loid)-Based Nanostructures. Microorganisms 2020, 8, 2027. https://doi.org/10.3390/microorganisms8122027
Presentato A, Piacenza E, Turner RJ, Zannoni D, Cappelletti M. Processing of Metals and Metalloids by Actinobacteria: Cell Resistance Mechanisms and Synthesis of Metal(loid)-Based Nanostructures. Microorganisms. 2020; 8(12):2027. https://doi.org/10.3390/microorganisms8122027
Chicago/Turabian StylePresentato, Alessandro, Elena Piacenza, Raymond J. Turner, Davide Zannoni, and Martina Cappelletti. 2020. "Processing of Metals and Metalloids by Actinobacteria: Cell Resistance Mechanisms and Synthesis of Metal(loid)-Based Nanostructures" Microorganisms 8, no. 12: 2027. https://doi.org/10.3390/microorganisms8122027
APA StylePresentato, A., Piacenza, E., Turner, R. J., Zannoni, D., & Cappelletti, M. (2020). Processing of Metals and Metalloids by Actinobacteria: Cell Resistance Mechanisms and Synthesis of Metal(loid)-Based Nanostructures. Microorganisms, 8(12), 2027. https://doi.org/10.3390/microorganisms8122027









