Evaluation of a kDNA-Based qPCR Assay for the Detection and Quantification of Old World Leishmania Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Leishmania Strains, Clinical Isolates and Clinical Samples
2.2. qPCR Assay
2.3. High-Resolution Melt (HRM) Analysis
2.4. PCR Product Sequencing
2.5. Statistical Analysis
3. Results
3.1. The qPCR-ML Assay Can Amplify All Medically Relevant Old World Leishmania Species
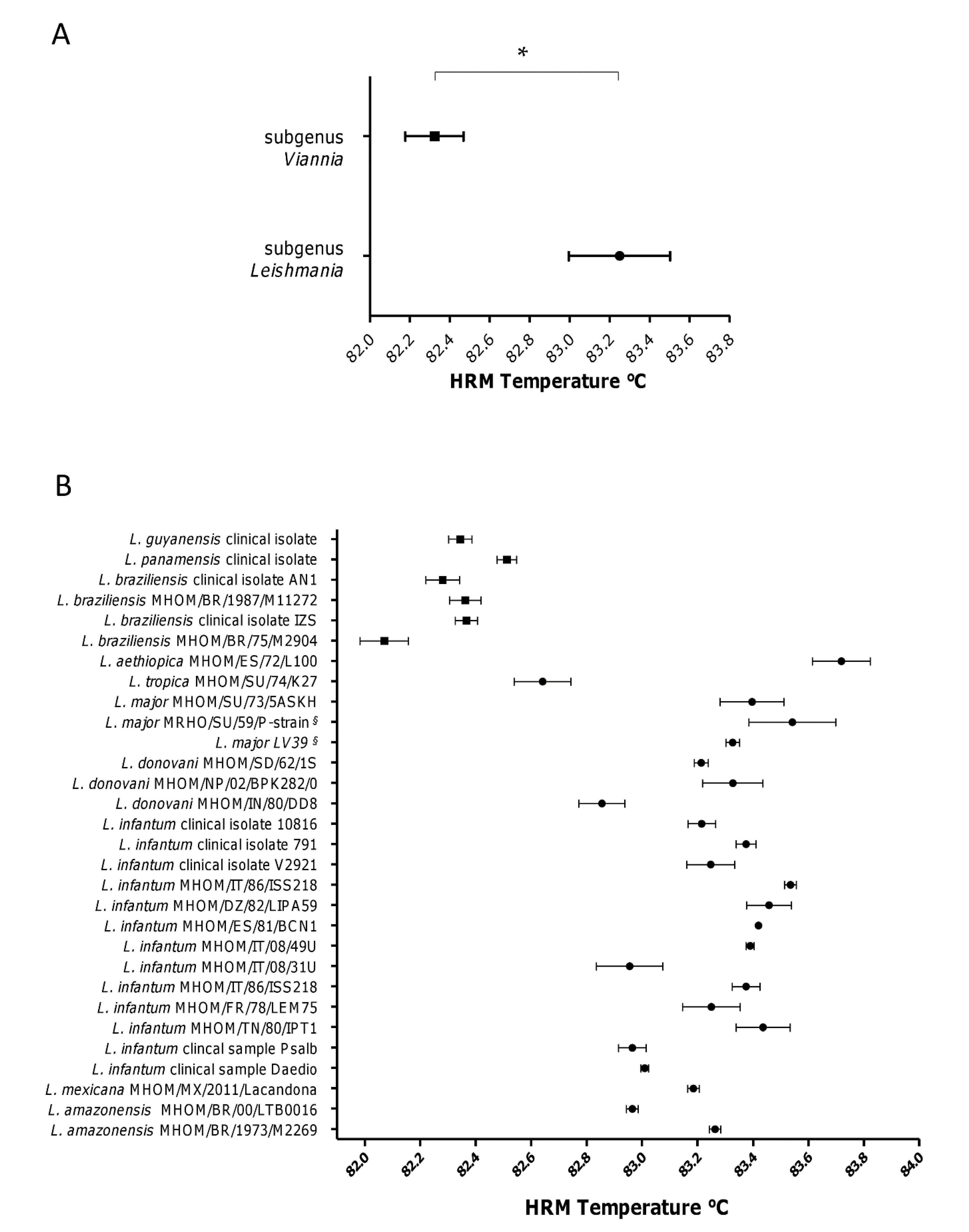
3.2. The subgenera Leishmania and Viannia Can Be Differentiated by HRM Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sereno, D.; Akhoundi, M.; Sayehmri, K.; Mirzaei, A.; Holzmuller, P.; Lejon, V.; Waleckx, E. Noninvasive Biological Samples to Detect and Diagnose Infections due to Trypanosomatidae Parasites: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2020, 21, 1684. [Google Scholar] [CrossRef]
- World Health Organization Leishmaniasis. Available online: http://www.who.int/mediacentre/factsheets/fs375/en/ (accessed on 16 March 2018).
- Shirian, S.; Oryan, A.; Hatam, G.R.; Daneshbod, Y. Three Leishmania/L. species—L. infantum, L. major, L. tropica—As causative agents of mucosal leishmaniasis in Iran. Pathog. Glob. Health 2013, 107, 267–272. [Google Scholar] [CrossRef]
- Rodriguez-Cortes, A.; Ojeda, A.; Francino, O.; Lopez-Fuertes, L.; Timon, M.; Alberola, J. Leishmania Infection: Laboratory Diagnosing in the Absence of a “Gold Standard”. Am. J. Trop. Med. Hyg. 2010, 82, 251–256. [Google Scholar] [CrossRef]
- Galluzzi, L.; Ceccarelli, M.; Diotallevi, A.; Menotta, M.; Magnani, M. Real-time PCR applications for diagnosis of leishmaniasis. Parasit. Vectors 2018, 11, 273. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Galluzzi, L.; Migliazzo, A.; Magnani, M. Detection and Characterization of Leishmania (Leishmania) and Leishmania (Viannia) by SYBR Green-Based Real-Time PCR and High Resolution Melt Analysis Targeting Kinetoplast Minicircle DNA. PLoS ONE 2014, 9, e88845. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Galluzzi, L.; Diotallevi, A.; Andreoni, F.; Fowler, H.; Petersen, C.; Vitale, F.; Magnani, M. The use of kDNA minicircle subclass relative abundance to differentiate between Leishmania (L.) infantum and Leishmania (L.) amazonensis. Parasit. Vectors 2017, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Diotallevi, A.; Buffi, G.; De Santi, M.; Fernández-Figueroa, E.A.; Rangel-Escareño, C.; Muñoz-Montero, S.A.; Becker, I.; Magnani, M.; Galluzzi, L. Differentiation of Leishmania (L.) infantum, Leishmania (L.) amazonensis and Leishmania (L.) mexicana Using Sequential qPCR Assays and High-Resolution Melt Analysis. Microorganisms 2020, 8, 818. [Google Scholar] [CrossRef] [PubMed]
- Diotallevi, A.; Buffi, G.; Ceccarelli, M.; Neitzke-Abreu, H.C.; Gnutzmann, L.V.; da Costa Lima, M.S.; Di Domenico, A.; De Santi, M.; Magnani, M.; Galluzzi, L. Real-time PCR to differentiate among Leishmania (Viannia) subgenus, Leishmania (Leishmania) infantum and Leishmania (Leishmania) amazonensis: Application on Brazilian clinical samples. Acta Trop. 2020, 201, 105178. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Diotallevi, A.; Andreoni, F.; Vitale, F.; Galluzzi, L.; Magnani, M. Exploiting genetic polymorphisms in metabolic enzymes for rapid screening of Leishmania infantum genotypes. Parasit. Vectors 2018, 11, 572. [Google Scholar] [CrossRef]
- White, H.; Potts, G. Mutation Scanning by High Resolution Melt Analysis. Evaluation of RotorGene 6000 (Corbett Life Science), HR1 and 384 Well LightScanner (Idaho Technology); National Genetics Reference Laboratory: Wessex, UK, 2006. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; Boer, M. den Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Mary, C.; Faraut, F.; Lascombe, L.; Dumon, H. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J. Clin. Microbiol. 2004, 42, 5249–5255. [Google Scholar] [CrossRef] [PubMed]
- de Paiva Cavalcanti, M.; Dantas-Torres, F.; da Cunha Gonçalves de Albuquerque, S.; Silva de Morais, R.C.; de Brito, M.E.F.; Otranto, D.; Brandão-Filho, S.P. Quantitative real time PCR assays for the detection of Leishmania (Viannia) braziliensis in animals and humans. Mol. Cell. Probes 2013, 27, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Pita-Pereira, D.; Lins, R.; Oliveira, M.P.; Lima, R.B.; Pereira, B.A.S.; Moreira, O.C.; Brazil, R.P.; Britto, C. SYBR Green-based real-time PCR targeting kinetoplast DNA can be used to discriminate between the main etiologic agents of Brazilian cutaneous and visceral leishmaniases. Parasit. Vectors 2012, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, L.; Prina, E.; Lang, T.; Milon, G. Real-Time PCR for Detection and Quantitation of Leishmania in Mouse Tissues. J. Clin. Microbiol. 2002, 40, 1666–1669. [Google Scholar] [CrossRef]
- De Monbrison, F.; Mihoubi, I.; Picot, S. Real-time PCR assay for the identification of cutaneous Leishmania parasite species in Constantine region of Algeria. Acta Trop. 2007, 102, 79–83. [Google Scholar] [CrossRef]
- Mansueto, P.; Seidita, A.; Vitale, G.; Cascio, A. Leishmaniasis in travelers: A literature review. Travel Med. Infect. Dis. 2014, 12, 563–581. [Google Scholar] [CrossRef]
- Pavli, A.; Maltezou, H.C. Leishmaniasis, an emerging infection in travelers. Int. J. Infect. Dis. 2010, 14, e1032–e1039. [Google Scholar] [CrossRef]
- Mody, R.M.; Lakhal-Naouar, I.; Sherwood, J.E.; Koles, N.L.; Shaw, D.; Bigley, D.P.; Co, E.M.A.; Copeland, N.K.; Jagodzinski, L.L.; Mukbel, R.M.; et al. Asymptomatic Visceral Leishmania infantum Infection in US Soldiers Deployed to Iraq. Clin. Infect. Dis. 2019, 68, 2036–2044. [Google Scholar] [CrossRef]
- Kniha, E.; Walochnik, J.; Poeppl, W.; Mooseder, G.; Obwaller, A.G. Leishmania spp. seropositivity in Austrian soldiers returning from the Kosovo. Wien. Klin. Wochenschr. 2020, 132, 47–49. [Google Scholar] [CrossRef]
- Bailey, M.S.; Langman, G. Misdiagnosis of cutaneous leishmaniasis and recurrence after surgical excision. J. R. Army Med. Corps 2013, 160, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Pareyn, M.; Hendrickx, R.; Girma, N.; Hendrickx, S.; Bockstal, L.V.; Houtte, N.V.; Shibru, S.; Maes, L.; Leirs, H.; Caljon, G. Evaluation of a pan—Leishmania SL RNA qPCR assay for parasite detection in laboratory-reared and field-collected sand flies and reservoir hosts. Parasit. Vectors 2020, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nateghi Rostami, M.; Darzi, F.; Farahmand, M.; Aghaei, M.; Parvizi, P. Performance of a universal PCR assay to identify different Leishmania species causative of Old World cutaneous leishmaniasis. Parasit. Vectors 2020, 13, 431. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.R.; Schoone, G.; Versteeg, I.; Gomez, M.A.; Diro, E.; Mori, Y.; Perlee, D.; Downing, T.; Saravia, N.; Assaye, A.; et al. Development and Evaluation of a Novel Loop-Mediated Isothermal Amplification Assay for Diagnosis of Cutaneous and Visceral Leishmaniasis. J. Clin. Microbiol. 2018, 56, 56. [Google Scholar] [CrossRef] [PubMed]
- Gow, I.; Millar, D.; Ellis, J.; Melki, J.; Stark, D. Semi-Quantitative, Duplexed qPCR Assay for the Detection of Leishmania spp. Using Bisulphite Conversion Technology. Trop. Med. Infect. Dis. 2019, 4, 135. [Google Scholar] [CrossRef]
- Krayter, L.; Schnur, L.F.; Schönian, G. The Genetic Relationship between Leishmania aethiopica and Leishmania tropica Revealed by Comparing Microsatellite Profiles. PLoS ONE 2015, 10, e0131227. [Google Scholar] [CrossRef]
- Schulz, A.; Mellenthin, K.; Schonian, G.; Fleischer, B.; Drosten, C. Detection, Differentiation, and Quantitation of Pathogenic Leishmania Organisms by a Fluorescence Resonance Energy Transfer-Based Real-Time PCR Assay. J. Clin. Microbiol. 2003, 41, 1529–1535. [Google Scholar] [CrossRef]
- Talmi-Frank, D.; Nasereddin, A.; Schnur, L.F.; Schönian, G.; Töz, S.Ö.; Jaffe, C.L.; Baneth, G. Detection and Identification of Old World Leishmania by High Resolution Melt Analysis. PLoS Negl. Trop. Dis. 2010, 4, e581. [Google Scholar] [CrossRef]
- Toz, S.O.; Culha, G.; Zeyrek, F.Y.; Ertabaklar, H.; Alkan, M.Z.; Vardarlı, A.T.; Gunduz, C.; Ozbel, Y. A Real-Time ITS1-PCR Based Method in the Diagnosis and Species Identification of Leishmania Parasite from Human and Dog Clinical Samples in Turkey. PLoS Negl. Trop. Dis. 2013, 7, e2205. [Google Scholar] [CrossRef]
- Weirather, J.L.; Jeronimo, S.M.B.; Gautam, S.; Sundar, S.; Kang, M.; Kurtz, M.A.; Haque, R.; Schriefer, A.; Talhari, S.; Carvalho, E.M.; et al. Serial Quantitative PCR Assay for Detection, Species Discrimination, and Quantification of Leishmania spp. in Human Samples. J. Clin. Microbiol. 2011, 49, 3892–3904. [Google Scholar] [CrossRef]


| Subgenus | Species | Strain/Isolate |
|---|---|---|
| Leishmania | L. infantum | MHOM/TN/80/IPT1 |
| Leishmania | L. infantum | MHOM/FR/78/LEM75 |
| Leishmania | L. infantum | Clinical isolate V2921 |
| Leishmania | L. infantum | MHOM/IT/08/31U |
| Leishmania | L. infantum | MHOM/IT/08/49U |
| Leishmania | L. infantum | Clinical isolate 10816 |
| Leishmania | L. infantum | Clinical isolate 791 |
| Leishmania | L. infantum | MHOM/DZ/82/LIPA59 |
| Leishmania | L. infantum | MHOM/ES/81/BCN1 |
| Leishmania | L. infantum | MHOM/IT/86/ISS218 |
| Leishmania | L. infantum | MHOM/IT/93/ISS822 |
| Leishmania | L. donovani | MHOM/IN/80/DD8 |
| Leishmania | L. donovani | MHOM/NP/02/BPK282/0cl4 |
| Leishmania | L. donovani | MHOM/SD/62/1S |
| Leishmania | L. major | MHOM/SU/73/5ASKH |
| Leishmania | L. major | MRHO/SU/59/P-strain * |
| Leishmania | L. major | SV39 * |
| Leishmania | L. tropica | MHOM/SU/74/K27 |
| Leishmania | L. aethiopica | MHOM/ET/72/L100 |
| Leishmania | L. amazonensis | MHOM/BR/00/LTB0016 |
| Leishmania | L. amazonensis | MHOM/BR/1973/M2269 |
| Leishmania | L. mexicana | MHOM/MX/2011/Lacandona |
| Leishmania | L. infantum | Clinical sample Psalb |
| Leishmania | L. infantum | Clinical sample Daedio |
| Viannia | L. panamensis | Clinical isolate |
| Viannia | L. guyanensis | Clinical isolate |
| Viannia | L. braziliensis | Clinical isolate AN1 |
| Viannia | L. braziliensis | Clinical isolate IZS |
| Viannia | L. braziliensis | MHOM/BR/1987/M11272 |
| Viannia | L. braziliensis | MHOM/BR/75/M2904 |
| Sauroleishmania | L. tarentolae | Parrot Tar II |
Publisher‘s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceccarelli, M.; Buffi, G.; Diotallevi, A.; Andreoni, F.; Bencardino, D.; Vitale, F.; Castelli, G.; Bruno, F.; Magnani, M.; Galluzzi, L. Evaluation of a kDNA-Based qPCR Assay for the Detection and Quantification of Old World Leishmania Species. Microorganisms 2020, 8, 2006. https://doi.org/10.3390/microorganisms8122006
Ceccarelli M, Buffi G, Diotallevi A, Andreoni F, Bencardino D, Vitale F, Castelli G, Bruno F, Magnani M, Galluzzi L. Evaluation of a kDNA-Based qPCR Assay for the Detection and Quantification of Old World Leishmania Species. Microorganisms. 2020; 8(12):2006. https://doi.org/10.3390/microorganisms8122006
Chicago/Turabian StyleCeccarelli, Marcello, Gloria Buffi, Aurora Diotallevi, Francesca Andreoni, Daniela Bencardino, Fabrizio Vitale, Germano Castelli, Federica Bruno, Mauro Magnani, and Luca Galluzzi. 2020. "Evaluation of a kDNA-Based qPCR Assay for the Detection and Quantification of Old World Leishmania Species" Microorganisms 8, no. 12: 2006. https://doi.org/10.3390/microorganisms8122006
APA StyleCeccarelli, M., Buffi, G., Diotallevi, A., Andreoni, F., Bencardino, D., Vitale, F., Castelli, G., Bruno, F., Magnani, M., & Galluzzi, L. (2020). Evaluation of a kDNA-Based qPCR Assay for the Detection and Quantification of Old World Leishmania Species. Microorganisms, 8(12), 2006. https://doi.org/10.3390/microorganisms8122006










