Establishment of a New PNA-FISH Method for Aspergillus fumigatus Identification: First Insights for Future Use in Pulmonary Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Maintenance and Growth Conditions
2.2. Design of a PNA Probe for the Specific Detection of A. fumigatus
2.3. Hybridization Conditions
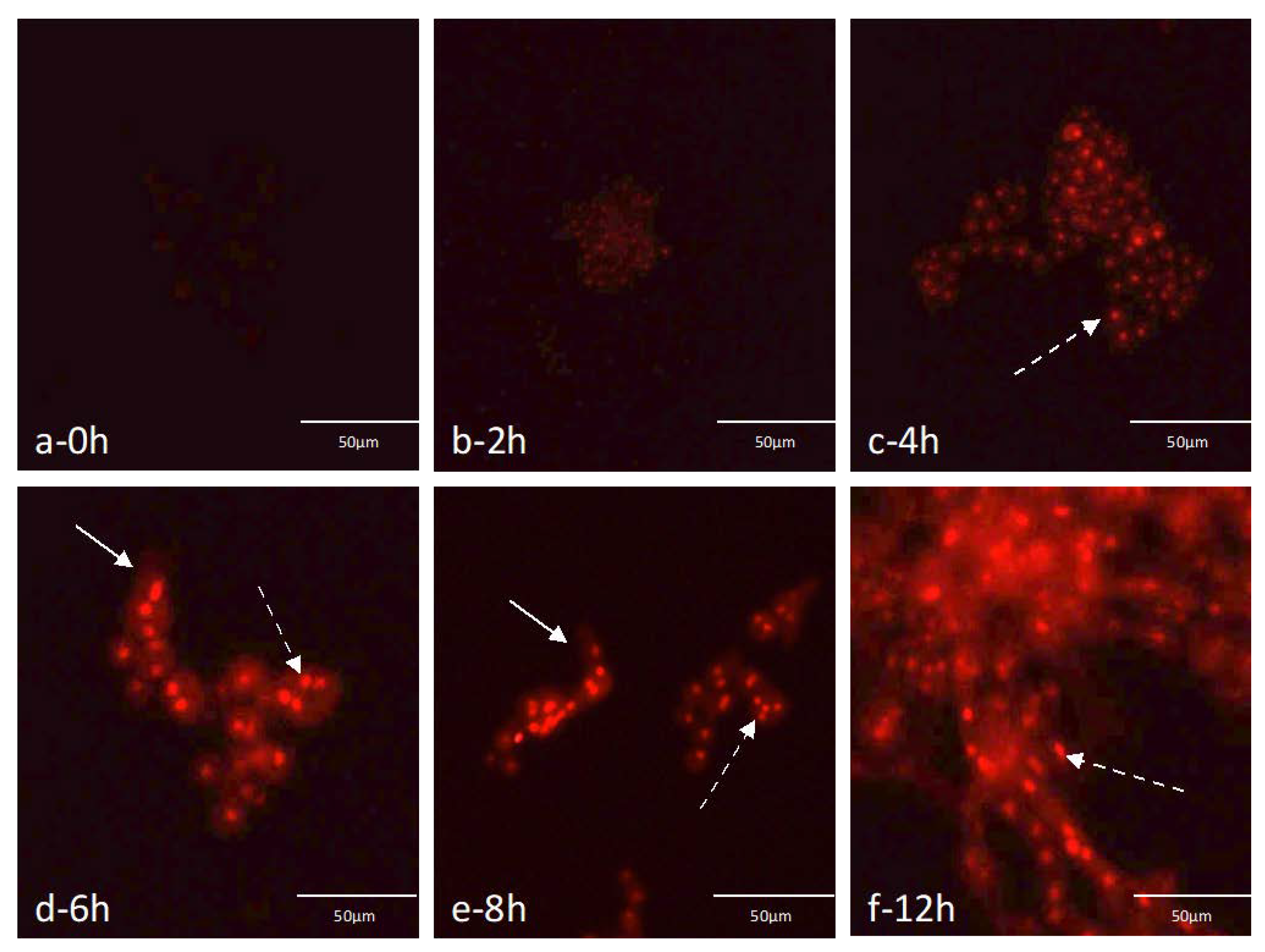
2.4. Germination Assays Using the Selected Probe
2.5. Detection of A. fumigatus in Artificial Sputum Media (ASM)
2.6. PNA-FISH Method Testing in Clinical Samples
2.7. Blind Study: Testing of PNA-FISH in Clinical Samples
2.8. Microscopy Visualization
2.9. Statistical Analysis
3. Results
3.1. Theoretical Specificity and Sensitivity Determination
3.2. PNA-FISH Performance
3.3. Detection of A. fumigatus in Artificial Sputum Medium Contaminated Samples
3.4. Optimization Assays Using Clinical Samples
3.5. Blind Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fang, W.; Latgé, J.P. Microbe Profile: Aspergillus fumigatus: A saprotrophic and opportunistic fungal pathogen. Microbiology (Reading) 2018, 164, 1009–1011. [Google Scholar] [CrossRef]
- Won-Chung, K.J.; Sugui, J.A. Aspergillus fumigatus—what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013, 9, e1003743. [Google Scholar] [CrossRef]
- Abad, A.; Fernandez-Molina, J.V.; Bikandi, J.; Ramirez, A.; Margareto, J.; Sendino, J.; Hernando, F.L.; Ponton, J.; Garaizar, J.; Rementeria, A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberam Micol. 2010, 27, 155–182. [Google Scholar] [CrossRef] [PubMed]
- Croft, C.; Culibrk, L.; Moore, M.; Tebbutt, S. Interactions of Aspergillus Fumigatus Conidia with Airway Epithelial Cells: A Critical Review. Front. Microbiol. 2016, 7, 472. [Google Scholar] [CrossRef]
- Berger, S.; El Chazli, Y.; Babu, A.F.; Coste, A.T. Azole Resistance in Aspergillus fumigatus: A Consequence of Antifungal Use in Agriculture? Front. Microbiol. 2017, 7, 1024. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin. Microbiol. 2009, 22, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Bandres, M.V.; Modi, P.; Sharma, S. Aspergillus fumigatus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, January 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK482464/ (accessed on 10 August 2020).
- Barton, R.C. Laboratory diagnosis of invasive aspergillosis: From diagnosis to prediction of outcome. Scientifica (Cairo) 2013, 2013, 459405. [Google Scholar] [CrossRef]
- Reyes-Montes, M.R.; Duarte-Escalante, E.; Frías-De-León, M.G.; Martínez-Herrera, E.O.; Acosta-Altamirano, G. Molecular Diagnosis of Invasive Aspergillosis. In Molecular Medicine; InTech Open: Rijeka, Croatia, 2018. [Google Scholar]
- Lamoth, F. Galactomannan and 1,3-β-d-Glucan Testing for the Diagnosis of Invasive Aspergillosis. J. Fungi 2016, 4, 22. [Google Scholar] [CrossRef]
- Bellanger, A.P.; Reboux, G.; Murat, J.B.; Bex, V.; Millon, L. Detection of Aspergillus fumigatus by quantitative polymerase chain reaction in air samples impacted on low-melt agar. Am. J. Infect. Control 2010, 38, 195–198. [Google Scholar] [CrossRef]
- Moura, S.; Cerqueira, L.; Almeida, A. Invasive pulmonary aspergillosis: Current diagnostic methodologies and a new molecular approach. Eur. J. Clin. Microbiol. Infect Dis. 2018, 37, 1393–1403. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Teertstra, W.R.; Lugones, L.G.; Wösten, H.A. In situ hybridisation in filamentous fungi using peptide nucleic acid probes. Fungal Genet Biol. 2004, 41, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, L.; Fernandes, R.M.; Ferreira, R.M.; Carneiro, F.; Dinis-Ribeiro, M.; Figueiredo, C.; Keevil, C.W.; Azevedo, N.F.; Vieira, M.J. PNA-FISH as a new diagnostic method for the determination of clarithromycin resistance of Helicobacter pylori. BMC Microbiol. 2011, 11, 101. [Google Scholar] [CrossRef] [PubMed]
- Economos, N.G.; Oyaghire, S.; Quijano, E.; Ricciardi, A.S.; Saltzman, W.M.; Glazer, P.M. Peptide Nucleic Acids and Gene Editing: Perspectives on Structure and Repair. Molecules 2020, 25, 735. [Google Scholar] [CrossRef]
- Sousa, J.M.; Rocha, R.; Cerqueira, L.; Almeida, C.; Azevedo, N.F.; Bastin, B.; Bird, P.; Benzinger, M.J., Jr.; Agin, J.; Goins, D.; et al. Validation of Biomode S.A. Probe4CronobacterTM for the Identification of Cronobacter spp. J. AOAC Int. 2019, 102, 855–864. [Google Scholar] [CrossRef]
- Rocha, R.; Sousa, J.M.; Cerqueira, L.; Vieira, M.J.; Almeida, C.; Azevedo, N.F. Development and application of Peptide Nucleic Acid Fluorescence in situ Hybridization for the specific detection of Listeria monocytogenes. Food Microbiol. 2019, 80, 1–8. [Google Scholar] [CrossRef]
- Lopes, S.P.; Carvalho, D.T.; Pereira, M.O.; Azevedo, N.F. Discriminating typical and atypical cystic fibrosis-related bacteria by multiplex PNA FISH. Biotechnol. Bioeng. 2017, 114, 355–367. [Google Scholar] [CrossRef]
- Mendes, L.; Rocha, R.; Azevedo, A.S.; Ferreira, C.; Henriques, M.; Pinto, M.G.; Azevedo, N.F. Novel strategy to detect and locate periodontal pathogens: The PNA-FISH technique. Microbiol. Res. 2016, 192, 185–191. [Google Scholar] [CrossRef]
- Almeida, C.; Sousa, J.M.; Rocha, R.; Cerqueira, L.; Fanning, S.; Azevedo, N.F.; Vieira, M.J. Detection of Escherichia coli O157 using PNA-FISH: Comparison to a standard culture method. Appl. Environ. Microbiol. 2013, AEM.01009-13. [Google Scholar] [CrossRef]
- Hall, L.; Le Febre, K.M.; Deml, S.M.; Wohlfiel, S.L.; Wengenack, N.L. Evaluation of the Yeast Traffic Light PNA FISH probes for identification of Candida species from positive blood cultures. J. Clin. Microbiol. 2012, 50, 1446–1448. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Lima, C.C.; Carvalho, D.; Meireles, F.; Guimarães, N.; Azevedo, N. Response surface methodology to optimize peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) in Saccharomyces cerevisiae. LWT 2017, 80, 27–31. [Google Scholar] [CrossRef]
- Shinozaki, M.; Okubo, Y.; Sasai, D.; Nakayama, H.; Murayama, S.Y.; Ide, T.; Wakayama, M.; Hiruta, N.; Shibuya, K. Identification of Fusarium species in formalin-fixed and paraffin-embedded sections by in situ hybridization using peptide nucleic acid probes. J. Clin. Microbiol. 2011, 49, 808–813. [Google Scholar] [CrossRef]
- Araujo, R.; Rodrigues, A.G.; Pina-Vaz, C. A fast, practical and reproducible procedure for the standardization of the cell density of an Aspergillus suspension. J. Med. Microbiol. 2004, 53 Pt 8, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Tarrand, J.J.; Han, X.Y.; Kontoyiannis, D.P.; May, G.S. Aspergillus hyphae in infected tissue: Evidence of physiologic adaptation and effect on culture recovery. J. Clin. Microbiol. 2005, 43, 382–386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, L.; Oliveira, J.A.; Nicolau, A.; Azevedo, N.F.; Vieira, M.J. Biofilm formation with mixed cultures of Pseudomonas aeruginosa/Escherichia coli on silicone using artificial urine to mimic urinary catheters. Biofouling 2013, 29, 829–840. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F.; Fernandes, R.M.; Keevil, C.W.; Vieira, M.J. Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples. Appl. Environ. Microbiol. 2010, 76, 4476–4485. [Google Scholar] [CrossRef]
- Cerqueira, L.; Azevedo, N.F.; Almeida, C.; Jardim, T.; Keevil, C.W.; Vieira, M.J. DNA mimics for the rapid identification of microorganisms by fluorescence in situ hybridization (FISH). Int. J. Mol. Sci. 2008, 9, 1944–1960. [Google Scholar] [CrossRef]
- Manavathu, E.K.; Cutright, J.; Chandrasekar, P.H. Comparative study of susceptibilities of germinated and ungerminated conidia of Aspergillus fumigatus to various antifungal agents. J. Clin. Microbiol. 1999, 37, 858–861. [Google Scholar] [CrossRef]
- Sriramulu, D.D.; Lünsdorf, H.; Lam, J.S.; Römling, U. Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 2005, 54 Pt 7, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Gonçalves Silva, D.; Rudnitskaya, A.; Almeida, A.; Rocha, S.M. Shedding light on Aspergillus niger volatile exometabolome. Sci. Rep. 2016, 6, 27441. [Google Scholar] [CrossRef][Green Version]
- Sabino, R.; Simões, H.; Veríssimo, C. Molecular Detection of Aspergillus: Application of a Real-Time PCR Multiplex Assay in Tissue Samples. J. Fungi (Basel) 2020, 6, 11. [Google Scholar] [CrossRef]
- Lamoth, F. Aspergillus fumigatus-Related Species in clinical Practice. Front. Microbiol. 2016, 7, 683. [Google Scholar] [CrossRef] [PubMed]
- Failmezger, J.; Ludwig, J.; Nieß, A.; Siemann-Herzberg, M. Quantifying ribosome dynamics in Escherichia coli using fluorescence. FEMS Microbiol. Lett. 2017, 364, fnx055. [Google Scholar] [CrossRef] [PubMed]
- Krijgsheld, P.; Bleichrodt, R.; Van Veluw, G.J.; Wang, F.; Muller, W.H.; Dijksterhuis, J.; Wosten, H.A.B. Development in Aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [CrossRef]
- Bhabhra, R.; Askew, D.S. Thermotolerance and virulence of Aspergillus fumigatus: Role of the fungal nucleolus. Med Mycol. 2005, 43, S87–S93. [Google Scholar] [CrossRef]
- Osherov, N. Conidial Germination in Aspergillus fumigatus. In Aspergillus fumigatus and Aspergillosis; Latgé, J., Steinbach, W., Eds.; ASM Press: Washington, DC, USA, 2009; pp. 131–142. [Google Scholar] [CrossRef]
- Herrero-Garcia, E.; Perez-de-Nanclares-Arregi, E.; Cortese, M.S.; Markina-Iñarrairaegui, A.; Oiartzabal-Arano, E.; Etxebeste, O.; Ugalde, U.; Espeso, E.A. Tip-to-nucleus migration dynamics of the asexual development regulator FlbB in vegetative cells. Mol. Microbiol. 2015, 98, 607–624. [Google Scholar] [CrossRef]
- Baltussen, T.J.H.; Zoll, J.; Verweij, P.E.; Melchers, W.J.G. Molecular Mechanisms of Conidial Germination in Aspergillus spp. Microbiol. Mol. Biol. Rev. 2019, 84, e00049-19. [Google Scholar] [CrossRef]
- Dague, E.; Alsteens, D.; Latgé, J.P.; Dufrêne, Y.F. High-resolution cell surface dynamics of germinating Aspergillus fumigatus conidia. Biophys. J. 2008, 94, 656–660. [Google Scholar] [CrossRef]
- Pandey, V.; Singh, P.; Singh, S.; Arora, N.; Quadir, N.; Singh, S.; Das, A.; Dudeja, M.; Kapur, P.; Ehtesham, N.Z. SeeTB: A novel alternative to sputum smear microscopy to diagnose tuberculosis in high burden countries. Sci. Rep. 2019, 9, 16371. [Google Scholar] [CrossRef]
- Kennedy, V.E.; Todd, J.L.; Palmer, S.M. Bronchoalveolar lavage as a tool to predict, diagnose and understand bronchiolitis obliterans syndrome. Am. J. Transplant. 2013, 13, 552–561. [Google Scholar] [CrossRef]
- Fontaine, T.; Beauvais, A.; Loussert, C.; Thevenard, B.; Fulgsang, C.C.; Ohno, N.; Clavaud, C.; Prevost, M.-C.; Latgé, J.-P. Cell wall alpha1-3glucans induce the aggregation of germinating conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2010, 47, 707–712. [Google Scholar] [CrossRef]
- Leite, G.M.; Magan, N.; Medina, A. Comparison of different bead-beating RNA extraction strategies: An optimizedmethod forfilamentous fungi. J. Microbiol. Methods 2012, 88, 413–418. [Google Scholar] [CrossRef]
- Francesconi, A.; Kasai, M.; Harrington, S.M.; Beveridge, M.G.; Petraitiene, R.; Petraitis, V.; Schaufele, R.L.; Walsh, T.J. Automated and manual methods of DNA extraction for Aspergillus fumigatus and Rhizopus oryzae analyzed by quantitative real-time PCR. J. Clin. Microbiol. 2008, 46, 1978–1984. [Google Scholar] [CrossRef]
- Rantakokko-Jalava, K.; Laaksonen, S.; Issakainen, J.; Vauras, J.; Nikoskelainen, J.; Viljanen, M.K.; Salonen, J. Semiquantitative detection by real-time PCR of Aspergillus fumigatus in bronchoalveolar lavage fluids and tissue biopsy specimens from patients with invasive aspergillosis. J. Clin. Microbiol. 2003, 41, 4304–4311. [Google Scholar] [CrossRef]

| Strains Tested | PNA−FISH Outcome |
|---|---|
| Aspergillus fumigatus MUM 02.24 | + |
| Aspergillus fumigatus MUM 07.05 | + |
| Aspergillus fumigatus MUM 9802 | + |
| Aspergillus fumigatus ATCC 46645 | + |
| Aspergillus fumigatus CECT 2071 | + |
| Aspergillus fumigatus CECT 20190 | + |
| Aspergillus fumigatus CECT 20228 | + |
| Aspergillus fumigatus CECT 20366 | + |
| Aspergillus ibericus MUM 03.49 | − |
| Aspergillus ochraceus MUM 9302 | − |
| Aspergillus clavatus MUM 9717 | − |
| Aspergillus versicolor MUM 00.20 | − |
| Aspergillus terreus MUM 9409 | − |
| Aspergillus tubingensis MUM 06.152 | − |
| Aspergillus oryzae MUM 10242 | − |
| Aspergillus flavus MUM 00.06 | − |
| Aspergillus flavus MUM 9201 | − |
| Aspergillus niger MUM 92.13 | − |
| Aspergillus niger MUM 01.01 | − |
| Emericella nidulans var. echinulata MUM 9832 | − |
| Neosartorya fisheri var. glabra MUM 9836 | − |
| Penicillium brevicompactum MUM 02.12 | − |
| Penicillium chrysogenum MUM 061.70 | − |
| Mucor hiemalis MUM 9732 | − |
| Trichoderma viride MUM 9754 | − |
| Candida parapsilosis ATCC 22019 | − |
| Candida tropicalis ATCC 750 | − |
| Candida glabrata ATCC 2001 | − |
| Candida albicans ATCC 1472 | − |
| Pseudomonas aeruginosa PAO1 | − |
| Pseudomonas aeruginosa CECT 111 | − |
| Escherichia coli K12 | − |
| Staphylococcus aureus CECT 239 | − |
| ASM | ||||
|---|---|---|---|---|
| Concentration (conidia·mL−1) | 6 h | 8 h | 12 h | 24 h |
| MUM 07.05 | ||||
| 1 × 104 | − | − | − | + |
| 1 × 103 | − | − | − | + |
| 1 × 102 | − | − | − | − |
| 1 × 101 | − | − | − | − |
| ATCC 46645 | ||||
| 1 × 104 | − | − | − | + |
| 1 × 103 | − | − | − | + |
| 1 × 102 | − | − | − | + |
| 1 × 101 | − | − | − | − |
| CECT 20366 | ||||
| 1 × 104 | − | − | − | + |
| 1 × 103 | − | − | − | + |
| 1 × 102 | − | − | − | + |
| 1 × 101 | − | − | − | − |
| CECT 2071 | ||||
| 1 × 104 | − | − | − | + |
| 1 × 103 | − | − | − | + |
| 1 × 102 | − | − | − | + |
| 1 × 101 | − | − | − | − |
| Sample | Culture | PNA−FISH Outcome | ||
|---|---|---|---|---|
| 8 h | 12 h | 24 h | ||
| Sputum | ||||
| 1 | + | − | + | + |
| 2 | + | − | − | + |
| 3 | + | − | + | + |
| BL | ||||
| 1 | + | − | + | + |
| 2 | + | − | + | + |
| 3 | + | + | + | + |
| BAL | ||||
| 1 | + | − | − | + |
| 2 | + | − | − | + |
| 3 | + | + | + | + |
| Sample | Inoculation | Culture | PNA−FISH Outcome |
|---|---|---|---|
| BAL | |||
| 1 | + | + | + |
| 2 | + | + | + |
| 3 | − | − | − |
| 4 | + | + | + |
| 5 | − | − | − |
| 6 | + | + | + |
| 7 | + | + | − |
| 8 | − | − | − |
| BL | |||
| 9 | − | − | − |
| 10 | + | + | + |
| 11 | − | − | − |
| 12 | + | + | + |
| 13 | − | − | − |
| 14 | + | + | − |
| Sputum | |||
| 15 | + | + | + |
| 16 | − | − | − |
| 17 | + | + | + |
| 18 | + | + | − |
| 19 | + | + | + |
| 20 | − | − | − |
| 21 | + | + | + |
| 22 | + | + | + |
| 23 | − | − | − |
| 24 | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerqueira, L.; Moura, S.; Almeida, C.; Vieira, M.J.; Azevedo, N.F. Establishment of a New PNA-FISH Method for Aspergillus fumigatus Identification: First Insights for Future Use in Pulmonary Samples. Microorganisms 2020, 8, 1950. https://doi.org/10.3390/microorganisms8121950
Cerqueira L, Moura S, Almeida C, Vieira MJ, Azevedo NF. Establishment of a New PNA-FISH Method for Aspergillus fumigatus Identification: First Insights for Future Use in Pulmonary Samples. Microorganisms. 2020; 8(12):1950. https://doi.org/10.3390/microorganisms8121950
Chicago/Turabian StyleCerqueira, Laura, Sara Moura, Carina Almeida, Maria João Vieira, and Nuno Filipe Azevedo. 2020. "Establishment of a New PNA-FISH Method for Aspergillus fumigatus Identification: First Insights for Future Use in Pulmonary Samples" Microorganisms 8, no. 12: 1950. https://doi.org/10.3390/microorganisms8121950
APA StyleCerqueira, L., Moura, S., Almeida, C., Vieira, M. J., & Azevedo, N. F. (2020). Establishment of a New PNA-FISH Method for Aspergillus fumigatus Identification: First Insights for Future Use in Pulmonary Samples. Microorganisms, 8(12), 1950. https://doi.org/10.3390/microorganisms8121950








