Comparative Genomics of Xanthomonas citri pv. citri A* Pathotype Reveals Three Distinct Clades with Varying Plasmid Distribution
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. DNA Extraction
2.3. Library Preparation and Sequencing with Illumina and Minion
2.4. Data Processing, Assembly, and Sequence Analysis
2.5. Identification of Type 3 Secreted Effectors
2.6. Phylogenetic Analysis and Pan-Genome
3. Results
3.1. Assembly Completion
3.2. General Features of Xanthomonas citri pv. citri A* Genomes
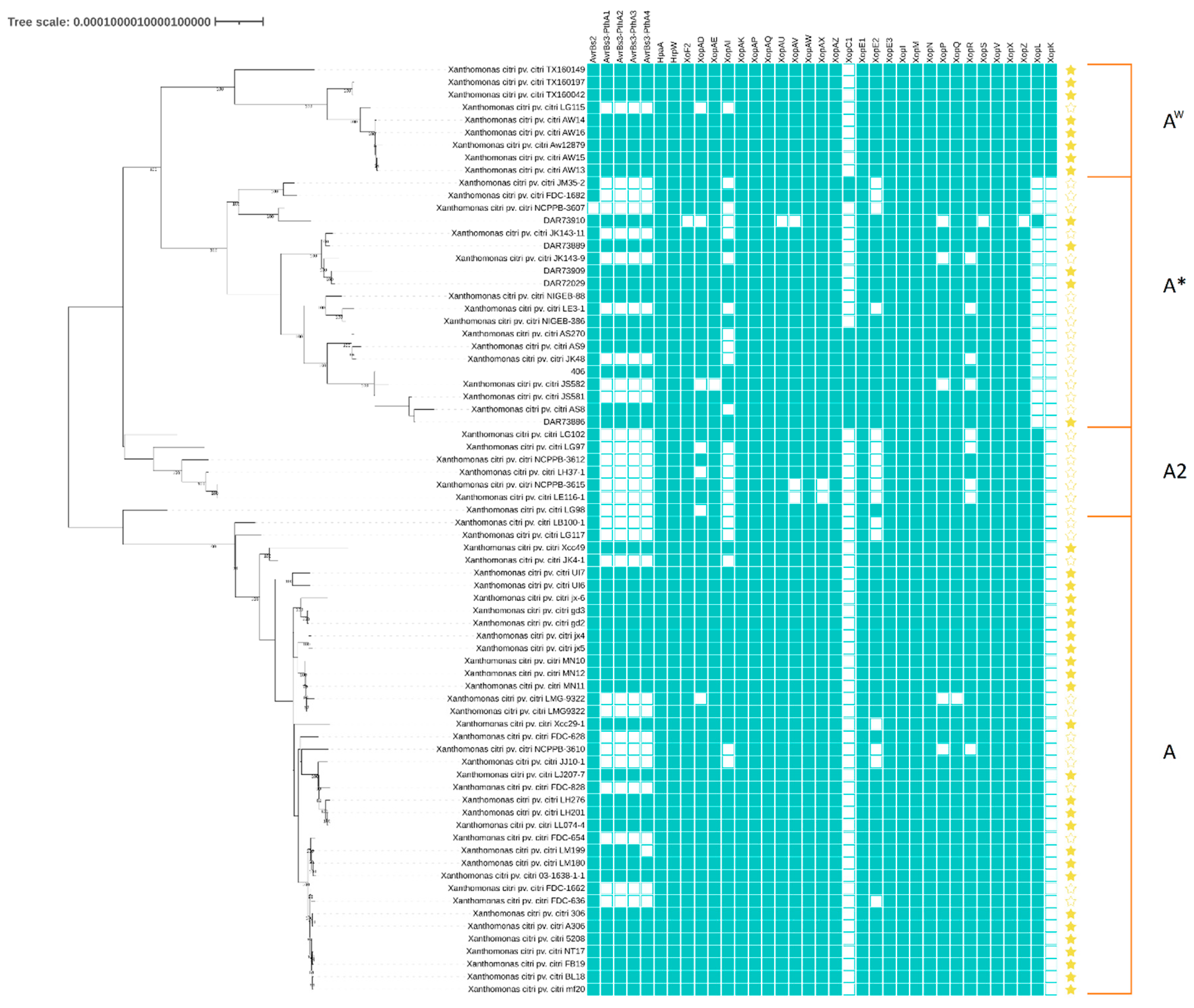
3.3. Placement of A* Strains amongst X. citri pv. citri Population
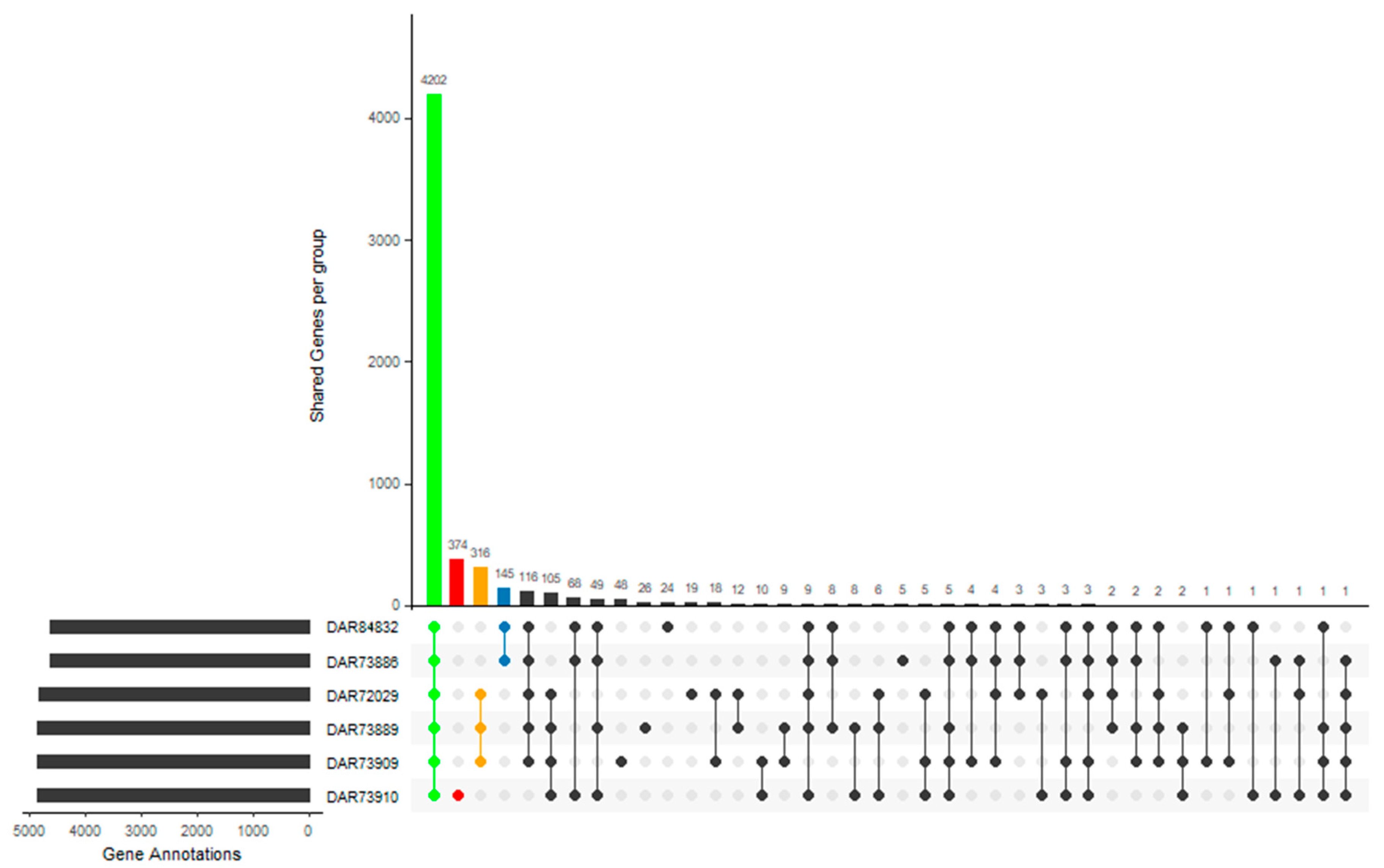
3.4. Pan-Genome Analysis
3.5. Type 3 Secreted Effectors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schubert, T.S.; Rizvi, S.A.; Sun, X.; Gottwald, T.R.; Graham, J.H.; Dixon, W.N. Meeting the challenge of eradicating citrus canker in Florida—Again. Plant Dis. 2001, 85, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Gambley, C.F.; Miles, A.K.; Ramsden, M.; Doogan, V.; Thomas, J.E.; Parmenter, K.; Whittle, P.J.L. The distribution and spread of citrus canker in Emerald, Australia. Australas. Plant Pathol. 2009, 38, 547–557. [Google Scholar] [CrossRef]
- Daniels, D. Australian Citrus Export Trade, on Citrus Australia. 2017. Available online: https://www.citrusaustralia.com.au/wp-content/uploads/David-Daniels_Citrus-Australia.pdf (accessed on 5 October 2020).
- Anonymous. Citrus Australia. Available online: https://citrusaustralia.com.au/ (accessed on 5 October 2020).
- Anonymous. Biosecurity Regulation, Queensland Government. Available online: https://www.legislation.qld.gov.au/view/pdf/inforce/2019-09-01/sl-2016-0075 (accessed on 25 November 2020).
- Civerolo, E. Bacterial canker disease of citrus (Xanthomonas campestris). J. Rio Gd. Val. Hortic. Soc. 1984, 27, 127–146. [Google Scholar]
- Rybak, M.; Minsavage, G.V.; Stall, R.E.; Jones, J.B. Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol. Plant Pathol. 2009, 10, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Vernière, C.; Hartung, J.; Pruvost, O.; Civerolo, E.; Alvarez, A.; Maestri, P.; Luisetti, J. Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 1998, 104, 477–487. [Google Scholar]
- Jalan, N.; Kumar, D.; Yu, F.; Jones, J.B.; Graham, J.H.; Wang, N. Complete genome sequence of Xanthomonas citri subsp. citri strain Aw12879, a restricted-host-Range citrus canker-causing bacterium. Genome Announc. 2013, 1, e00235-13. [Google Scholar] [CrossRef]
- ordon, J.L.; Lefeuvre, P.; Escalon, A.; Barbe, V.; Cruveiller, S.; Gagnevin, L.; Pruvost, O. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genom. 2015, 16, 1098. [Google Scholar]
- Salehzadeh, A.; Alavi, S.M.; Sangtarash, M.H. Comparative genomic analysis of wide and narrow host range strains of Xanthomonas citri subsp. citri, showing differences in the genetic content of their pathogenicity and virulence factors. Australas. Plant Pathol. 2017, 46, 49–61. [Google Scholar]
- Zhang, Y.; Jalan, N.; Zhou, X.; Goss, E.; Jones, J.B.; Setubal, J.C.; Deng, X.; Wang, N. Positive selection is the main driving force for evolution of citrus canker-causing Xanthomonas. ISME J. 2015, 9, 2128–2138. [Google Scholar] [CrossRef]
- Patané, J.S.L.; Martins, J.; Rangel, L.T.; Belasque, J.; Digiampietri, L.A.; Facincani, A.P.; Ferreira, R.M.; Jaciani, F.J.; Zhang, Y.; Varani, A.M.; et al. Origin and diversification of Xanthomonas citri subsp. citri pathotypes revealed by inclusive phylogenomic, dating, and biogeographic analyses. BMC Genom. 2019, 20, 700. [Google Scholar]
- Pruvost, O.; Magne, M.; Boyer, K.; LeDuc, A.; Tourterel, C.; Drevet, C.; Ravigné, V.; Gagnevin, L.; Guérin, F.; Chiroleu, F.; et al. A MLVA genotyping scheme for global surveillance of the citrus pathogen Xanthomonas citri pv. citri suggests a worldwide geographical expansion of a single genetic lineage. PLoS ONE 2014, 9, 98129. [Google Scholar] [CrossRef]
- Büttner, D.; Bonas, U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 2010, 34, 107–133. [Google Scholar] [CrossRef] [PubMed]
- cheibner, F.; Hartmann, N.; Hausner, J.; Lorenz, C.; Hoffmeister, A.-K.; Büttner, D. The type III secretion chaperone hpab controls the translocation of effector and noneffector proteins from Xanthomonas campestris pv. vesicatoria. Mol. Plant Microbe Interact. 2018, 31, 61–74. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- 1acques, M.-A.; Arlat, M.; Boulanger, A.; Boureau, T.; Carrère, S.; Cesbron, S.; Chen, N.W.G.; Cociancich, S.; Darrasse, A.; Denancé, N.; et al. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu. Rev. Phytopathol. 2016, 54, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ji, C.; Huguet-Tapia, J.C.; White, F.F.; Dong, H.; Yang, B. An efficient method to clone TAL effector genes from Xanthomonas oryzae using Gibson assembly. Mol. Plant Pathol. 2019, 20, 1453–1462. [Google Scholar] [CrossRef]
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I Cs LOB 1 promoter using Cas9/sg RNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4: DCs LOB 1.3 infection. Plant Biotechnol. J. 2016, 14, 1291–1301. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, N. High-throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol. Plant Microbe Interact. 2012, 25, 69–84. [Google Scholar] [CrossRef]
- Swarup, S.; Yang, Y.; Kingsley, M.T.; Gabriel, D.W. An Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant Microbe Interact. 1992, 5, 204–213. [Google Scholar] [CrossRef]
- Duan, S.; Jia, H.; Pang, Z.; Teper, D.; White, F.; Jones, J.; Zhou, C.Y.; Wang, N. Functional characterization of the citrus canker susceptibility gene CsLOB1. Mol. Plant Pathol. 2018, 19, 1908–1916. [Google Scholar] [CrossRef]
- Alegria, M.C.; Souza, D.P.; Andrade, M.O.; Docena, C.; Khater, L.; Ramos, C.H.I.; Da Silva, A.C.R.; Farah, C.S. Identification of new protein-protein interactions involving the products of the chromosome-and plasmid-encoded type iv secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J. Bacteriol. 2005, 187, 2315–2325. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.G.; Christie, P.J. The Agrobacterium VirB/VirD4 T4SS: Mechanism and Architecture Defined Through In Vivo Mutagenesis and Chimeric Systems, Agrobacterium Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 233–260. [Google Scholar]
- Llosa, M.; Gomis-Ruth, F.X.; Coll, M.; Cruz, F. Bacterial conjugation: A two-step mechanism for DNA transport. Mol. Microbiol. 2002, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cenens, W.; Andrade, M.O.; Llontop, E.; Alvarez-Martinez, C.E.; Sgro, G.G.; Farah, C.S. Bactericidal type IV secretion system homeostasis in Xanthomonas citri. PLoS Pathog. 2020, 16, 1008561. [Google Scholar] [CrossRef] [PubMed]
- Sgro, G.G.; Oka, G.U.; Souza, D.P.; Cenens, W.; Bayer-Santos, E.; Matsuyama, B.Y.; Bueno, N.F.; Dos Santos, T.R.; Alvarez-Martinez, C.E.; Salinas, R.K.; et al. Bacteria-killing type IV secretion systems. Front. Microbiol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Souza, D.P.; Okam, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.; Salinas, R.K.; et al. Bacterial killing via a type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef]
- Qian, W.; Jia-Xun, F.; Ren, S.X.; He, Y.Q.; Feng, J.X.; Lu, L.F.; Sun, Q.; Yingchuan, T.; Tang, D.J.; Tang, H.; et al. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 2005, 15, 757–767. [Google Scholar] [CrossRef]
- Loman, N.J.; Constantinidou, C.; Chan, J.Z.M.; Halachev, M.R.; Sergeant, M.J.; Penn, C.W.; Robinson, E.R.; Pallen, M.J. High-throughput bacterial genome sequencing: An embarrassment of choice, a world of opportunity. Nat. Rev. Genet. 2012, 10, 599–606. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; McDonald, B.A. The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 2008, 46, 75–100. [Google Scholar] [CrossRef]
- McCann, H.C. Skirmish or war: The emergence of agricultural plant pathogens. Curr. Opin. Plant Biol. 2020, 56, 147–152. [Google Scholar] [CrossRef]
- Vinatzer, B.A.; Monteil, C.L.; Clarke, C.R. Harnessing population genomics to understand how bacterial pathogens emerge, adapt to crop hosts, and disseminate. Annu. Rev. Phytopathol. 2014, 52, 19–43. [Google Scholar] [CrossRef]
- Behlau, F.; Canteros, B.I.; Minsavage, G.V.; Jones, J.B.; Graham, J.H. Molecular characterization of copper resistance genes from Xanthomonas citri subsp.citriand Xanthomonas alfalfae subsp. citrumelonis. Appl. Environ. Microbiol. 2011, 77, 4089–4096. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, E.; De Pascale, F.; Vezzi, A.; Hübner, S.; Aharonovich, D.; Sher, D. Why close a bacterial genome? The plasmid of Alteromonas macleodii HOT1A3 is a vector for inter-specific transfer of a flexible genomic Island. Front. Microbiol. 2016, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Yan, C.; Wu, J.; Pan, X.; Yan, N. Revisiting the TALE repeat. Protein Cell 2014, 5, 297–306. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bogema, D.; Micallef, M.L.; Liu, M.; Padula, M.P.; Djordjevic, S.P.; Darling, A.E.; Jenkins, C. Analysis of Theileria orientalis draft genome sequences reveals potential species-level divergence of the Ikeda, Chitose and Buffeli genotypes. BMC Genom. 2018, 19, 298. [Google Scholar] [CrossRef] [PubMed]
- Wick, R. Porechop. Available online: https://github.com/rrwick/Porechop (accessed on 12 February 2019).
- Wick, R. Filtlong. Available online: https://github.com/rrwick/FiltLong (accessed on 12 February 2019).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, 1005595. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated Tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, 112963. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.J.B. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Snippy: Rapid haploid variant calling and core SNP phylogeny. Available online: https://github.com/tseemann/snippy (accessed on 18 February 2020).
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J.; et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020, 21, 1–21. [Google Scholar] [CrossRef]
- Cubero, J.; Graham, J.H. Genetic Relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 2002, 68, 1257–1264. [Google Scholar] [CrossRef]
- Timilsina, S.; Pereira-Martin, J.A.; Minsavage, G.V.; Iruegas-Bocardo, F.; Abrahamian, P.; Potnis, N.; Kolaczkowski, B.; Vallad, G.; Goss, E.M.; Jones, J.B. Multiple recombination events drive the current genetic structure of Xanthomonas perforans in Florida. Front. Microbiol. 2019, 10, 448. [Google Scholar] [CrossRef]
- Jibrin, M.O.; Potnis, N.; Timilsina, S.; Minsavage, G.V.; Vallad, G.E.; Roberts, P.D.; Jones, J.B.; Goss, E.M. Genomic Inference of Recombination-Mediated Evolution in Xanthomonas euvesicatoria and X. perforans. Appl. Environ. Microbiol. 2018, 84, e00136-18. [Google Scholar] [CrossRef]
- Didelot, X.; Wilson, D.J. ClonalFrameML: Efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 2015, 11, 1004041. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Harris, S.R.; Fraser, C.; Quail, M.A.; Burton, J.; Van Der Linden, M.; McGee, L.; Von Gottberg, A.; Song, J.H.; Ko, K.S.; et al. Rapid pneumococcal evolution in response to clinical interventions. Science 2011, 331, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Wallden, K.; Rivera-Calzada, A.; Waksman, G. Microreview: Type IV secretion systems: Versatility and diversity in function. Cell. Microbiol. 2010, 12, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.C.R.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Van Sluys, M.A.; Almeida, N.F.; Alves, L.M.C.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nat. Cell Biol. 2002, 417, 459–463. [Google Scholar] [CrossRef]
- Richard, D.; Ravigné, V.; Rieux, A.; Facon, B.; Boyer, C.; Boyer, K.; Grygiel, P.; Javegny, S.; Terville, M.; Canteros, B.I.; et al. Adaptation of genetically monomorphic bacteria: Evolution of copper resistance through multiple horizontal gene transfers of complex and versatile mobile genetic elements. Mol. Ecol. 2017, 26, 2131–2149. [Google Scholar] [CrossRef]
- Colombi, E.; Straubm, C.; Künzelm, S.; Templeton, M.D.; McCann, H.C.; Rainey, P.B. Evolution of copper resistance in the kiwifruit pathogen Pseudomonas syringae pv. actinidiae through acquisition of integrative conjugative elements and plasmids. Environ. Microbiol. 2017, 19, 819–832. [Google Scholar] [CrossRef]
- Graham, M. 2017–2018 Florida Citrus Production Guide; University of Florida: Gainesville, FL, USA, 2017. [Google Scholar]
- Timmer, L.; Graham, J.; Chamberlain, H. Fundamentals of Citrus Canker Management; University of Florida: Gainesville, FL, USA, 2006. [Google Scholar]
- Yan, X.; Tao, J.; Luo, H.L.; Tan, L.T.; Rong, W.; Li, H.P.; He, C.Z. A type III effector XopLXcc8004 is vital for Xanthomonas campestris pathovar campestris to regulate plant immunity. Res. Microbiol. 2019, 170, 138–146. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- chwessinger, B.; Zipfel, C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008, 11, 389–395. [Google Scholar] [CrossRef]
- Singer, A.U.; Schulze, S.; Skarina, T.; Xu, X.; Cui, H.; Eschen-Lippold, L.; Egler, M.; Srikumar, T.; Raught, B.; Lee, J.; et al. A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 2013, 9, 1003121. [Google Scholar] [CrossRef]
- Malamud, F.; Torres, P.S.; Roeschlin, R.; Rigano, L.A.; Enrique, R.; Bonomi, H.R.; Castagnaro, A.P.; Marano, M.R.; Vojnov, A.A. The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 2011, 157, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Hajri, A.; Brin, C.; Hunault, G.; Lardeux, F.; Lemaire, C.; Manceau, C.; Boureau, T.; Poussier, S. A “repertoire for repertoire” hypothesis: Repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS ONE 2009, 4, 6632. [Google Scholar] [CrossRef]

| Isolate | Location | Year |
|---|---|---|
| DAR73910 | International intercept from India at Sydney Airport | 2000 |
| DAR73909 | Thailand | 2000 |
| DAR73886 | Iran | Unknown |
| DAR73889 | International intercept from Thailand at Sydney Airport | Unknown |
| DAR72029 | International intercept from Singapore at Sydney Airport | 1997 |
| DAR84832 | Southwest Asia | Unknown |
| Size (bp) | GC Content | Total CDS | tRNA | rRNA | |
|---|---|---|---|---|---|
| DAR84832 | |||||
| Chromosome | 5,223,652 | 64.7 | 4429 | 58 | 6 |
| Plasmid 1 (pXAC64) | 61,273 | 61.4 | 67 | 0 | 0 |
| Plasmid 2 (pXAS47) | 47,525 | 60.6 | 54 | 0 | 0 |
| Plasmid 3 (pXAS28) | 28,106 | 60.5 | 35 | 0 | 0 |
| Plasmid 4 (pXAS25) | 25,259 | 63.4 | 31 | 0 | 0 |
| DAR73886 | |||||
| Chromosome | 5,208,945 | 64.7 | 4407 | 58 | 6 |
| Plasmid 1 (pXAC64) | 61,267 | 61.4 | 67 | 0 | 0 |
| Plasmid 2 (pXAS47) | 47,520 | 60.6 | 52 | 0 | 0 |
| Plasmid 3 (pXAS28) | 28,110 | 60.5 | 35 | 0 | 0 |
| Plasmid 4 (pXAS25) | 25,259 | 63.4 | 31 | 0 | 0 |
| DAR73909 | |||||
| Chromosome | 5,178,469 | 64.9 | 4424 | 59 | 6 |
| Plasmid 1 (pXAS220) | 219,634 | 62.8 | 230 | 0 | 0 |
| Plasmid 2(pXAS56) | 56,537 | 61.5 | 67 | 0 | 0 |
| Plasmid 3 (pXAS42) | 42,682 | 61.1 | 47 | 0 | 0 |
| Plasmid 4 (pXAS27) | 27,107 | 62.9 | 32 | 0 | 0 |
| Plasmid 5 (pXAS24) | 24,453 | 62.6 | 21 | 0 | 0 |
| DAR72029 | |||||
| Chromosome | 5,178,793 | 64.9 | 4390 | 59 | 6 |
| Plasmid 1 (pXAS220) | 220,762 | 62.8 | 230 | 0 | 0 |
| Plasmid 2 (pXAS56) | 56,053 | 61.5 | 66 | 0 | 0 |
| Plasmid 3 (pXAS42) | 42,682 | 61.1 | 48 | 0 | 0 |
| Plasmid 4 (pXAS27) | 27,107 | 62.9 | 33 | 0 | 0 |
| Plasmid 5 (pXAS24) | 24,453 | 62.6 | 22 | 0 | 0 |
| DAR73889 | |||||
| Chromosome | 5,222,784 | 64.8 | 4446 | 59 | 6 |
| Plasmid 1 (pXAS220) | 219,409 | 62.8 | 229 | 0 | 0 |
| Plasmid 2 (pXAS56) | 56,563 | 61.5 | 65 | 0 | 0 |
| Plasmid 3 (pXAS42) | 42,717 | 61.1 | 46 | 0 | 0 |
| Plasmid 4 (pXAS30) | 30,179 | 62.6 | 37 | 0 | 0 |
| Plasmid 5 (pXAS24) | 24,453 | 62.6 | 22 | 0 | 0 |
| DAR73910 | |||||
| Chromosome | 5,326,504 | 64.8 | 4851 | 59 | 6 |
| Plasmid 1 (plasmid pF) | 116,539 | 63.3 | 142 | 0 | 0 |
| Plasmid 2 (pXAS38) | 38,592 | 62.2 | 38 | 0 | 0 |
| Plasmid 3 (pXAS28-2) | 28,174 | 63.0 | 37 | 0 | 0 |
| Isolate | PthA1 | Trunc-PthA1 | PthA2 | Trunc-PthA2 | PthA3 | Trunc-PthA3 | PthA4 | Trunc-PthA4 |
|---|---|---|---|---|---|---|---|---|
| DAR73886 | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| DAR84832 | 1 | 0 | 2 | 0 | 2 | 0 | 1 | 0 |
| DAR72029 | 0 | 1N | 1 | 1N,1C | 1 | 1N | 0 | 1N |
| DAR73889 | 1 | 1N | 1 | 1N | 1 | 1N | 1 | 1N |
| DAR73909 | 0 | 1N | 0 | 1N,1C | 0 | 1N | 0 | 1N |
| DAR73910 | 1 | 1N | 1 | 1N | 1 | 1N | 1 | 1N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Webster, J.; Bogema, D.; Chapman, T.A. Comparative Genomics of Xanthomonas citri pv. citri A* Pathotype Reveals Three Distinct Clades with Varying Plasmid Distribution. Microorganisms 2020, 8, 1947. https://doi.org/10.3390/microorganisms8121947
Webster J, Bogema D, Chapman TA. Comparative Genomics of Xanthomonas citri pv. citri A* Pathotype Reveals Three Distinct Clades with Varying Plasmid Distribution. Microorganisms. 2020; 8(12):1947. https://doi.org/10.3390/microorganisms8121947
Chicago/Turabian StyleWebster, John, Daniel Bogema, and Toni A. Chapman. 2020. "Comparative Genomics of Xanthomonas citri pv. citri A* Pathotype Reveals Three Distinct Clades with Varying Plasmid Distribution" Microorganisms 8, no. 12: 1947. https://doi.org/10.3390/microorganisms8121947
APA StyleWebster, J., Bogema, D., & Chapman, T. A. (2020). Comparative Genomics of Xanthomonas citri pv. citri A* Pathotype Reveals Three Distinct Clades with Varying Plasmid Distribution. Microorganisms, 8(12), 1947. https://doi.org/10.3390/microorganisms8121947





