Metabolic Engineering for Unusual Lipid Production in Yarrowia lipolytica
Abstract
1. Introduction
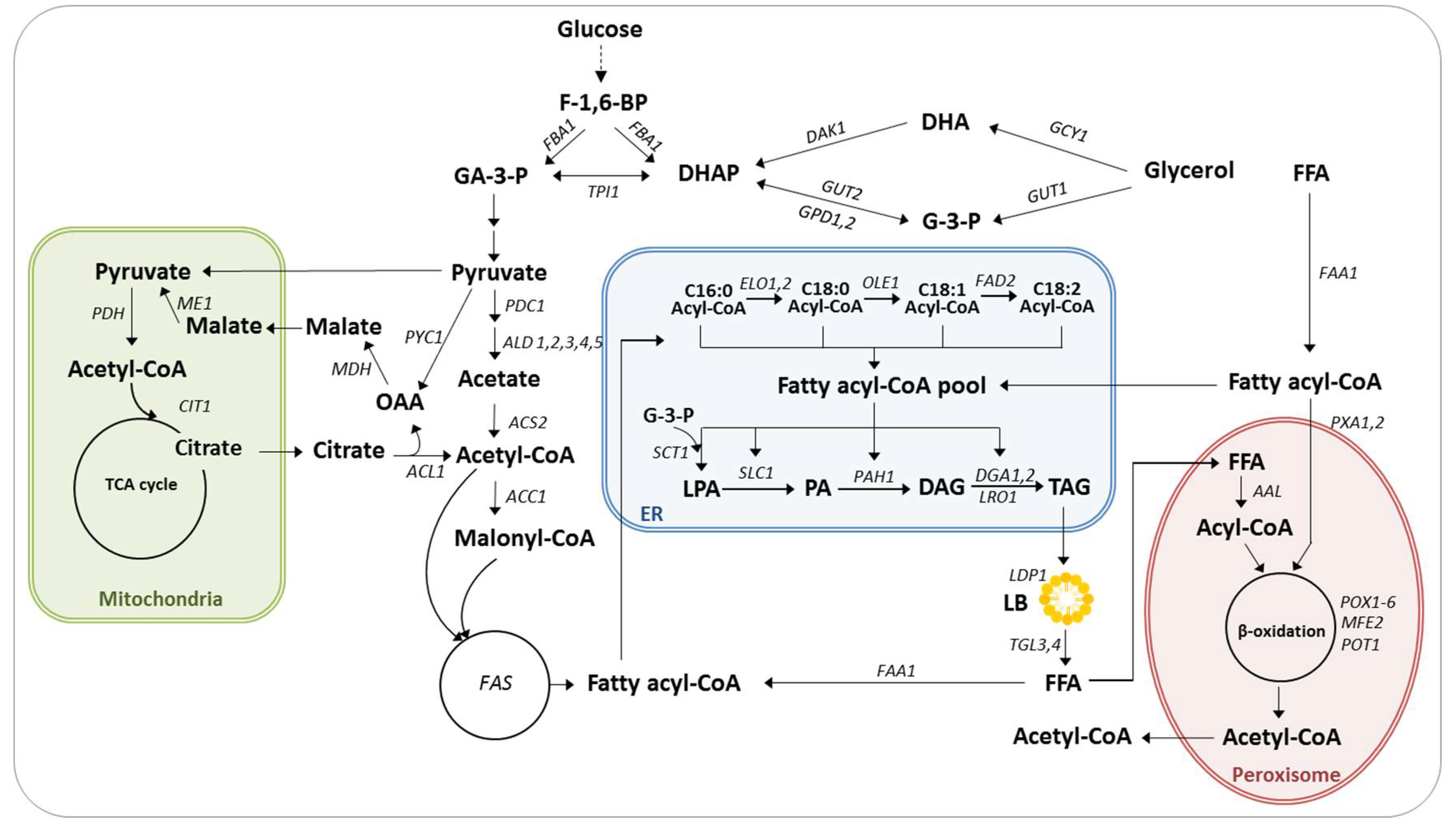
2. Lipid Production in Yarrowia lipolytica
2.1. Lipid Metabolism
2.2. Metabolic Engineering to Improve Lipid Production
2.2.1. Increasing the Size of Precursor Pools
2.2.2. Increasing Lipogenic Metabolic Flux
2.2.3. Inhibiting Lipid Remobilization and Degradation
2.2.4. Engineering Redox Metabolism
2.2.5. Removing Competing Byproducts
3. Unusual Lipids: Fatty Acids with Tailored Chain Lengths
3.1. Medium-Chain Fatty Acids
3.2. Odd-Chain Fatty Acids
3.3. Very-Long-Chain Fatty Acids
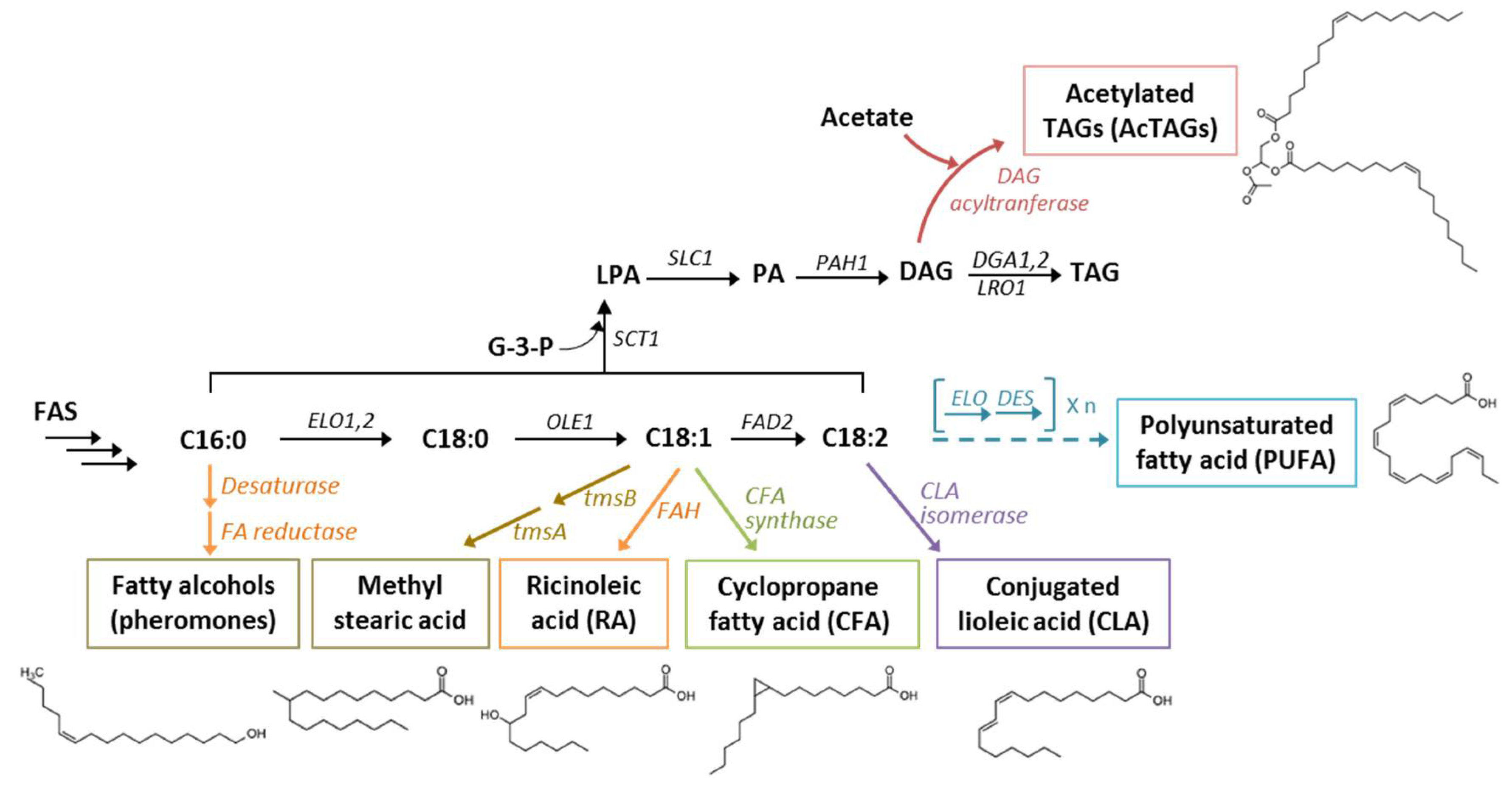
4. Unusual Lipids: Fatty Acid Derivatives
4.1. Conjugated Linoleic Acids
4.2. Cyclopropane Fatty Acids
4.3. Ricinoleic Acid
4.4. Polyunsaturated Fatty Acids
4.5. Emerging Target Compounds
4.5.1. Acetylated Triacylglycerols
4.5.2. Methylated Fatty Acids
4.5.3. Pheromones (Fatty Alcohols)
5. Conclusions and Perspectives
| Product | Strain Genotype | Strategy | Production Details | % of Total Lipids | Reference |
|---|---|---|---|---|---|
| MCFA | po1f pTEF-UcACPT | Introduction of heterologous acyl-ACP thioesterases | C8-FA | 14 | [54] |
| MCFA | po1f pTEF-CpaACPT | Introduction of heterologous acyl-ACP thioesterases | C10-FA | 57 | [54] |
| MCFA | po1g hFAS-EcTesA ′1 | Introduction of heterologous acyl-ACP thioesterases and hybrid fatty acid synthase-thioesterase | C14-FA: 29.2% of DCW | [25] | |
| MCFA | po1d pox1-6∆ fas1-I1220W | Engineering of native FAS system | C14-FA: 2.02% of DCW | 11.6 | [55] |
| MCFA | po1d pox1-6∆ elo1∆ fas1-I1220W pTEF-egDGAT | Inhibition of native elongation; introduction of heterologous diacylglycerol acyltransferase | C14-FA: 1.25 g/L | 45 | [56] |
| OCFA | po1d phd1Δ mfe1∆ tgl4Δ pTEF-DGA2 pTEF-GPD1 | Inhibition of precursor catabolism (propionyl-CoA); high levels of lipid accumulation | 0.57 g/L | 41.9 | [66] |
| OCFA | po1d phd1Δ mfe1∆ tgl4Δ pTEF-DGA2 pTEF-GPD1 hp4d-LDP1 pTEF-Repct pTEF-RebktB | Increase in precursor pool size; high levels of lipid accumulation 2 | 1.87 g/L | 62.1 | [67] |
| OCFA | po1d pox1-6∆ tgl4Δ pTEF-DGA2 pTEF-GPD1 pTEF-AAT2 pTEF-THR1 pTEF-THR4 pTEF-ILV1 pTEF-HOM3 pTEF-HOM2 pTEF-HOM6 | Overexpression of threonine synthesis pathway; high levels of lipid accumulation 2 | 0.36 g/L | 5.6 | [68] |
| VLCFA | po1f pex10Δ F1::UT-MaELO3 UT-AtKCS UT-CraKCS UT-EcAldh | Introduction of heterologous β-ketoacyl-CoA synthases and heterologous elongase; inhibition of peroxiome biogenesis; increase in precursor pool size | 280 mg/L | [74] | |
| VLCFA | po1d fad2Δ pTEF-FAE1 | Inhibition of Δ12 desaturation; introduction of heterologous elongase | C22:1-FA: 887 mg/L | 9 | [73] |
| CLA | polh (hp4d-oPAI) × 24 | Introduction of heterologous CLA-producing isomerase with large numbers of gene copies | 5.9 | [79] | |
| CLA | po1h (hp16d-oPAI) × 8 hp4d-MaFAD2 | Introduction of heterologous CLA-producing isomerase and desaturase | 30% of DCW | 44 | [80] |
| CLA | po1d pox1-6Δ dga1Δ dga2Δ lro1Δ fad2Δ pTEF-FAD2 pTEF-oPAI-LEU2ex pTEF-oPAI-URA3ex | Introduction of heterologous CLA-producing isomerase; overexpression of FAD2; high levels of lipid accumulation 2 | 302 mg/L | 6.5 | [81] |
| CLA | po1h hp4d-PAI hp4d-MA12D hp4d-DGA1 | Introduction of heterologous desaturase, dicylglycerol acyltransferase, and isomerase | 132.6 mg/L | 5.2 | [82] |
| CFA | po1f pex10Δ mfe1Δ UAS1B16-TEF-DGA1 (UAS1B16-TEF-ycoCFA) × 2 | Introduction of heterologous CFA synthase; high levels of lipid accumulation 2 | C19CP: 3.13 g/L | 32.7 | [86] |
| CFA | po1d pox1–6Δ tgl4Δ Hp8d-CFAs | Introduction of heterologous CFA synthase; high levels of lipid accumulation 2 | 2.3 g/L | 45 | [87] |
| CFA | po1d pox1-6Δ tgl4Δ pTEF-GPD1 pTEF-DGA2 hp8d-CFA pTEF-LRO1 | Introduction of heterologous CFA synthase; overexpression of native LRO1; optimization of carbon source | 7.5 g/L | 19.6 | [88] |
| RA | po1d pox1-6Δ dga1Δ lro1Δ dga2Δ fad2ΔpTEF-CpFAH12 (pTEF-CpFAH12) × 2 pTEF-LRO1 | Introduction of heterologous fatty acid hydroxylase; increase in precursor pool size | 12 g/L | 60 | [98] |
| PUFA | ATCC 20362 (Δ12 DES) × 3 (Δ6 DES) × 2 (C18/20 ELO) × 4 (Δ5 DES) × 5 (Δ17 DES) × 3 (C16/18 ELO) × 2 | Introduction of desaturases and elongases; push and pull of carbon into the engineered pathway | EPA | 40 | [101] |
| PUFA | Introduction of desaturases and elongases with multiple gene copies; inhibition of the competitive pathway | EPA:21.3% of DCW | 57 | [106] | |
| PUFA | Po1h hp4d-Pfa hp4d-PptAf4 | Introduction of multifunctional polyketide synthase (PKS)-like PUFA synthases from Aetherobacter fasciculatus and Minicystis rosea | DHA: 350 mg/L | 16.8 | [107] |
| acTAG | Po1d pTEF-EeDAcT | Introduction of heterologous diacylglycerol acetylransferase | 4.06% of DCW | 20 | [108] |
| meTAG | Introduction of heterologous methyltransferase and reductase | 10-methylstearic acid | 20 | [109] | |
| Pheromones | Y-17536 ku70Δ Cas9 hfd1Δ hfd4Δ pex10Δ fao1Δ PGPAT100bpPr (AtrΔ11)x3 HsuFAR (HarFAR) × 2 | Introduction of heterologous desaturase and reductase with multiple gene copies | Z11-16OH: 2.57 g/L | [110] | |
| Pheromones | po1d Δpox1-6 Δtgl4+pXPR2-SUC2 pTEF-DGA2 pTEF-GPD1 8UAS-pTEF-BlucFAR1 | Introduction of heterologous fatty acyl-CoA reductases | 166.6 mg/L | [111] |
Funding
Conflicts of Interest
Abbreviations
| ACPT | acyl-ACP thioesterase |
| AcTAGs | acetylated TAGs |
| CFA | cyclopropane fatty acid |
| CLA | conjugated linoleic acid |
| C/N ratio | carbon-to-nitrogen ratio |
| DacT | diacylglycerol acetyltransferase |
| DAG | diacylglycerol |
| DCW | dry cell weight |
| DHA | docosahexaenoic acid |
| DPA | docosapentaenoic acid |
| ECFA | even-chain fatty acid |
| EPA | eicosapentaenoic acid |
| ER | endoplasmic reticulum |
| FA | fatty acid |
| FAEE | fatty acid ethyl ester |
| FAME | fatty acid methyl ester |
| FAS | fatty acid synthase |
| FFA | free fatty acid |
| KAS | beta-ketoacyl-ACP synthase |
| KS | ketoacyl synthase |
| LA | linoleic acid |
| LB | lipid body |
| LCFA | long-chain fatty acid |
| LPA | lysophosphatidic acid |
| MAG | monoacylglycerol |
| MCFA | medium-chain fatty acid |
| MFE | multifunctional enzyme |
| MPT | malonyl/palmitoyl transacylase |
| OCFA | odd-chain fatty acids |
| PA | phosphatidic acid |
| PCT | propionyl-CoA transferase |
| PKT | polyketide synthase |
| PPP | pentose phosphate pathway |
| PUFA | polyunsaturated fatty acid |
| RA | ricinoleic acid |
| SCFA | short-chain fatty acid |
| TAG | triacylglycerol |
| TALEN | transcription activator-like effector nuclease |
| VLCFA | very-long-chain fatty acid |
| WT | wild type |
References
- Beopoulos, A.; Nicaud, J.M. Yeast: A new oil producer? OCL—Ol. Corps Gras Lipides 2012, 19, 22–28. [Google Scholar] [CrossRef]
- André, A.; Chatzifragkou, A.; Diamantopoulou, P.; Sarris, D.; Philippoussis, A.; Galiotou-Panayotou, M.; Komaitis, M.; Papanikolaou, S. Biotechnological conversions of bio-dieselderived crude glycerol by strains. Eng. Life Sci. 2009, 9, 468–478. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Nicaud, J.M. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef]
- Lazar, Z.; Liu, N.; Stephanopoulos, G. Holistic Approaches in Lipid Production by Yarrowia lipolytica. Trends Biotechnol. 2018, 36, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- van de Loo, F.J.; Fox, B.G.; Somerville, C. Unusual fatty acids. In Lipid Metabolism in Plants; Moore, T., Ed.; CRC Press: Boca Raton, FL, USA, 2018; pp. 91–126. [Google Scholar]
- Napier, J.A. The Production of Unusual Fatty Acids in Transgenic Plants. Annu. Rev. Plant Biol. 2007, 58, 295–319. [Google Scholar] [CrossRef]
- Xie, D.; Jackson, E.N.; Zhu, Q. Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: From fundamental research to commercial production. Appl. Microbiol. Biotechnol. 2015, 99, 1599–1610. [Google Scholar] [CrossRef]
- Aznar-Moreno, J.A.; Durrett, T.P. Simultaneous targeting of multiple gene homeologs to alter seed oil production in Camelina sativa. Plant Cell Physiol. 2017, 58, 1260–1267. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Markham, K.A.; Palmer, C.M.; Liu, N.; Stephanopoulos, G.; Alper, H.S. Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 2018, 50, 192–208. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Lazar, Z.; Rakicka, M.; Guo, Z.; Fouchard, F.; Le Coq, A.M.C.; Nicaud, J.M. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 2016, 38, 115–124. [Google Scholar] [CrossRef]
- Qiao, K.; Wasylenko, T.M.; Zhou, K.; Xu, P.; Stephanopoulos, G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 2017, 35, 173–177. [Google Scholar] [CrossRef]
- Friedlander, J.; Tsakraklides, V.; Kamineni, A.; Greenhagen, E.H.; Consiglio, A.L.; MacEwen, K.; Crabtree, D.V.; Afshar, J.; Nugent, R.L.; Hamilton, M.A.; et al. Engineering of a high lipid producing Yarrowia lipolytica strain. Biotechnol. Biofuels 2016, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Larroude, M.; Rossignol, T.; Nicaud, J.M.; Ledesma-Amaro, R. Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 2018, 36, 2150–2164. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, V.; Spagnuolo, M.; Agrawal, A.; Smith, S.; Gao, D.; Blenner, M. Advances and opportunities in gene editing and gene regulation technology for Yarrowia lipolytica. Microb. Cell Fact. 2019, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vorapreeda, T.; Thammarongtham, C.; Cheevadhanarak, S.; Laoteng, K. Alternative routes of acetyl-CoA synthesis identified by comparative genomic analysis: Involvement in the lipid production of oleaginous yeast and fungi. Microbiology 2012, 158, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Kamineni, A.; Shaw, J. Engineering triacylglycerol production from sugars in oleaginous yeasts. Curr. Opin. Biotechnol. 2020, 62, 239–247. [Google Scholar] [CrossRef]
- Jenni, S.; Leibundgut, M.; Boehringer, D.; Frick, C.; Mikolásek, B.; Ban, N. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 2007, 316, 254–261. [Google Scholar] [CrossRef]
- Lomakin, I.B.; Xiong, Y.; Steitz, T.A. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 2007, 129, 319–332. [Google Scholar] [CrossRef]
- Beopoulos, A.; Haddouche, R.; Kabran, P.; Dulermo, T.; Chardot, T.; Nicaud, J.M. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA: Diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 2012, 93, 1523–1537. [Google Scholar] [CrossRef]
- Athenstaedt, K.; Daum, G. Lipid storage: Yeast we can! Eur. J. Lipid Sci. Technol. 2011, 113, 1188–1197. [Google Scholar] [CrossRef]
- Dulermo, T.; Tréton, B.; Beopoulos, A.; Gnankon, A.P.K.; Haddouche, R.; Nicaud, J.M. Characterization of the two intracellular lipases of Yarrowia lipolytica encoded by TGL3 and TGL4 genes: New insights into the role of intracellular lipases and lipid body organisation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 1486–1495. [Google Scholar] [CrossRef]
- Dulermo, R.; Gamboa-Meléndez, H.; Dulermo, T.; Thevenieau, F.; Nicaud, J.M. The fatty acid transport protein Fat1p is involved in the export of fatty acids from lipid bodies in Yarrowia lipolytica. FEMS Yeast Res. 2014, 14, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, R.; Gamboa-Meléndez, H.; Ledesma-Amaro, R.; Thévenieau, F.; Nicaud, J.M. Unraveling fatty acid transport and activation mechanisms in Yarrowia lipolytica. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 1202–1217. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Le Dall, M.-T.; Waché, Y.; Laroche, C.; Belin, J.M.; Nicaud, J.M. Cloning, sequencing, and characterization of five genes coding for acyl-CoA oxidase isozymes in the yeast Yarrowia lipolytica. Cell Biochem. Biophys. 1999, 31, 165–174. [Google Scholar] [CrossRef]
- Xu, P.; Qiao, K.; Ahn, W.S.; Stephanopoulos, G. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. USA 2016, 113, 10848–10853. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, T.; Lazar, Z.; Dulermo, R.; Rakicka, M.; Haddouche, R.; Nicaud, J.M. Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim. Biophys. Acta 2015, 1851, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Imam Abidi, S.H.; Liu, H.; Zhang, H.; Chakraborty, S.; Watson, N.; Kumaran Ajikumar, P.; Stephanopoulos, G. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gajdoš, P.; Ledesma-Amaro, R.; Nicaud, J.M.; Čertík, M.; Rossignol, T. Overexpression of diacylglycerol acyltransferase in Yarrowia lipolytica affects lipid body size, number and distribution. FEMS Yeast Res. 2016, 16, fow062. [Google Scholar] [CrossRef]
- Dulermo, T.; Nicaud, J.M. Involvement of the G3P shuttle and Β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011, 13, 482–491. [Google Scholar] [CrossRef]
- Beopoulos, A.; Mrozova, Z.; Thevenieau, F.; Le Dall, M.T.; Hapala, I.; Papanikolaou, S.; Chardot, T.; Nicaud, J.M. Control of Lipid Accumulation in the Yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2008, 74, 7779–7789. [Google Scholar] [CrossRef] [PubMed]
- Lazar, Z.; Dulermo, T.; Neuvéglise, C.; Crutz-Le Coq, A.M.; Nicaud, J.M. Hexokinase-A limiting factor in lipid production from fructose in Yarrowia lipolytica. Metab. Eng. 2014, 26, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Sagnak, R.; Cochot, S.; Molina-Jouve, C.; Nicaud, J.M.; Guillouet, S.E. Modulation of the Glycerol Phosphate availability led to concomitant reduction in the citric acid excretion and increase in lipid content and yield in Yarrowia lipolytica. J. Biotechnol. 2018, 265, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Sharpe, P.L.; Hong, S.P.; Yadav, N.S.; Xie, D.; Short, D.R.; Damude, H.G.; Rupert, R.A.; Seip, J.E.; Wang, J. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 2013, 31, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, R.; Dulermo, T.; Gamboa-Meléndez, H.; Thevenieau, F.; Nicaud, J.M. Role of Pex11p in Lipid Homeostasis in Yarrowia lipolytica. Eukaryot. Cell 2015, 14, 511–525. [Google Scholar] [CrossRef]
- Liu, L.; Pan, A.; Spofford, C.; Zhou, N.; Alper, H.S. An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab. Eng. 2015, 29, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Wynn, J.P.; Hamid, A.A.; Li, Y.; Ratledge, C. Biochemical events leading to the diversion of carbon into storage lipids in the oleaginous fungi Mucor circinelloides and Mortierella alpina. Microbiology 2001, 147, 2857–2864. [Google Scholar] [CrossRef]
- Zhang, S.; Ito, M.; Skerker, J.M.; Arkin, A.P.; Rao, C.V. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl. Microbiol. Biotechnol. 2016, 100, 9393–9405. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Chen, H.; Chen, Y.Q.; Ratledge, C.; Song, Y.; Chen, W. Regulatory properties of malic enzyme in the oleaginous yeast, Yarrowia lipolytica, and its non-involvement in lipid accumulation. Biotechnol. Lett. 2013, 35, 2091–2098. [Google Scholar] [CrossRef]
- Wasylenko, T.M.; Ahn, W.S.; Stephanopoulos, G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 2015, 30, 27–39. [Google Scholar] [CrossRef]
- Silverman, A.M.; Qiao, K.; Xu, P.; Stephanopoulos, G. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2016, 100, 3781–3798. [Google Scholar] [CrossRef] [PubMed]
- Yuzbasheva, E.Y.; Agrimi, G.; Yuzbashev, T.V.; Scarcia, P.; Vinogradova, E.B.; Palmieri, L.; Shutov, A.V.; Kosikhina, I.M.; Palmieri, F.; Sineoky, S.P. The mitochondrial citrate carrier in Yarrowia lipolytica: Its identification, characterization and functional significance for the production of citric acid. Metab. Eng. 2019, 54, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolski, A.; Mirończuk, A.M. The influence of transketolase on lipid biosynthesis in the yeast Yarrowia lipolytica. Microb. Cell Fact. 2020, 19, 138. [Google Scholar] [CrossRef] [PubMed]
- Bhutada, G.; Kavšcek, M.; Ledesma-Amaro, R.; Thomas, S.; Rechberger, G.N.; Nicaud, J.M.; Natter, K. Sugar versus fat: Elimination of glycogen storage improves lipid accumulation in Yarrowia lipolytica. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Biermann, U.; Bornscheuer, U.; Meier, M.A.R.; Metzger, J.O.; Schäfer, H.J. Oils and fats as renewable raw materials in chemistry. Angew. Chemie Int. Ed. 2011, 50, 3854–3871. [Google Scholar] [CrossRef]
- Sarria, S.; Kruyer, N.S.; Peralta-Yahya, P. Microbial synthesis of medium-chain chemicals from renewables. Nat. Biotechnol. 2017, 35, 1158–1166. [Google Scholar] [CrossRef]
- Lennen, R.M.; Braden, D.J.; West, R.A.; Dumesic, J.A.; Pfleger, B.F. A process for microbial hydrocarbon synthesis: Overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol. Bioeng. 2010, 106, 193–202. [Google Scholar] [CrossRef]
- Hernández Lozada, N.J.; Lai, R.Y.; Simmons, T.R.; Thomas, K.A.; Chowdhury, R.; Maranas, C.D.; Pfleger, B.F. Highly Active C 8 -Acyl-ACP thioesterase variant isolated by a synthetic selection strategy. ACS Synth. Biol. 2018, 7, 2205–2215. [Google Scholar] [CrossRef]
- Jing, F.; Cantu, D.C.; Tvaruzkova, J.; Chipman, J.P.; Nikolau, B.J.; Yandeau-Nelson, M.D.; Reilly, P.J. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011, 12, 1–16. [Google Scholar] [CrossRef]
- Val, D.; Banu, G.; Seshadri, K.; Lindqvist, Y.; Dehesh, K. Re-engineering ketoacyl synthase specificity. Structure 2000, 8, 565–566. [Google Scholar] [CrossRef]
- Gajewski, J.; Pavlovic, R.; Fischer, M.; Boles, E.; Grininger, M. Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Hu, Y.; Teixeira, P.G.; Pereira, R.; Chen, Y.; Siewers, V.; Nielsen, J. Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids. Nat. Catal. 2020, 3, 64–74. [Google Scholar] [CrossRef]
- Rutter, C.D.; Zhang, S.; Rao, C.V. Engineering Yarrowia lipolytica for production of medium-chain fatty acids. Appl. Microbiol. Biotechnol. 2015, 99, 7359–7368. [Google Scholar] [CrossRef] [PubMed]
- Rigouin, C.; Gueroult, M.; Croux, C.; Dubois, G.; Borsenberger, V.; Barbe, S.; Marty, A.; Daboussi, F.; André, I.; Bordes, F. Production of medium chain fatty acids by Yarrowia lipolytica: Combining molecular design and TALEN to engineer the fatty acid synthase. ACS Synth. Biol. 2017, 6, 1870–1879. [Google Scholar] [CrossRef] [PubMed]
- Rigouin, C.; Croux, C.; Borsenberger, V.; Ben Khaled, M.; Chardot, T.; Marty, A.; Bordes, F. Increasing medium chain fatty acids production in Yarrowia lipolytica by metabolic engineering. Microb. Cell Fact. 2018, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Degwert, J. Use of cis-9-heptadecenoic Acid for Treating Psoriasis and Allergies. U.S. Patent 5708028A, 1994. [Google Scholar] [CrossRef]
- Avis, T.J. Synthesis and biological characterization of (Z)-9-heptadecenoic and (Z)-6-methyl-9-heptadecenoic acids: Fatty acids with antibiotic activity produced by Pseudozyma flocculosa. J. Chem. Ecol. 2000, 26, 987–1000. [Google Scholar] [CrossRef]
- Clausen, C.A.; Coleman, R.D.; Yang, V.W. Fatty acid–based formulations for wood protection against mold and sapstain. For. Prod. J. 2010, 60, 301–304. [Google Scholar] [CrossRef]
- Köckritz, A.; Blumenstein, M.; Martin, A. Catalytic cleavage of methyl oleate or oleic acid. Eur. J. Lipid Sci. Technol. 2010, 112, 58–63. [Google Scholar] [CrossRef]
- Fitton, A.; Goa, K.L. Azelaic Acid: A Review of its pharmacological properties and therapeutic efficacy in acne and hyperpigmentary skin disorders. Drugs 1991, 41, 780–798. [Google Scholar] [CrossRef]
- Ingram, L.O.; Chevalier, L.S.; Gabbay, E.J. Priopionate-induced synthesis of odd-chain-length fatty acids by Escherichia coli. J. Bacteriol. 1977, 131, 1023–1025. [Google Scholar] [CrossRef]
- Wu, H.; San, K.Y. Efficient odd straight medium chain free fatty acid production by metabolically engineered Escherichia coli. Biotechnol. Bioeng. 2014, 111, 2209–2219. [Google Scholar] [CrossRef] [PubMed]
- Fontanille, P.; Kumar, V.; Christophe, G.; Nouaille, R.; Larroche, C. Bioconversion of volatile fatty acids into lipids by the oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 2012, 114, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Kolouchová, I.; Schreiberová, O.; Sigler, K.; Masák, J.; Řezanka, T. Biotransformation of volatile fatty acids by oleaginous and non-oleaginous yeast species. FEMS Yeast Res. 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Dulermo, T.; Ledesma-Amaro, R.; Nicaud, J.M. Optimization of odd chain fatty acid production by Yarrowia lipolytica. Biotechnol. Biofuels 2018, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Bordes, F.; Letisse, F.; Nicaud, J.M. Engineering precursor pools for increasing production of odd-chain fatty acids in Yarrowia lipolytica. bioRxiv 2020. [Google Scholar] [CrossRef]
- Park, Y.K.; Ledesma-Amaro, R.; Nicaud, J.M. De novo biosynthesis of odd-chain fatty acids in Yarrowia lipolytica enabled by modular pathway engineering. Front. Bioeng. Biotechnol. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Wisniak, J. Potential uses of jojoba oil and meal—A review. Ind. Crops Prod. 1994, 3, 43–68. [Google Scholar] [CrossRef]
- Jannin, V.; Cuppok, Y. Hot-melt coating with lipid excipients. Int. J. Pharm. 2013, 457, 480–487. [Google Scholar] [CrossRef]
- Wenning, L.; Yu, T.; David, F.; Nielsen, J.; Siewers, V. Establishing very long-chain fatty alcohol and wax ester biosynthesis in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2017, 114, 1025–1035. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, Y.J.; Wenning, L.; Liu, Q.; Krivoruchko, A.; Siewers, V.; Nielsen, J.; David, F. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Gajdoš, P.; Hambalko, J.; Slaný, O.; Čertík, M. Conversion of waste materials into very long chain fatty acids by the recombinant yeast Yarrowia lipolytica. FEMS Microbiol. Lett. 2019, 367, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, J.L.; Zhao, X.R.; Liu, S.C.; Liu, Z.J.; Wei, L.J.; Hua, Q. Yarrowia lipolytica as a metabolic engineering platform for the production of very-long-chain wax esters. J. Agric. Food Chem. 2020, 68, 10730–10740. [Google Scholar] [CrossRef] [PubMed]
- Crumb, D.; Vattem, D. Conjugated linoleic acid (CLA)-An overview. Int. J. Appl. Res. Nat. Prod. 2011, 4, 12–15. [Google Scholar]
- Kishino, S.; Ogawa, J.; Omura, Y.; Matsumura, K.; Shimizu, S. Conjugated linoleic acid production from linoleic acid by lactic acid bacteria. J. Am. Oil Chem. Soc. 2002, 79, 159–163. [Google Scholar] [CrossRef]
- Ando, A.; Ogawa, J.; Kishino, S.; Shimizu, S. Conjugated linoleic acid production from castor oil by Lactobacillus plantarum JCM 1551. Enzyme Microb. Technol. 2004, 35, 40–45. [Google Scholar] [CrossRef]
- Hornung, E.; Krueger, C.; Pernstich, C.; Gipmans, M.; Porzel, A.; Feussner, I. Production of (10E,12Z)-conjugated linoleic acid in yeast and tobacco seeds. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2005, 1738, 105–114. [Google Scholar] [CrossRef]
- Zhang, B.; Rong, C.; Chen, H.; Song, Y.; Zhang, H.; Chen, W. De novo synthesis of trans-10, cis-12 conjugated linoleic acid in oleaginous yeast Yarrowia lipolytica. Microb. Cell Fact. 2012, 11, 51. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, H.; Li, M.; Gu, Z.; Song, Y.; Ratledge, C.; Chen, Y.Q.; Zhang, H.; Chen, W. Genetic engineering of Yarrowia lipolytica for enhanced production of trans-10, cis-12 conjugated linoleic acid. Microb. Cell Fact. 2013, 12, 70. [Google Scholar] [CrossRef]
- Imatoukene, N.; Verbeke, J.; Beopoulos, A.; Idrissi Taghki, A.; Thomasset, B.; Sarde, C.O.; Nonus, M.; Nicaud, J.M. A metabolic engineering strategy for producing conjugated linoleic acids using the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2017, 101, 4605–4616. [Google Scholar] [CrossRef]
- Wang, X.; Xia, Q.; Wang, F.; Zhang, Y.; Li, X. Modulating heterologous pathways and optimizing culture conditions for biosynthesis of trans-10, cis-12 conjugated linoleic acid in Yarrowia lipolytica. Molecules 2019, 24, 1753. [Google Scholar] [CrossRef]
- Svensson, L.; Hansson, U.; Gronowitz, S.; Klingstedt, T. The relationship between the structure of monoalkyl branched saturated triacylglycerols and some physical properties. Lipids 1997, 32, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Cronan, J.E.; Reed, R.; Taylor, F.R.; Jackson, M.B. Properties and biosynthesis of cyclopropane fatty acids in Escherichia coli. J. Bacteriol. 1979, 138, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Schmid, K.M. Cyclopropane Fatty Acid Expression in Plants. U.S. Patent 5936139A, 1999. [Google Scholar]
- Markham, K.A.; Alper, H.S. Engineering Yarrowia lipolytica for the production of cyclopropanated fatty acids. J. Ind. Microbiol. Biotechnol. 2018, 45, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Czerwiec, Q.; Idrissitaghki, A.; Imatoukene, N.; Nonus, M.; Thomasset, B.; Nicaud, J.M.; Rossignol, T. Optimization of cyclopropane fatty acids production in Yarrowia lipolytica. Yeast 2019, 36, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Imatoukene, N.; Back, A.; Nonus, M.; Thomasset, B.; Rossignol, T.; Nicaud, J.M. Fermentation process for producing CFAs using Yarrowia lipolytica. J. Ind. Microbiol. Biotechnol. 2020, 47, 403–412. [Google Scholar] [CrossRef]
- Mutlu, H.; Meier, M.A.R. Castor oil as a renewable resource for the chemical industry. Eur. J. Lipid Sci. Technol. 2010, 112, 10–30. [Google Scholar] [CrossRef]
- Da Silva, N.L.; Maciel, M.R.W.; Batistella, C.B.; Filho, R.M. Optimization of biodiesel production from castor oil. J. Environ. Chem. Eng. 2018, 6, 2684–2695. [Google Scholar] [CrossRef]
- Ogunniyi, D.S. Castor oil: A vital industrial raw material. Bioresour. Technol. 2006, 97, 1086–1091. [Google Scholar] [CrossRef]
- Broun, P.; Somerville, C. Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol. 1997, 113, 933–942. [Google Scholar] [CrossRef][Green Version]
- Lu, C.; Fulda, M.; Wallis, J.G.; Browse, J. A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 2006, 45, 847–856. [Google Scholar] [CrossRef]
- Burgal, J.; Shockey, J.; Lu, C.; Dyer, J.; Larson, T.; Graham, I.; Browse, J. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol. J. 2008, 6, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Holic, R.; Yazawa, H.; Kumagai, H.; Uemura, H. Engineered high content of ricinoleic acid in fission yeast Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 2012, 95, 179–187. [Google Scholar] [CrossRef]
- Yazawa, H.; Ogiso, M.; Kumagai, H.; Uemura, H. Suppression of ricinoleic acid toxicity by ptl2 overexpression in fission yeast Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 2014, 98, 9325–9337. [Google Scholar] [CrossRef] [PubMed]
- Meesapyodsuk, D.; Chen, Y.; Ng, S.H.; Chen, J.; Qiu, X. Metabolic engineering of Pichia pastoris to produce ricinoleic acid, a hydroxy fatty acid of industrial importance. J. Lipid Res. 2015, 56, 2102–2109. [Google Scholar] [CrossRef] [PubMed]
- Beopoulos, A.; Verbeke, J.; Bordes, F.; Guicherd, M.; Bressy, M.; Marty, A.; Nicaud, J.M. Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2014, 98, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.; Gueroult, M.; Cheikhrouhou, R.; Guichard, M.; Borsenberger, V.; Marty, A.; Bordes, F. Identification of a crucial amino acid implicated in the hydroxylation/desaturation ratio of CpFAH12 bifunctional hydroxylase. Biotechnol. Bioeng. 2019, 116, 2451–2462. [Google Scholar] [CrossRef]
- Guo, Z.P.; Robin, J.; Duquesne, S.; O’Donohue, M.J.; Marty, A.; Bordes, F. Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose. Biotechnol. Biofuels 2018, 11, 141. [Google Scholar] [CrossRef]
- Zhu, Q.; Xue, Z.; Yadav, N.; Damude, H.; Pollak, D.W.; Rupert, R.; Seip, J.; Hollerbach, D.; Macool, D.; Zhang, H. Metabolic Engineering of an Oleaginous Yeast for the Production of Omega-3 Fatty Acids, 2nd ed.; Academic Press and AOCS Press: Urbana, IL, USA, 2010; pp. 51–73. [Google Scholar] [CrossRef]
- Domingo, J.L. Omega-3 fatty acids and the benefits of fish consumption: Is all that glitters gold? Environ. Int. 2007, 33, 993–998. [Google Scholar] [CrossRef]
- Martins, D.A.; Custódio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K.M. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef]
- Maccol, D.J.; Xue, Z.; Zhu, Q.Q. Mortierella Alpina C16/18 Fatty Acid Elongase. U.S. Patent 7470532B2, 2008. [Google Scholar]
- Narendra, S.; Yadav, Q.Q.; Zhu, H.Z. Δ12 Desaturases Suitable for Altering Levels of Polyunsaturated Fatty Acids in Oleaginous Yeast. U.S. Patent 7504259B, 2009. [Google Scholar]
- Hong, S.P.; Sharpe, P.L.; Xue, Z.; Yadav, N.S. Improved Optimized Strains of Yarrowia lipolytica for High Eicosapentaenoic Acid Production. U.S. Patent 7645604, 2010. [Google Scholar]
- Gemperlein, K.; Dietrich, D.; Kohlstedt, M.; Zipf, G.; Bernauer, H.S.; Wittmann, C.; Wenzel, S.C.; Müller, R. Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Gajdoš, P.; Hambalko, J.; Nicaud, J.M.; Čertík, M. Overexpression of diacylglycerol acetyltransferase from Euonymus europaeus in Yarrowia lipolytica leads to the production of single-cell oil enriched with 3-acetyl-1,2-diacylglycerols. Yeast 2020, 37, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.J.; Blitzblau, H.; Crabtree, D.V. Heterologous Production of 10-methylstearic Acid. U.S. Patent 20200123579A1, 2020. [Google Scholar]
- Holkenbrink, C.; Ding, B.J.; Wang, H.L.; Dam, M.I.; Petkevicius, K.; Kildegaard, K.R.; Wenning, L.; Sinkwitz, C.; Lorántfy, B.; Koutsoumpeli, E. Production of moth sex pheromones for pest control by yeast fermentation. Metab. Eng. 2020, 62, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Hambalko, J.; Gajdoš, P.; Nicaud, J.M.; Ledesma-Amaro, R.; Tupec, M.; Pichová, I.; Čertík, M. Expression of bumble bee reductases BlucFAR1 and BlapFAR4 in Yarrowia lipolytica. Front. Bioeng. Biotechnol. under revision.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.-K.; Nicaud, J.-M. Metabolic Engineering for Unusual Lipid Production in Yarrowia lipolytica. Microorganisms 2020, 8, 1937. https://doi.org/10.3390/microorganisms8121937
Park Y-K, Nicaud J-M. Metabolic Engineering for Unusual Lipid Production in Yarrowia lipolytica. Microorganisms. 2020; 8(12):1937. https://doi.org/10.3390/microorganisms8121937
Chicago/Turabian StylePark, Young-Kyoung, and Jean-Marc Nicaud. 2020. "Metabolic Engineering for Unusual Lipid Production in Yarrowia lipolytica" Microorganisms 8, no. 12: 1937. https://doi.org/10.3390/microorganisms8121937
APA StylePark, Y.-K., & Nicaud, J.-M. (2020). Metabolic Engineering for Unusual Lipid Production in Yarrowia lipolytica. Microorganisms, 8(12), 1937. https://doi.org/10.3390/microorganisms8121937





