Hydrocarbon Degradation and Enzyme Activities of Aspergillus oryzae and Mucor irregularis Isolated from Nigerian Crude Oil-Polluted Sites
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Sampling
2.2. Fungal Strains
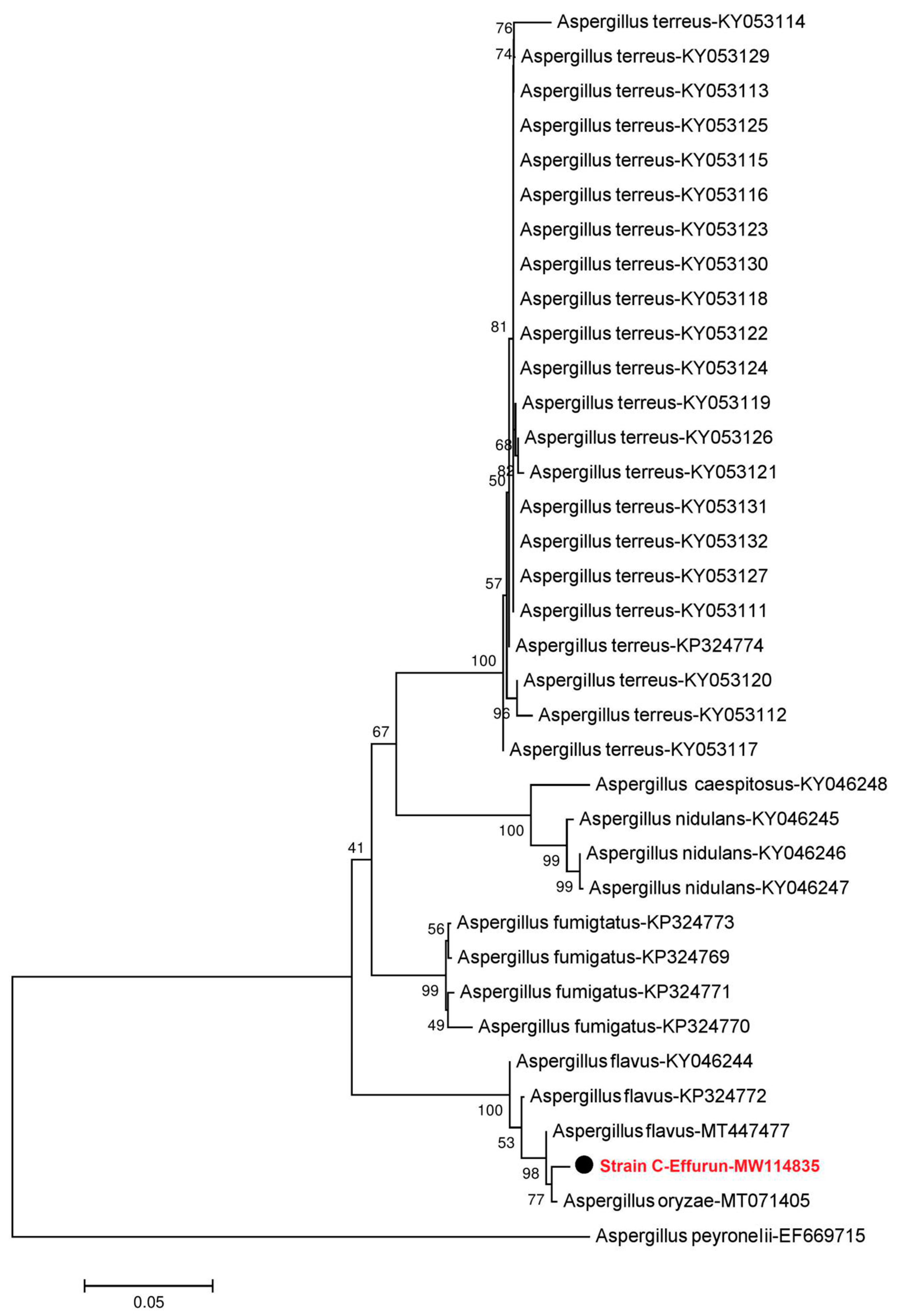
2.3. Morphology and Molecular Identification of Dominant Strains
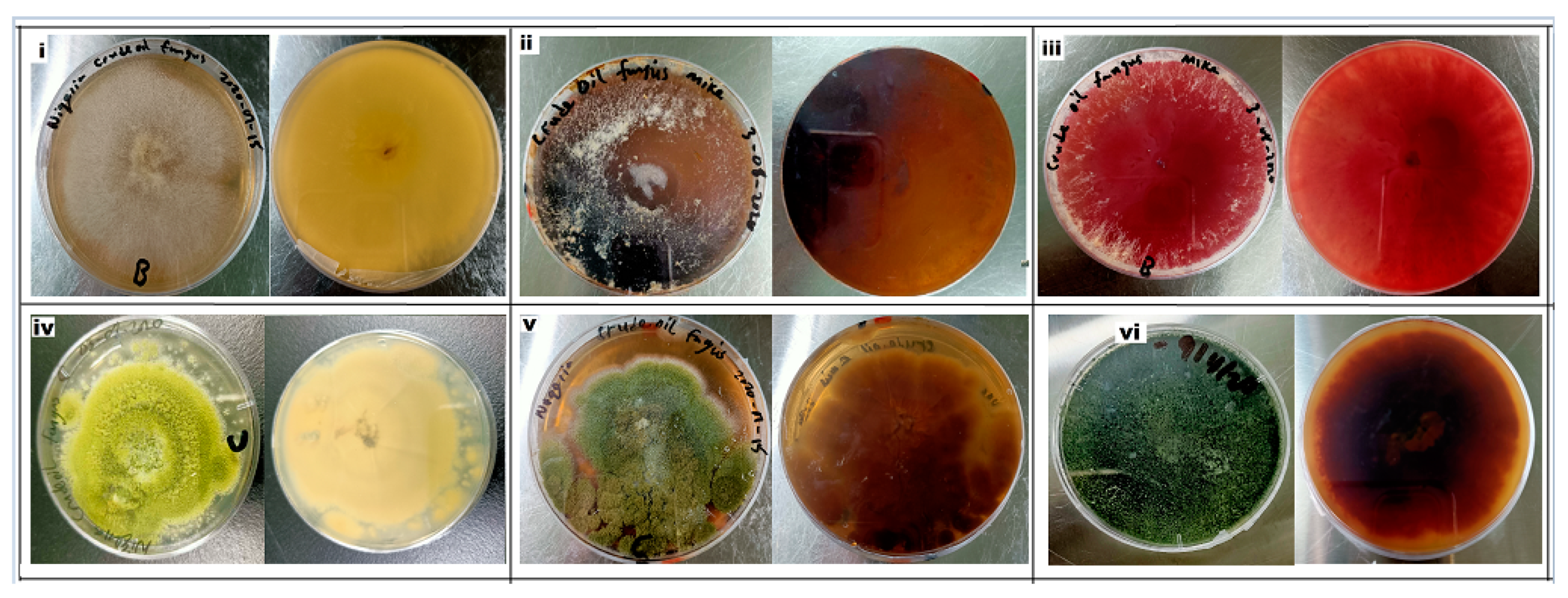
2.4. Hydrocarbon Tolerance Test
2.5. Enzyme Activities of the Selected Dominant Fungi
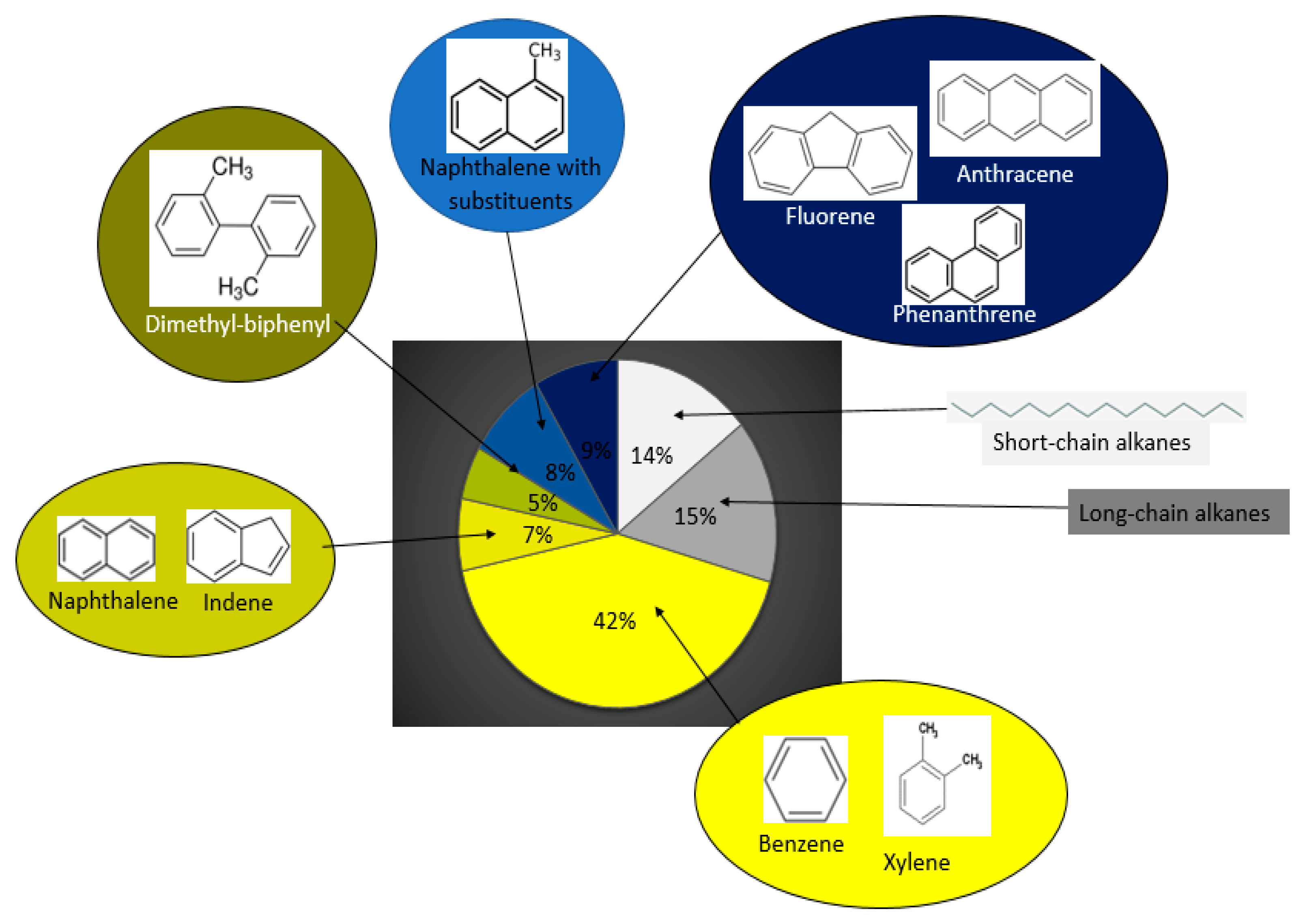
2.6. Hydrocarbon Degradation Analysis
3. Results
3.1. Identification of Fungi Isolated from Crude Oil Polluted Sites
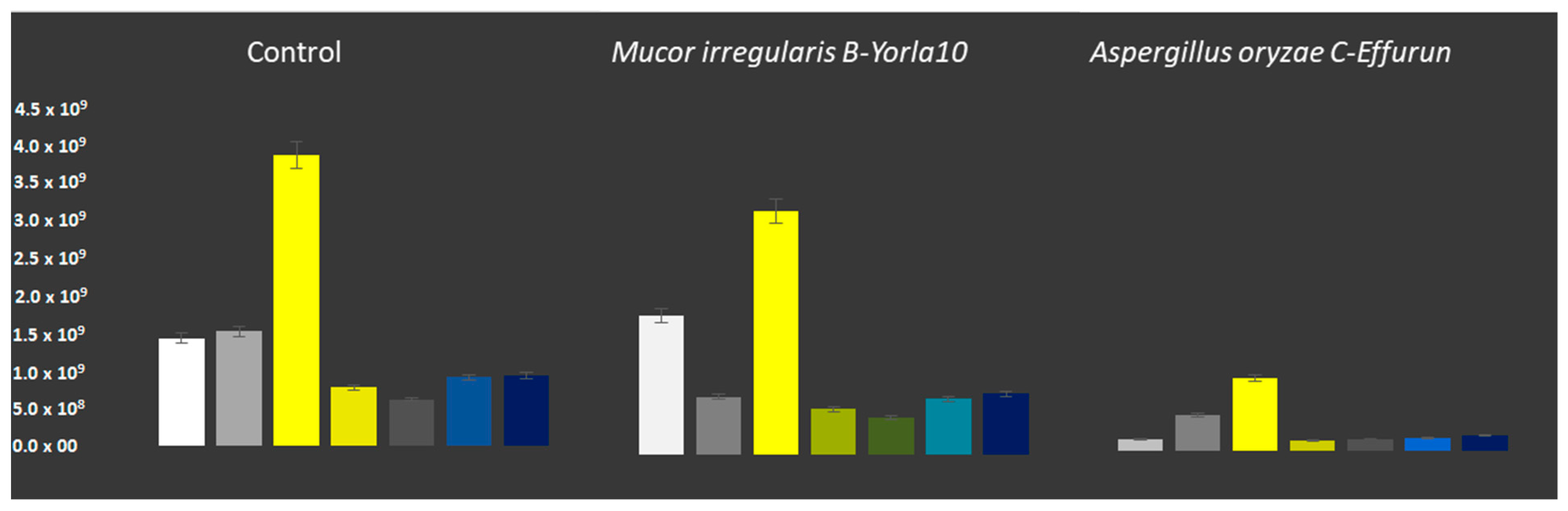
3.2. Assay and Activities of Enzymes Secreted by Selected Fungal Strains
3.3. Hydrocarbon Degradation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Voutsa, D.; Hartmann, P.; Schaffner, C.; Giger, W. Benzotriazoles, alkylphenols and bisphenol A in municipal wastewaters and in the Glatt River, Switzerland. Environ. Sci. Pollut. Res. Int. 2006, 13, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Desbiolles, F.; Malleret, L.; Tiliacos, C.; Wong-Wah-Chung, P.; Laffont-Schwob, I. Occurrence and ecotoxicological assessment of pharmaceuticals: Is there a risk for the Mediterranean aquatic environment? Sci. Total Environ. 2018, 639, 1334–1348. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.H.; Hoang, L.; Nghiem, L.D.; Nguyen, N.M.H.; Ngo, H.H.; Guo, W.; Trinh, Q.T.; Mai, N.H.; Chen, H.; Nguyen, D.D.; et al. Occurrence and risk assessment of multiple classes of antibiotics in urban canals and lakes in Hanoi. Vietnam. Sci. Total Environ. 2019, 692, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Dutta, A.; Pal, S.; Gupta, A.; Sarkar, J.; Chatterjee, A.; Saha, A.; Sarkar, P.; Sar, P.; Kazy, S.K. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour. Technol. 2018, 253, 22–32. [Google Scholar] [CrossRef]
- Priya, A.; Mandal, A.K.; Ball, A.S.; Manefield, M.; Lal, B.; Sarma, P.M. Mass culture strategy for bacterial yeast co-culture for degradation of petroleum hydrocarbons in marine environment. Mar. Pollut. Bull. 2015, 100, 191–199. [Google Scholar] [CrossRef]
- Turner, N.R.; Renegar, D.A. Petroleum hydrocarbon toxicity to corals: A review. Mar. Pollut. Bull. 2017, 119, 1–16. [Google Scholar] [CrossRef]
- Sarma, P.M.; Bhattacharya, D.; Krishnan, S.; Lal, B. Degradation of polycyclic aromatic hydrocarbons by a newly discovered enteric bacterium, Leclercia adecarboxylata. Appl. Environ. Microbiol. 2004, 70, 3163–3166. [Google Scholar] [CrossRef]
- Steliga, T.; Jakubowicz, P.; Kapusta, P. Changes in toxicity during in situ bioremediation of weathered drill wastes contaminated with petroleum hydrocarbons. Bioresour. Technol. 2012, 125, 1–10. [Google Scholar] [CrossRef]
- Adeniyi, A.; Afolabi, J. Determination of total petroleum hydrocarbons and heavy metals in soils within the vicinity of facilities handling refined petroleum products in Lagos metropolis. Environ. Int. 2002, 28, 79–82. [Google Scholar] [CrossRef]
- Antizar-Ladislao, B.; Lopez-Real, J.M.; Beck, A.J. Bioremediation of polycyclic aromatic hydrocarbon (PAH)-contaminated waste using composting approaches. Crit. Rev. Environ. Sci. Technol. 2004, 34, 249–289. [Google Scholar] [CrossRef]
- Tang, J.; Wang, M.; Wang, F.; Sun, Q.; Zhou, Q. Eco-toxicity of petroleum hydrocarbon contaminated soil. J. Environ. Sci. 2011, 23, 845–851. [Google Scholar] [CrossRef]
- Chen, M.; Xua, P.; Zeng, G.; Yang, C.; Huang, D.; Zhang, J. Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 2015, 33, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Guarino, C.; Spada, V.; Sciarrillo, R. Assessment of three approaches of bioremediation (natural attenuation, landfarming and bioagumentation—Assistited land farming) for a petroleum hydrocarbons contaminated soil. Chemosphere 2017, 170, 10–16. [Google Scholar] [CrossRef] [PubMed]
- United States Wnvironmental Protection Agency (SEPA). National Contingency Plan Product Schedule (March 2015). Available online: http://epa.gov/ncer/rfa/2015/2015_star_gradfellow.html (accessed on 17 March 2017).
- Agency for Toxic Substances and Disease Registry (ATSDR). Public Health Statement Total Petroleum Hydrocarbons. Available online: http://www.atsdr.cdc.gov/ToxProfiles/tp123-c1-b.pdf (accessed on 14 September 2018).
- Tang, G.; Sun, J.; Wu, F.; Sun, Y.; Zhu, X.; Geng, Y.; Wang, Y. Organic composition of gasoline and its potential effects on air pollution in North China. Sci. China Chem. 2015, 58, 1416–1425. [Google Scholar] [CrossRef]
- Jesubunmi, C.O. Isolation of oil—Degrading microorganisms in spent engine oil—Contaminated soil. J. Biol. Agric. Health 2014, 4, 191–195. [Google Scholar]
- Wang, Z.; Fingas, M.; Blenkinsopp, S.; Sergy, G.; Landriault, M. Comparison of oil composition changes due to biodegradation and physical weathering in different oils. J. Chromatogr. A 1998, 809, 89–107. [Google Scholar] [CrossRef]
- Kadali, K.K.; Simons, K.L.; Skuza, P.P.; Moore, R.B.; Ball, A.S. A complementary approach to identifying and assessing the remediation potential of hydrocarbonoclastic bacteria. J. Microbiol. Methods 2012, 88, 348–355. [Google Scholar] [CrossRef]
- Cappello, S.; Calogero, R.; Santisi, S.; Genovese, M.; Denaro, R.; Genovese, L.; Giuliano, L.; Mancini, G.; Yakimov, M.M. Bioremediation of oil polluted marine sediments: A bio-engineering treatment. Int. Microbiol. 2015, 18, 127–134. [Google Scholar]
- Asemoloye, M.D.; Jonathan, S.G.; Jayeola, A.A.; Ahmad, R. Mediational influence of spent mushroom compost on phytoremediation of black-oil hydrocarbon polluted soil and response of Megathyrsus maximus Jacq. J. Environ. Manag. 2017, 200, 253–262. [Google Scholar] [CrossRef]
- Hamdi, H.; Benzarti, S.; Manusadzianas, L.; Aoyama, I.; Jedidi, N. Solid-phase bioassays and soil microbial activities to evaluate PAH-spiked soil ecotoxicity after a long-term bioremediation process simulating landfarming. Chemosphere 2007, 70, 135–143. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Ahmad, R.; Jonathan, S.G. Synergistic action of rhizospheric fungi with Megathyrsus maximus root speeds up hydrocarbon degradation kinetics in oil polluted soil. Chemosphere 2017, 184, 1–12. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Ahmad, R.; Jonathan, S.G. Synergistic rhizosphere degradation of g-hexachlorocyclohexane (Lindane) through the combinatorial plant-fungal action. PLoS ONE 2017, 12, e0183373. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Chukwuka, K.S.; Jonathan, S.G. Spent mushroom compost enhances plant response and phytoremediation of heavy metal polluted soil. J. Plant Nutr. Soil Sci. 2020, 183, 492–499. [Google Scholar] [CrossRef]
- Daccò, C.; Girometta, C.; Asemoloye, M.D.; Carpani, G.; Picco, A.M.; Tosi, S. Key fungal degradation patterns, enzymes and their applications for the removal of aliphatic hydrocarbons in polluted soils: A review. Int. Biodeterior. Biodegrad. 2020, 147. [Google Scholar] [CrossRef]
- Ferreira, A.M.R.; Rangel, A.O.S.S.; Lima, J.L.F.C. Flow injection system for elemental soil analysis determination. Commun. Soil Sci. Plant Anal. 1998, 29, 327–336. [Google Scholar] [CrossRef]
- Jonathan, S.G.; Adeniyi, M.A.; Asemoloye, M.D. Nutrient value, fungal biodeterioration, and aflatoxin contamination of suya spices a Novel Nigerian Indigenous Snacks. Hindawi Sci. 2016, 2, 1–12. [Google Scholar]
- Jonathan, S.G.; Oghodero, O.; Asemoloye, M.D. Incidence of molds in treated and untreated drinking water of selected local governments in Ibadan, South-Western Nigeria. Res. Rev. Biosc. 2016, 11, 108–118. [Google Scholar]
- Asemoloye, M.D.; Ahmad, R.; Jonathan, S.G. Transcriptomic responses of catalase, peroxidase and laccase encoding genes and enzymatic activities of oil spill inhabiting rhizospheric fungal strains. Environ. Pollut. 2018, 230, 150–160. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi; Webmaster Atlas: Utrecht, The Netherlands, 2014; pp. 34–37. [Google Scholar]
- Möller, E.M.; Bahnweg, G.; Sandermann, H.; Geiger, H.H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992, 20, 6115–6116. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Jung, H.Y.; Lee, Y.S.; Voigt, K.; Lee, H.B. Phylogenetic Status of Two Undescribed Zygomycete Species from Korea: Actinomucor elegans and Mucor minutus. Mycobiology 2017, 45, 344–352. [Google Scholar] [CrossRef]
- Zhang, J.H.; Xue, Q.H.; Gao, H. Degradation of crude oil by fungal enzyme preparations from Aspergillus spp. for potential use in enhanced oil recovery. J. Chem. Technol. Biotechnol. 2016, 91, 865–875. [Google Scholar] [CrossRef]
- Mohammadian, E.; Arzanlou, M.; Babai-Ahari, A. Diversity of culturable fungi inhabiting petroleum-contaminated soils in Southern Iran. Antonie van Leeuwenhoek 2017, 110, 903–923. [Google Scholar] [CrossRef] [PubMed]
- Krimitzas, A.; Pyrri, I.; Kouvelis, V.N. A Phylogenetic Analysis of Greek Isolates of Aspergillus Species Based on Morphology and Nuclear and Mitochondrial Gene Sequences. Biomed. Res. Int. 2013, 1, 260395. [Google Scholar]
- Jonathan, S.G.; Oyetunji, O.J.; Olawuyi, O.J.; Asemoloye, M.D. Growth responses of Corchorus olitorius Lin. (Jute) to the application of SMC as an organic fertilizer. Academ. Arena 2012, 4, 48–56. [Google Scholar]
- Asemoloye, M.D.; Jonathan, S.G.; Ahmad, R. Degradation of 2, 2- Dichlorovinyldimethylphosphate (Dichlorvos) through plant-fungi-SMC interactions in the rhizosphere of Panicum maximus Jacq. Chemosphere 2019, 221, 403–411. [Google Scholar] [CrossRef]
- Castillo, M.P.; Torstensson, L. Effect of biobed composition, moisture, and temperature on the degradation of pesticides. J. Agric. Food Chem. 2007, 55, 5725–5733. [Google Scholar] [CrossRef]
- Ameen, F.; Moslem, M.; Hadi, S.; Al-Sabri, A.E. Biodegradation of diesel fuel hydrocarbons by mangrove fungi from Red Sea Coast of Saudi Arabia. Saudi J. Biol. Sci. 2016, 23, 211–218. [Google Scholar] [CrossRef]
- Tien, M.; Kirk, T.K. Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium Burds. Science 1983, 221, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.K.; Morgan, M.A.; Mayfield, M.B.; Kuwahara, M.; Gold, M.H. An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 1983, 114, 1077–1083. [Google Scholar] [CrossRef]
- Paszcymski, A.; Crawford, R.; Huynh, V.B. Manganese peroxidase of Phanerochaete chrysosporium: Purification. Meth. Enzymol. 1988, 161, 264–271. [Google Scholar]
- Novotny, C.; Erbanova, P.; Sasek, V.; Kubatova, A.; Cajthaml, T.; Lang, E.; Krahl, J.; Zadrazil, F. Extracellular oxidative enzyme production and PAH removal in soil by exploratory mycelium of white rot fungi. Biodegradation 1999, 10, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, A.; Naidu, R. Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: A review of the microbial degradation of Benzo[a]pyrene. Int. Biodeterior. Biodegrad. 2000, 45, 57–88. [Google Scholar] [CrossRef]
- Aban, J.L.; Hipo, R.M.; Balangco, T.D. Diversity and Phylogenetic Relationships among Isolated Root Symbiotic Fungi from Drynaria quercifolia L. in La Union, Philippines. Manila J. Sci. 2017, 10, 87–100. [Google Scholar]
- Aban, J.L.; Hipo, R.M. Isolated root symbiotic fungi (RSF) from Drynaria quercifolia L. show various stress tolerance effects on PSB RC10 (Pagsanjan) rice (Oryza sativa L.) exposed to Snap-peg 8000 mild drought. Int. J. Agric. Environ. Res. 2017, 3, 352–362. [Google Scholar]
- Chukwura, E.I.; Nwando, M.; Ojiegbu, A.; Nwankwegu, S. Hydrocarbon Degradation Potentials of Fungi Associated with oil-Contaminated Soil from Selected Mechanic Workshops in Awka, Anambra State, Nigeria. Front. Environ. Microbiol. 2016, 2, 38–44. [Google Scholar] [CrossRef]
- Li, G.; Cao, B.; Liu, W. Isolation and Identification of Endophytic Fungi in Kernels of Coix lachrymal-jobi L. Cultivars. Curr. Microbiol. 2020, 77, 1448–1456. [Google Scholar] [CrossRef]
- Fall, R.R.; Brown, J.L.; Schaeffer, T.L. Enzyme recruitment allows the biodegradation of recalcitrant–branched hydrocarbons by Pseudomonas citronellolis. Appl. Environ. Microbiol. 1979, 38, 715–722. [Google Scholar] [CrossRef]
- Rusansky, S.; Avigad, R.; Michaeli, S.; Gutnick, D.L. Involvement of a plasmid in growth on and dispersion of crude oil by Acinetobacter calcoaceticus RA57. Appl. Environ. Microbiol. 1987, 53, 1918–1923. [Google Scholar] [CrossRef]
- Xue, J.; Yu, Y.; Bai, Y.; Wang, L.; Wu, Y. Marine oil degrading microorganisms and biodegradation process of petroleum hydrocarbon in marine environments: A review. Curr. Microbiol. 2015, 71, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Barnes, N.M.; Khodse, V.B.; Lotlikar, N.P.; Meena, R.M.; Damare, S.R. Bioremediation potential of hydrocarbon-utilizing fungi from select marine niches of India. 3 Biotech 2018, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.A.R.; Dutta, S.; Mondal, S. Bioremediation of hydrocarbon a review. Int. J. Adv. Res. 2016, 4, 1303–1313. [Google Scholar] [CrossRef]
- Obuekwe, C.O.; Badrudeen, A.M.; Al-Saleh, E.; Mulder, J.L. Growth and hydrocarbon degradation by three desert fungi under conditions of simultaneous temperature and salt stress. Int. Biodeterior. Biodegrad. 2005, 56, 197–205. [Google Scholar] [CrossRef]
- Ye, J.S.; Yin, H.; Qiang, J.; Peng, H.; Qin, H.M.; Zhang, N.; He, B.Y. Biodegradation of anthracene by Aspergillus fumigatus. J. Hazard. Mater. 2011, 185, 174–181. [Google Scholar] [CrossRef]
- Ali, M.I.A.; Khalil, N.M.; El-Ghany, M.N.A. Biodegradation of some polycyclic aromatic hydrocarbons by Aspergillus terreus. Afr. J. Microbiol. Res. 2012, 6, 3783–3790. [Google Scholar] [CrossRef]
- Baniasadi, M.; Mousavi, S.M.; Zilouei, H.; Shojaosadati, S.A.; Rastegar, S.O. Statistical evaluation and process optimization of bioremediation of polycyclic aromatic hydrocarbon in a bioreactor. Environ. Eng. Manag. J. 2018, 17, 1782–1790. [Google Scholar]
- Capotorti, G.; Digianvincenzo, P.; Cesti, P.; Bernardi, A.; Guglielmetti, G. Pyrene and benzo(a) pyrene metabolism by Aspergillus terreus strain isolated from a polycyclic aromatic hydrocarbons polluted soil. Biodegradation 2004, 15, 79–85. [Google Scholar] [CrossRef]
- AI-Jawhari, I.H. Bioremediation of anthracene by Aspergillus niger and Penicillium funiculosum. Int. Res. J. Biol. Sci. 2016, 5, 1–13. [Google Scholar]
- AI-Jawhari, I.H. Ability of some fungi isolated from a sediment of Suq-Al Shuyukh marshes on biodegradation of crude oil. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 19–32. [Google Scholar]
- Mahmoud, G.A.; Koutb, M.M.M.; Morsy, F.M.; Bagy, M.M.K. Mycoflora isolated from Mazot and solar polluted soils in upper Egypt. Egypt. J. Soil. Sci. 2015, 55, 15–30. [Google Scholar] [CrossRef]
- Mahmoud, G.A.; Koutb, M.M.M.; Morsy, F.M.; Bagy, M.M.K. Characterization of lipase enzyme produced by hydrocarbons utilizing fungus Aspergillus terreus. Eur. J. Biol. Res. 2015, 5, 70–77. [Google Scholar]
- Passarini, M.R.; Rodrigues, M.V.; Da Silva, M.; Sette, L.D. Marine-derived filamentous fungi and their potential application for polycyclic aromatic hydrocarbon bioremediation. Mar. Pollut. Bull. 2011, 62, 364–370. [Google Scholar] [CrossRef]
- Al-Nasrawi, H. Biodegradation of crude oil by fungi isolated from Gulf of Mexico. J. Bioremed. Biodegar. 2012, 3, 147. [Google Scholar] [CrossRef]
- Cerniglia, C.E.; Perry, J.J. Crude oil degradation by microorganisms isolated from the marine environment. J. Basic Microbiol. 1973, 13, 299–306. [Google Scholar]
- Walker, J.D.; Colwell, R.R. Microbial degradation of model petroleum at low temperatures. Microb. Ecol. 1974, 1, 63–95. [Google Scholar] [CrossRef]
- Walker, J.D.; Austin, H.F.; Colwell, R.R. Utilization of mixed hydrocarbon substrate by petroleum-degrading microorganisms. J. Gen. Appl. Microbiol. 1975, 21, 27–39. [Google Scholar] [CrossRef]
- Bossert, I.; Bartha, R. The treatment and disposal of petroleum wastes. In Petroleum Microbiology; Atlas, R.M., Ed.; Macmillan: New York, NY, USA, 1984; pp. 553–578. [Google Scholar]
- Da Silva, M.; Cerniglia, C.E.; Pothuluri, J.V.; Canhos, V.P.; Esposito, E. Screening filamentous fungi isolated from estuarine sediments for the ability to oxidize polycyclic aromatic hydrocarbons. World J. Microbiol. Biotechnol. 2003, 19, 399–405. [Google Scholar] [CrossRef]
- Nievas, M.L.; Commendatore, M.G.; Esteves, J.L.; Bucala, V. Biodegradation pattern of hydrocarbons from a fuel oil-type complex residue by an emulsifier-producing microbial consortium. J. Hazard. Mater. 2008, 154, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Li, X.; Allinson, G.; Liu, C.; Gong, Z. Composition and morphology characterization of exopolymeric substances produced by the PAH-degrading fungus of Mucor mucedo. Environ. Sci. Pollut. Res. 2016, 23, 8421–8430. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Viraraghavan, T. Oil removal from water by fungal biomass: A factorial design analysis. J. Hazard. Mater. 2010, 175, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Viraraghavan, T. Oil removal from water using biomaterials. Bioresour. Technol. 2010, 101, 6594–6600. [Google Scholar] [CrossRef]
- Marchut-Mikolajczyk, O.; Kwapisz, E.; Wieczorek, D.; Antczak, T. Biodegradation of diesel oil hydrocarbons enhanced with Mucor circinelloides enzyme preparation. Int. Biodeterior. Biodegrad. 2015, 104, 142–148. [Google Scholar] [CrossRef]
- Marchut-Mikolajczyk, O.; Drozdzynski, P.; Struszczyk-Swita, K. Biodegradation of slop oil by endophytic Bacillus cereus EN18 coupled with lipase from Rhizomucor miehei (Palatase®). Chemosphere 2020, 250, 126203. [Google Scholar] [CrossRef]
- Marchut-Mikolajczyk, O.; Drozdzynski, P.; Pietrzyk, D.; Antczak, T. Biosurfactant production and hydrocarbon degradation activity of endophytic bacteria isolated from Chelidonium majus L. Microb. Cell Factories 2018, 17, 171. [Google Scholar] [CrossRef]
- Ramirez, D.A.; Munoz, S.V.; Atehortua, L.; Michel, F.C. Effects of different wavelengths of light on lignin peroxidase production by the white-rot fungi Phanerochaete chrysosporium grown in submerged cultures. Biol. Technol. 2010, 101, 9213–9220. [Google Scholar] [CrossRef]
- Pribnow, D.; Mayfield, M.B.; Nipper, V.J.; Brown, J.A.; Gold, M.H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 1989, 264, 5036–5040. [Google Scholar]
- Ronne, H. Glucose repression in fungi. Trends. Genet. 1995, 11, 12–17. [Google Scholar] [CrossRef]
- Janusz, G.; Katarzyna, H.; Kucharzykb, A.P.; Magdalena, S.A.; Paszczynski, J. Fungal laccase, manganese peroxidase and lignin peroxidase: Gene expression and regulation. Enzym. Micro. Technol. 2013, 52, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda, R.H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Roux, I.; Woodcraft, C.; Hu, J.; Wolters, R.; Gilchrist, C.L.M.; Chooi, Y.H. CRISPR-Mediated Activation of Biosynthetic Gene Clusters for Bioactive Molecule Discovery in Filamentous Fungi. ACS Synth. Biol. 2020, 9, 1843–1854. [Google Scholar] [CrossRef]

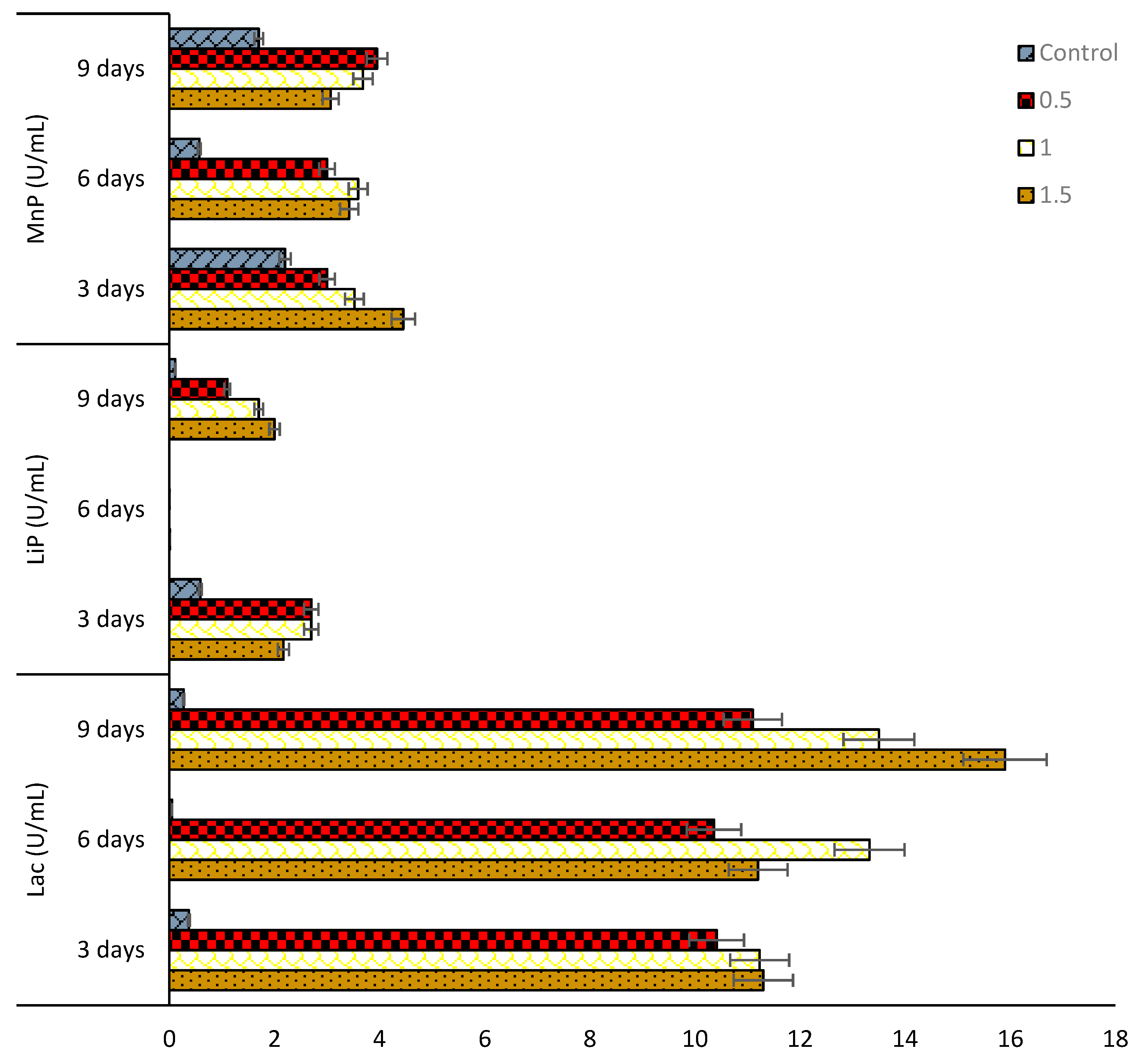

| Strain Code | B-Yorla10 | C-Effurun | ||
|---|---|---|---|---|
| Incidence (N = 30) | % Incidence | Incidence (N = 30) | % Incidence | |
| A | 9 | 30 | 1 | 3.33 |
| B | 20 | 66.67 | 13 | 43.33 |
| C | 13 | 43.33 | 17 | 56.67 |
| D | 0 | 0.00 | 0 | 0.00 |
| E | 5 | 16.67 | 11 | 36.67 |
| F | 3 | 10.00 | 2 | 6.67 |
| G | 0 | 0.00 | 3 | 13.33 |
| H | 11 | 36.67 | 0 | 0.00 |
| I | 1 | 3.33 | 1 | 3.33 |
| J | 5 | 16.66 | 11 | 36.67 |
| K | 2 | 6.67 | 3 | 10.00 |
| N1 | 9 | 30.00 | 0 | 0.00 |
| N2 | 6 | 20.00 | 5 | 16.67 |
| N3 | 11 | 36.67 | 0 | 0.00 |
| Fungal Strain | Oil Concentration | Growth Rate (cm/day) | DIR (%) |
|---|---|---|---|
| B | 0% | 4.82 | 100.00 |
| B | 5% | 3.64 | 75.52 |
| B | 10% | 2.89 | 59.96 |
| B | 15% | 2.47 | 51.24 |
| B | 20% | 2.21 | 45.85 |
| C | 0% | 3.02 | 100.00 |
| C | 5% | 1.63 | 53.97 |
| C | 10% | 1.41 | 46.69 |
| C | 15% | 1.18 | 39.07 |
| C | 20% | 1.19 | 39.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asemoloye, M.D.; Tosi, S.; Daccò, C.; Wang, X.; Xu, S.; Marchisio, M.A.; Gao, W.; Jonathan, S.G.; Pecoraro, L. Hydrocarbon Degradation and Enzyme Activities of Aspergillus oryzae and Mucor irregularis Isolated from Nigerian Crude Oil-Polluted Sites. Microorganisms 2020, 8, 1912. https://doi.org/10.3390/microorganisms8121912
Asemoloye MD, Tosi S, Daccò C, Wang X, Xu S, Marchisio MA, Gao W, Jonathan SG, Pecoraro L. Hydrocarbon Degradation and Enzyme Activities of Aspergillus oryzae and Mucor irregularis Isolated from Nigerian Crude Oil-Polluted Sites. Microorganisms. 2020; 8(12):1912. https://doi.org/10.3390/microorganisms8121912
Chicago/Turabian StyleAsemoloye, Michael Dare, Solveig Tosi, Chiara Daccò, Xiao Wang, Shihan Xu, Mario Andrea Marchisio, Wenyuan Gao, Segun Gbolagade Jonathan, and Lorenzo Pecoraro. 2020. "Hydrocarbon Degradation and Enzyme Activities of Aspergillus oryzae and Mucor irregularis Isolated from Nigerian Crude Oil-Polluted Sites" Microorganisms 8, no. 12: 1912. https://doi.org/10.3390/microorganisms8121912
APA StyleAsemoloye, M. D., Tosi, S., Daccò, C., Wang, X., Xu, S., Marchisio, M. A., Gao, W., Jonathan, S. G., & Pecoraro, L. (2020). Hydrocarbon Degradation and Enzyme Activities of Aspergillus oryzae and Mucor irregularis Isolated from Nigerian Crude Oil-Polluted Sites. Microorganisms, 8(12), 1912. https://doi.org/10.3390/microorganisms8121912








