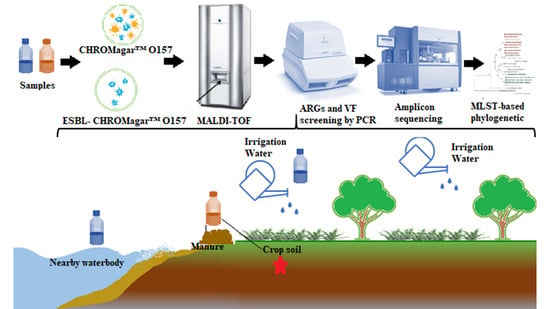
Shared Extended-Spectrum β-Lactamase-Producing Salmonella Serovars between Agricultural and Aquatic Environments Revealed through invA Amplicon Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site Description and Sample Collection
2.2. Media and Sample Preparation
2.3. Detection and Isolation of ESBL-Producing Salmonella spp. and Salmonella spp.
2.4. Bacterial Confirmation and Identification
2.5. Serogrouping of ESBL-Producing Salmonella spp. Isolates
2.6. DNA Extraction
2.7. Detection of ARG in ESBL-Producing Salmonella spp. Using PCR
2.8. Detection of VF in ESBL-Producing Salmonella spp. Using PCR
2.9. InvA Amplicon Sequencing and Analysis
2.10. Bioinformatic Analysis of the invA Sequences
2.11. Data Analysis
3. Results
3.1. Prevalence of Nonresistant and ESBL-Producing Salmonella spp.
3.2. Bacterial Confirmation and Identification
3.3. Serotyping of ESBL-Producing Salmonella spp. Isolates
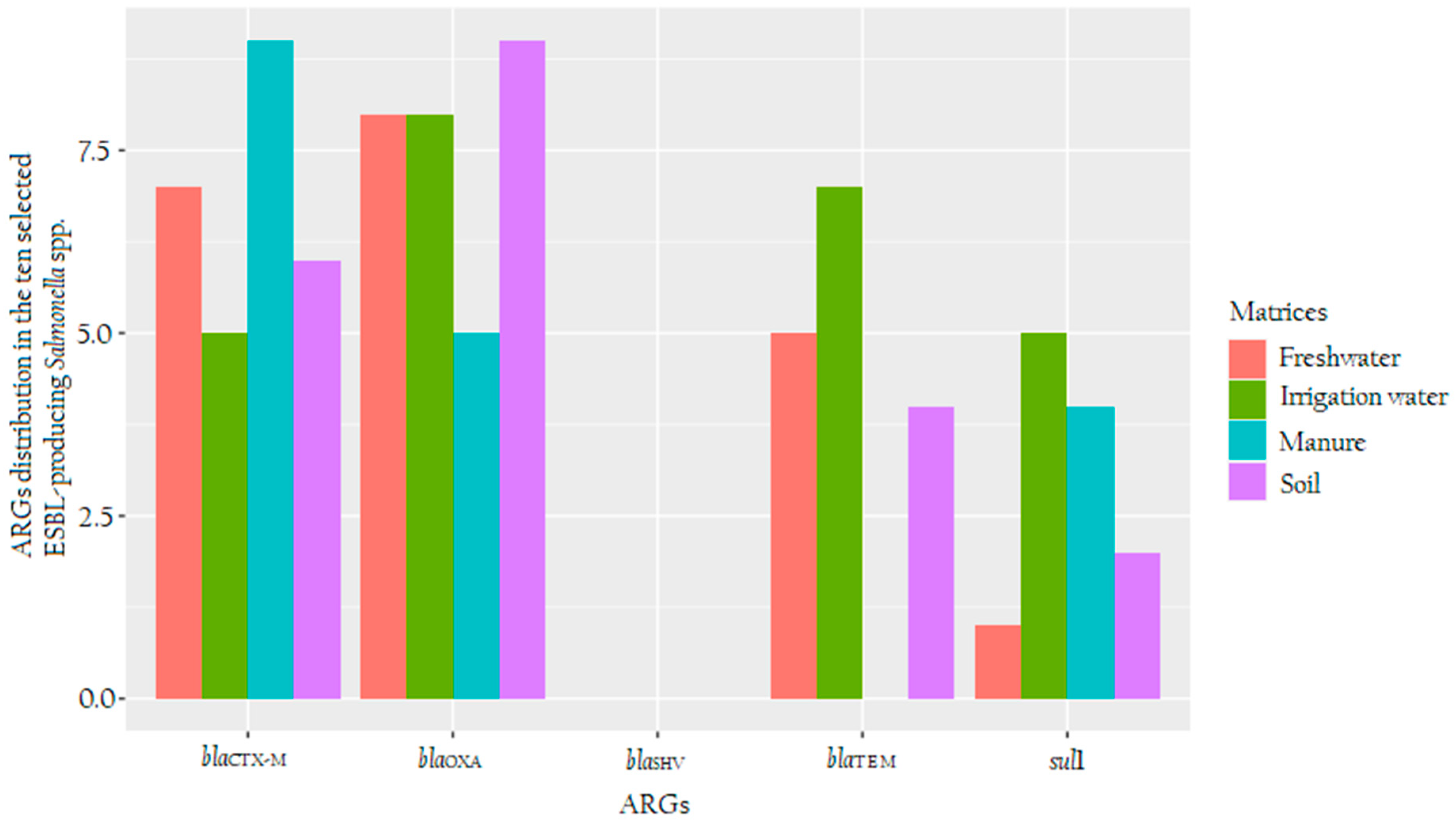
3.4. Detection of ARG in ESBL-Producing Salmonella spp.
3.5. Detection of VF in ESBL-Producing Salmonella spp.
3.6. InvA Amplicon Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bhatta, D.R.; Bangtrakulnonth, A.; Tishyadhigama, P.; Saroj, S.D.; Bandekar, J.R.; Hendriksen, R.S.; Kapadnis, B.P. Serotyping, PCR, phage-typing and antibiotic sensitivity testing of Salmonella serovars isolated from urban drinking water supply systems of Nepal. Lett. Appl. Microbiol. 2007, 44, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Amagliani, G.; Brandi, G.; Schiavano, G.F. Incidence and role of Salmonella in seafood safety. Food Res. Int. 2012, 45, 780–788. [Google Scholar] [CrossRef]
- Stanaway, J.D.; Parisi, A.; Sarkar, K.; Blacker, B.F.; Reiner, R.C.; Hay, S.I.; Nixon, M.R.; Dolecek, C.; James, S.L.; Mokdad, A.H.; et al. The global burden of non-typhoidal salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 1312–1324. [Google Scholar] [CrossRef]
- Yang, Y.T.; Swinburne, M. New Produce Safety Regulations. Public Health Rep. 2016, 131, 754–757. [Google Scholar] [CrossRef]
- Jaja, I.F.; Bhembe, N.L.; Green, E.; Oguttu, J.; Muchenje, V. Molecular characterisation of antibiotic-resistant Salmonella enterica isolates recovered from meat in South Africa. Acta Trop. 2019, 190, 129–136. [Google Scholar] [CrossRef]
- Hanning, I.B.; Nutt, J.D.; Ricke, S.C. Salmonellosis Outbreaks in the United States Due to Fresh Produce: Sources and Potential Intervention Measures. Foodborne Pathog. Dis. 2009, 6, 635–648. [Google Scholar] [CrossRef]
- Fearnley, E.; Raupach, J.; Lagala, F.; Cameron, S. Salmonella in chicken meat, eggs and humans; Adelaide, South Australia, 2008. Int. J. Food Microbiol. 2011, 146, 219–227. [Google Scholar] [CrossRef]
- M’ikanatha, N.M.; Sandt, C.H.; Localio, A.R.; Tewari, D.; Rankin, S.C.; Whichard, J.M.; Altekruse, S.F.; Lautenbach, E.; Folster, J.P.; Russo, A.; et al. Multidrug-Resistant Salmonella Isolates from Retail Chicken Meat Compared with Human Clinical Isolates. Foodborne Pathog. Dis. 2010, 7, 929–934. [Google Scholar] [CrossRef]
- Ifeoma Stella, E. Evaluation of Salmonella Species in Water Sources in Two Local Government Areas of Anambra State. Cohesive J. Microbiol. Infect. Dis. 2018, 1. [Google Scholar] [CrossRef]
- Gelband, H.; Molly Miller, P.; Pant, S.; Gandra, S.; Levinson, J.; Barter, D.; White, A.; Laxminarayan, R. The state of the world’s antibiotics 2015. Wound Health South. Afr. 2015, 8, 30–34. [Google Scholar]
- Akinyemi, K.O.; Iwalokun, B.; Alafe, O.; Mudashiru, S.; Fakorede, C. blaCTX-M-I group extended spectrum beta lactamase-producing Salmonella typhi from hospitalised patients in Lagos, Nigeria. Infect. Drug Resist. 2015, 99. [Google Scholar] [CrossRef][Green Version]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Thottathil, S.E.; Newman, T.B. Antibiotics Overuse in Animal Agriculture: A Call to Action for Health Care Providers. Am. J. Public Health 2015, 105, 2409–2410. [Google Scholar] [CrossRef] [PubMed]
- Choukr-Allah, R. Wastewater Recycling and Reuse in Mediterranean Region as a Potential Resources for Water Saving and Sustainable Agriculture. In Proceedings of the Symposium International” Agriculture Durable en Region Mediterranean (AGDUMED), Rabat, Maroc, 14–16 May 2009; pp. 14–15. [Google Scholar]
- Parisi, A.; Crump, J.A.; Glass, K.; Howden, B.P.; Furuya-Kanamori, L.; Vilkins, S.; Gray, D.J.; Kirk, M.D. Health Outcomes from Multidrug-Resistant Salmonella Infections in High-Income Countries: A Systematic Review and Meta-Analysis. Foodborne Pathog. Dis. 2018, 15, 428–436. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Nicolau, D.P.; Carmeli, Y.; Crank, C.W.; Goff, D.A.; Graber, C.J.; Lima, A.L.L.; Goldstein, E.J. Carbapenem stewardship: Does ertapenem affect Pseudomonas susceptibility to other carbapenems? A review of the evidence. Int. J. Antimicrob. Agents 2012, 39, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Ndugulile, F.; Jureen, R.; Harthug, S.; Urassa, W.; Langeland, N. Extended Spectrum β-Lactamases among Gram-negative bacteria of nosocomial origin from an Intensive Care Unit of a tertiary health facility in Tanzania. BMC Infect. Dis. 2005, 5, 86. [Google Scholar] [CrossRef]
- Seong, W.-J.; Kwon, H.-J.; Kim, T.-E.; Lee, D.-Y.; Park, M.-S.; Kim, J.-H. Molecular serotyping of Salmonella enterica by complete rpoB gene sequencing. J. Microbiol. 2012, 50, 962–969. [Google Scholar] [CrossRef]
- Blaak, H.; van Hoek, A.H.A.M.; Veenman, C.; Docters van Leeuwen, A.E.; Lynch, G.; van Overbeek, W.M.; de Roda Husman, A.M. Extended spectrum ß-lactamase- and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 2014, 168–169, 8–16. [Google Scholar] [CrossRef]
- Reuland, E.A.; al Naiemi, N.; Raadsen, S.A.; Savelkoul, P.H.M.; Kluytmans, J.A.J.W.; Vandenbroucke-Grauls, C.M.J.E. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1843–1846. [Google Scholar] [CrossRef]
- Veldman, K.; Kant, A.; Dierikx, C.; van Essen-Zandbergen, A.; Wit, B.; Mevius, D. Enterobacteriaceae resistant to third-generation cephalosporins and quinolones in fresh culinary herbs imported from Southeast Asia. Int. J. Food Microbiol. 2014, 177, 72–77. [Google Scholar] [CrossRef]
- Raseala, C.M.; Ekwanzala, M.D.; Momba, M.N.B. Multilocus-based phylogenetic analysis of extended-spectrum beta-lactamase Escherichia coli O157:H7 uncovers related strains between agriculture and nearby water sources. J. Infect. Public Health 2020. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Poppe, C.; Martin, L.; Muckle, A.; Archambault, M.; McEwen, S.; Weir, E. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can. J. Vet. Res. 2006, 70, 105. [Google Scholar] [PubMed]
- Aarestrup, F.M. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J. Antimicrob. Chemother. 2003, 52, 715–718. [Google Scholar] [CrossRef]
- Zishiri, O.T.; Mkhize, N.; Mukaratirwa, S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J. Vet. Res. 2016, 83. [Google Scholar] [CrossRef]
- Fakhr, M.K.; Nolan, L.K.; Logue, C.M. Multilocus Sequence Typing Lacks the Discriminatory Ability of Pulsed-Field Gel Electrophoresis for Typing Salmonella enterica Serovar Typhimurium. J. Clin. Microbiol. 2005, 43, 2215–2219. [Google Scholar] [CrossRef][Green Version]
- Abakpa, G.O.; Umoh, V.J.; Ameh, J.B.; Yakubu, S.E.; Kwaga, J.K.P.; Kamaruzaman, S. Diversity and antimicrobial resistance of Salmonella enterica isolated from fresh produce and environmental samples. Environ. Nanotechnol. Monit. Manag. 2015, 3, 38–46. [Google Scholar] [CrossRef]
- Kadry, M.; Nader, S.M.; Dorgham, S.M.; Kandil, M.M. Molecular diversity of the invA gene obtained from human and egg samples. Vet. World 2019, 12, 1033–1038. [Google Scholar] [CrossRef]
- Buehler, A.J.; Wiedmann, M.; Kassaify, Z.; Cheng, R.A. Evaluation of invA Diversity among Salmonella Species Suggests Why Some Commercially Available Rapid Detection Kits May Fail To Detect Multiple Salmonella Subspecies and Species. J. Food Prot. 2019, 82, 710–717. [Google Scholar] [CrossRef]
- Blankenberg, D.; Gordon, A.; von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010, pp. 175–176. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 September 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F. Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Bioinformatics 2015, 516, 517. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Baraniak, A.; Sadowy, E.; Hryniewicz, W.; Gniadkowski, M. Two Different Extended-Spectrum -Lactamases (ESBLs) in One of the First ESBL-Producing Salmonella Isolates in Poland. J. Clin. Microbiol. 2002, 40, 1095–1097. [Google Scholar] [CrossRef][Green Version]
- Rotimi, V.O.; Jamal, W.; Pal, T.; Sovenned, A.; Albert, M.J. Emergence of CTX-M-15 type extended-spectrum β-lactamase-producing Salmonella spp. in Kuwait and the United Arab Emirates. J. Med Microbiol. 2008, 57, 881–886. [Google Scholar] [CrossRef]
- Sjölund, M.; Yam, J.; Schwenk, J.; Joyce, K.; Medalla, F.; Barzilay, E.; Whichard, J.M. Human Salmonella Infection Yielding CTX-M β-Lactamase, United States. Emerg. Infect. Dis. 2008, 14, 1957–1959. [Google Scholar] [CrossRef] [PubMed]
- Usha, G.; Chunderika, M.; Prashini, M.; Willem, S.A.; Yusuf, E.S. Characterization of extended-spectrum β-lactamases in Salmonella spp. at a tertiary hospital in Durban, South Africa. Diagn. Microbiol. Infect. Dis. 2008, 62, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Yates, C.; Amyes, S. Extended-spectrum β-lactamases in non-typhoidal Salmonella spp. isolated in the UK are now a reality: Why the late arrival? J. Antimicrob. Chemother. 2005, 56, 262–264. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Raskin, L.; Zilles, J.L. Macrolide Resistance in Microorganisms at Antimicrobial-Free Swine Farms. Appl. Environ. Microbiol. 2009, 75, 5814–5820. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Okubo, T.; Usui, M.; Yokota, S.; Izumiyama, S.; Tamura, Y. Association of Veterinary Third-Generation Cephalosporin Use with the Risk of Emergence of Extended-Spectrum-Cephalosporin Resistance in Escherichia coli from Dairy Cattle in Japan. PLoS ONE 2014, 9, e96101. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, C.S.; Bech, T.B. Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res. Int. 2012, 45, 557–566. [Google Scholar] [CrossRef]
- Adzitey, F.; Ashiagbor, C.N.K.; Abu, H. Prevalence and antibiotic susceptibility of Salmonella spp. from water sources in Tamale, Ghana. Int. J. One Health 2016, 2, 24–28. [Google Scholar] [CrossRef][Green Version]
- Qiao, J.; Zhang, Q.; Alali, W.Q.; Wang, J.; Meng, L.; Xiao, Y.; Yang, H.; Chen, S.; Cui, S.; Yang, B. Characterization of extended-spectrum β-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int. J. Food Microbiol. 2017, 248, 72–81. [Google Scholar] [CrossRef]
- Benagli, C.; Rossi, V.; Dolina, M.; Tonolla, M.; Petrini, O. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for the Identification of Clinically Relevant Bacteria. PLoS ONE 2011, 6, e16424. [Google Scholar] [CrossRef]
- Abdel-Maksoud, M.; Abdel-Khalek, R.; El-Gendy, A.; Gamal, R.F.; Abdelhady, H.M.; House, B.L. Genetic characterisation of multidrug-resistant Salmonella enterica serotypes isolated from poultry in Cairo, Egypt. Afr. J. Lab. Med. 2015, 4. [Google Scholar] [CrossRef]
- Roy, P.; Dhillon, A.S.; Lauerman, L.H.; Schaberg, D.M.; Bandli, D.; Johnson, S. Results of Salmonella isolation from poultry products, poultry, poultry environment, and other characteristics. Avian Dis. 2002, 46, 17–24. [Google Scholar] [CrossRef]
- Maraki, S.; Papadakis, I.S. Serotypes and Antimicrobial Resistance of Human Nontyphoidal Isolates of Salmonella enterica from Crete, Greece. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, G. Prevalence of human Salmonellosis in Ethiopia: A systematic review and meta-analysis. BMC Infect. Dis. 2014, 14, 88. [Google Scholar] [CrossRef] [PubMed]
- Kwambana-Adams, B.; Darboe, S.; Nabwera, H.; Foster-Nyarko, E.; Ikumapayi, U.N.; Secka, O.; Betts, M.; Bradbury, R.; Wegmüller, R.; Lawal, B.; et al. Salmonella Infections in The Gambia, 2005–2015. Clin. Infect. Dis. 2015, 61, S354–S362. [Google Scholar] [CrossRef][Green Version]
- Donaldson, S.C.; Straley, B.A.; Hegde, N.V.; Sawant, A.A.; DebRoy, C.; Jayarao, B.M. Molecular Epidemiology of Ceftiofur-Resistant Escherichia coli Isolates from Dairy Calves. Appl. Environ. Microbiol. 2006, 72, 3940–3948. [Google Scholar] [CrossRef]
- Frye, J.G.; Fedorka-Cray, P.J. Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int. J. Antimicrob. Agents 2007, 30, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Guerri, M. Detection of integrons and antibiotic-resistance genes in Salmonella enterica serovar Typhimurium isolates with resistance to ampicillin and variable susceptibility to amoxicillin-clavulanate. Int. J. Antimicrob. Agents 2004, 24, 327–333. [Google Scholar] [CrossRef]
- Binh, C.T.T.; Heuer, H.; Gomes, N.C.M.; Kaupenjohann, M.; Smalla, K. Similar bacterial community structure and high abundance of sulfonamide resistance genes in field-scale manures. Manure Manag. Uses Environ. Impacts. Hauppauge: Nova Sci. Publ. 2010, 141–166. Available online: http://www.novapublishers.org/catalog/downloadOA.php?order=1&access=true (accessed on 22 August 2020).
- Randall, L.P. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004, 53, 208–216. [Google Scholar] [CrossRef]
- Durso, L.M.; Miller, D.N.; Wienhold, B.J. Distribution and Quantification of Antibiotic Resistant Genes and Bacteria across Agricultural and Non-Agricultural Metagenomes. PLoS ONE 2012, 7, e48325. [Google Scholar] [CrossRef] [PubMed]
- Nesme, J.; Cécillon, S.; Delmont, T.O.; Monier, J.-M.; Vogel, T.M.; Simonet, P. Large-Scale Metagenomic-Based Study of Antibiotic Resistance in the Environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, I.H. Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals. Trop. Anim. Health Prod. 2015, 47, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Zhang, X.-X.; Miao, Y.; Zhao, Y.; Ye, L.; Li, B.; Zhang, T. Fate of antibiotic resistance genes and their associations with bacterial community in livestock breeding wastewater and its receiving river water. Water Res. 2017. [Google Scholar] [CrossRef]
- Falagas, M.E.; Karageorgopoulos, D.E. Extended-spectrum β-lactamase-producing organisms. J. Hosp. Infect. 2009, 73, 345–354. [Google Scholar] [CrossRef]
- Foote, A.D.; Thomsen, P.F.; Sveegaard, S.; Wahlberg, M.; Kielgast, J.; Kyhn, L.A.; Salling, A.B.; Galatius, A.; Orlando, L.; Gilbert, M.T.P. Investigating the Potential Use of Environmental DNA (eDNA) for Genetic Monitoring of Marine Mammals. PLoS ONE 2012, 7, e41781. [Google Scholar] [CrossRef]
- Adelowo, O.O.; Caucci, S.; Banjo, O.A.; Nnanna, O.C.; Awotipe, E.O.; Peters, F.B.; Fagade, O.E.; Berendonk, T.U. Extended Spectrum Beta-Lactamase (ESBL)-producing bacteria isolated from hospital wastewaters, rivers and aquaculture sources in Nigeria. Environ. Sci. Pollut. Res. 2018, 25, 2744–2755. [Google Scholar] [CrossRef]
- Liu, J.; Dan, X.; Lu, G.; Shen, J.; Wu, D.; Yan, Z. Investigation of pharmaceutically active compounds in an urban receiving water: Occurrence, fate and environmental risk assessment. Ecotoxicol. Environ. Saf. 2018, 154, 214–220. [Google Scholar] [CrossRef]
- Eckert, C.; Gautier, V.; Arlet, G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2006, 57, 14–23. [Google Scholar] [CrossRef]
- Hughes, L.A.; Shopland, S.; Wigley, P.; Bradon, H.; Leatherbarrow, A.H.; Williams, N.J.; Bennett, M.; de Pinna, E.; Lawson, B.; Cunningham, A.A.; et al. Characterisation of Salmonella enterica serotype Typhimurium isolates from wild birds in northern England from 2005–2006. BMC Vet. Res. 2008, 4, 4. [Google Scholar] [CrossRef]
- Galán, J.E.; Ginocchio, C.; Costeas, P. Molecular and functional characterisation of the Salmonella invasion gene invA: Homology of InvA to members of a new protein family. J. Bacteriol. 1992, 174, 4338–4349. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-M.; Lin, W.; Van, K.T.; Phan, L.; Tran, N.N.; Farmer, D. Rapid Detection of Salmonella in Foods Using Real-Time PCR. J. Food Prot. 2008, 71, 2436–2441. [Google Scholar] [CrossRef] [PubMed]
- Al Arafat, T.; Mahmud, M.R.; Tanim, M.T.I.; Chowdhury, M.M.K.; Rahaman, M.M.; Rahman, S.R.; Rahman, M.M. Genetic Diversity of Salmonella enterica Strains Isolated from Sewage Samples of Different Hospitals in Bangladesh. Bangladesh J. Microbiol. 2019, 35, 57–60. [Google Scholar] [CrossRef]
- Zhao, S.; White, D.G.; Friedman, S.L.; Glenn, A.; Blickenstaff, K.; Ayers, S.L.; Abbott, J.W.; Hall-Robinson, E.; McDermott, P.F. Antimicrobial Resistance in Salmonella enterica Serovar Heidelberg Isolates from Retail Meats, Including poultry, from 2002 to 2006. Appl. Environ. Microbiol. 2008, 74, 6656–6662. [Google Scholar] [CrossRef] [PubMed]
- Kidanemariam, A.; Engelbrecht, M.; Picard, J. Retrospective study on the incidence of Salmonella isolations in animals in South Africa, 1996 to 2006. J. South Afr. Vet. Assoc. 2010, 81. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bisi-Johnson, M.; Obi, C. Detection of Carbapenem Resistance in Salmonella Species from a Tertiary Hospital in Eastern Cape, South Africa. Br. Microbiol. Res. J. 2015, 10, 1–6. [Google Scholar] [CrossRef]
- Dekker, D.; Krumkamp, R.; Sarpong, N.; Frickmann, H.; Boahen, K.; Frimpong, M.; Asare, R.; Larbi, R.; Hagen, R.; Poppert, S.; et al. Drinking Water from Dug Wells in Rural Ghana—Salmonella Contamination, Environmental Factors, and Genotypes. Int. J. Environ. Res. Public Health 2015, 12, 3535–3546. [Google Scholar] [CrossRef]
- Traoré, O.; Nyholm, O.; Siitonen, A.; Bonkoungou, I.J.O.; Traoré, A.S.; Barro, N.; Haukka, K. Prevalence and diversity of Salmonella enterica in water, fish and lettuce in Ouagadougou, Burkina Faso. BMC Microbiol. 2015, 15, 151. [Google Scholar] [CrossRef]




| Genes | Nucleotide Sequences (5′-3′) | Target Size | TAnnealing (°C) | Reference |
|---|---|---|---|---|
| Salmonella spp. ARG | ||||
| blaCTX | F:ATGTGCAGYACCAGTAARGTKATGGC R:TGGGTRAARTARGTSACCAGAAYCAGCGG | 593 | 60 | [24] |
| blaSHV | F:TTCGCCTGTGTATTATCTCCCTG R:TTAGCGTTGCCAGTGYTCG | 854 | 50 | [24] |
| blaOXA-1 | F:ATGAAAAACACAATACATATCAACTTCGC R:GTGTGTTTAGAATGGTGATCGCATT | 820 | 58 | [24] |
| sul1 | F:GCGCGGCGTGGGCTACCT R:GATTTCCGCGACACCGAGACAA | 350 | 65 | [25] |
| blaTEM | F:ATGAGTATTCAACATTTCCG R:ACCAATGCTTAATCAGTGAG | 859 | 53 | [26] |
| Salmonella spp. VF | ||||
| spiC | F:CCTGGATAATGACTATTGAT R:AGTTTATGGTGATTGCGTAT | 309 | 54 | [27] |
| misL | F:GTCGGCGAATGCCGCGAATA R:GCGCTGTTAACGCTAATAGT | 400 | 58 | [27] |
| pipD | F:CGGCGATTCATGACTTTGAT R:CGTTATCATTCGGATCGTAA | 350 | 56 | [27] |
| spaM | F:CGCTGTACGGTATTTCATT R:CTGACTCGGCCTCTTCCTG | 394 | 55 | [28] |
| orfL | F:GGAGTATCGATAAAGATGTT R:GCGCGTAACGTCAGAATCAA | 550 | 58 | [27] |
| invA | F:GTGAAATTATCGCCACGTTCGGGCAA R:TCATCGCACCGTCAAAGGAACC | 284 | 45 | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raseala, C.M.; Ekwanzala, M.D.; Momba, M.N.B. Shared Extended-Spectrum β-Lactamase-Producing Salmonella Serovars between Agricultural and Aquatic Environments Revealed through invA Amplicon Sequencing. Microorganisms 2020, 8, 1898. https://doi.org/10.3390/microorganisms8121898
Raseala CM, Ekwanzala MD, Momba MNB. Shared Extended-Spectrum β-Lactamase-Producing Salmonella Serovars between Agricultural and Aquatic Environments Revealed through invA Amplicon Sequencing. Microorganisms. 2020; 8(12):1898. https://doi.org/10.3390/microorganisms8121898
Chicago/Turabian StyleRaseala, Cecilia Mahlatse, Mutshiene Deogratias Ekwanzala, and Maggy Ndombo Benteke Momba. 2020. "Shared Extended-Spectrum β-Lactamase-Producing Salmonella Serovars between Agricultural and Aquatic Environments Revealed through invA Amplicon Sequencing" Microorganisms 8, no. 12: 1898. https://doi.org/10.3390/microorganisms8121898
APA StyleRaseala, C. M., Ekwanzala, M. D., & Momba, M. N. B. (2020). Shared Extended-Spectrum β-Lactamase-Producing Salmonella Serovars between Agricultural and Aquatic Environments Revealed through invA Amplicon Sequencing. Microorganisms, 8(12), 1898. https://doi.org/10.3390/microorganisms8121898