Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota
Abstract
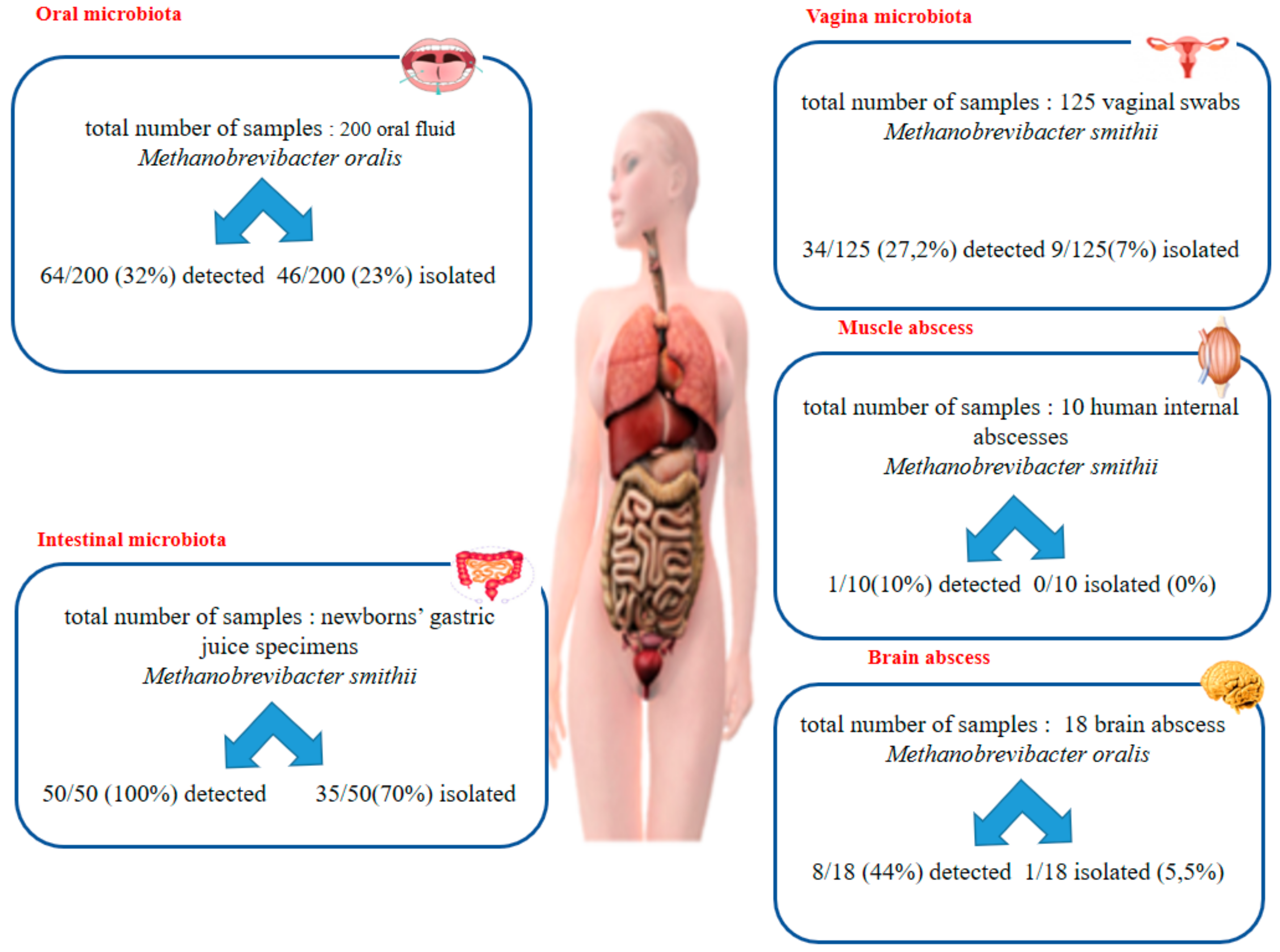
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Molecular Detection of Methanogens
2.3. Isolation and Culture
2.4. Fluorescent In Situ Hybridization
3. Results
3.1. Molecular Detection
3.1.1. PCR-Sequencing
3.1.2. Real-Time PCR
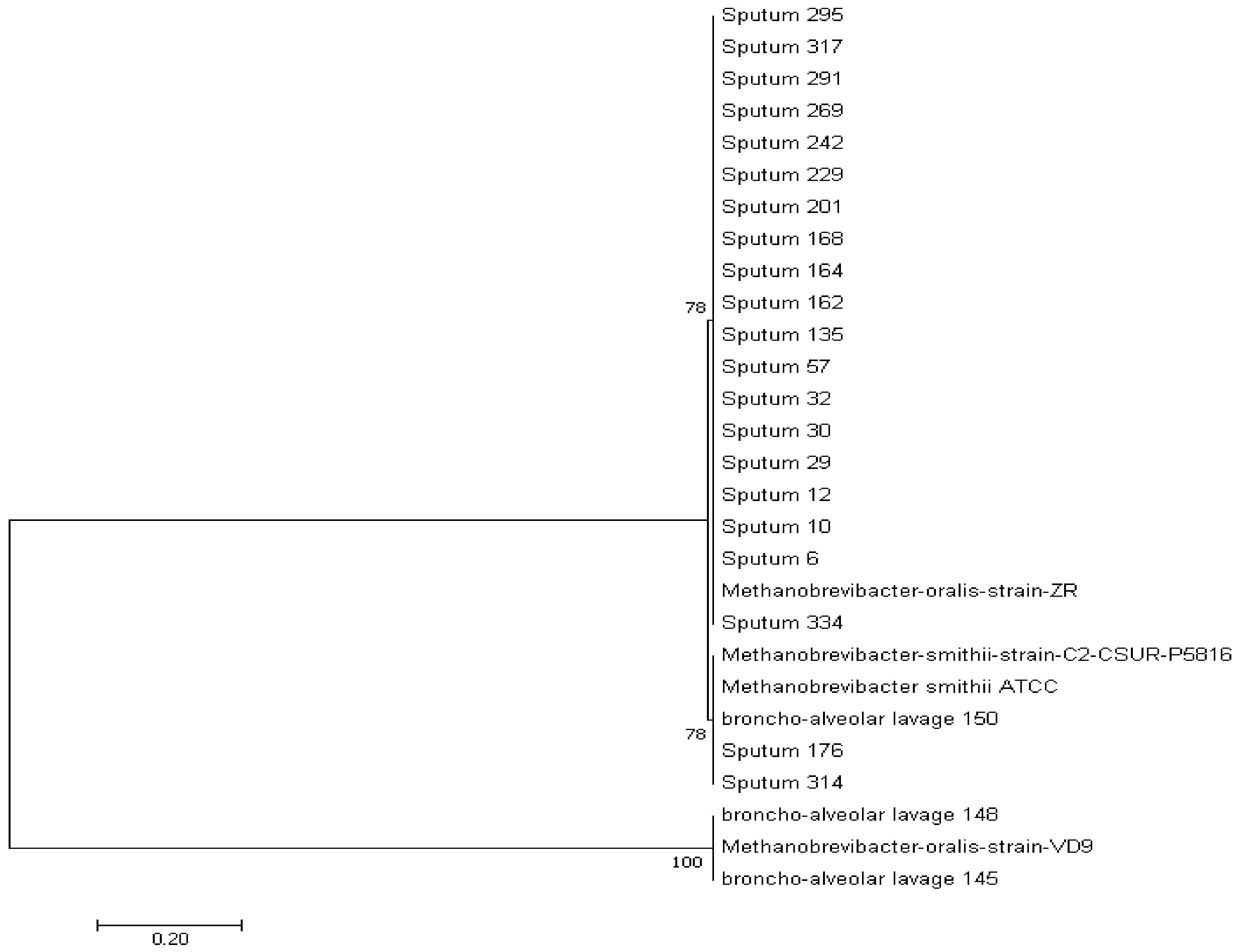
3.1.3. Phylogenetic Analysis
3.2. Isolation and Culture
3.3. Fluorescent In Situ Hybridization
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grine, G.; Terrer, E.; Boualam, M.A.; Aboudharam, G.; Chaudet, H.; Ruimy, R.; Drancourt, M. Tobacco-smoking-related prevalence of methanogens in the oral fluid microbiota. Sci. Rep. 2018, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Hieu, T.; Khelaifia, S.; Aboudharam, G.; Drancourt, M. Methanogenic archaea in subgingival sites: A review. APMIS 2012, 121, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.T.T.; Pignoly, M.; Drancourt, M.; Aboudharam, G. A new methanogen “Methanobrevibacter massiliense” isolated in a case of severe periodontitis. BMC Res. Notes 2017, 10, 657. [Google Scholar] [CrossRef] [PubMed]
- Grine, G.; Boualam, M.A.; Drancourt, M. Methanobrevibacter smithii, a methanogen consistently colonising the newborn stomach. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2449–2455. [Google Scholar] [CrossRef]
- Nkamga, V.D.; Henrissat, B.; Drancourt, M. Archaea: Essential inhabitants of the human digestive microbiota. Hum. Microbiome J. 2017, 3, 1–8. [Google Scholar] [CrossRef]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef]
- Grine, G.; Lotte, R.; Chirio, D.; Chevalier, A.; Raoult, D.; Drancourt, M.; Ruimy, R. Co-Culture of Methanobrevibacter Smithii with Enterobacteria During Urinary Infection. SSRN Electron. J. 2019, 43, 333–337. [Google Scholar] [CrossRef]
- Huynh, H.T.T.; Pignoly, M.; Nkamga, V.D.; Drancourt, M.; Aboudharam, G. The Repertoire of Archaea Cultivated from Severe Periodontitis. PLoS ONE 2015, 10, e0121565. [Google Scholar] [CrossRef]
- Belkacemi, S.; Mazel, A.; Tardivo, D.; Tavitian, P.; Stephan, G.; Bianca, G.; Terrer, E.; Drancourt, M.; Aboudharam, G. Peri-implantitis-associated methanogens: A preliminary report. Sci. Rep. 2018, 8, 1–6. [Google Scholar] [CrossRef]
- Sogodogo, E.; Drancourt, M.; Grine, G. Methanogens as emerging pathogens in anaerobic abscesses. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 811–818. [Google Scholar] [CrossRef]
- Grine, G.; Drouet, H.; Fenollar, F.; Bretelle, F.; Raoult, D.; Drancourt, M. Detection of Methanobrevibacter smithii in vaginal samples collected from women diagnosed with bacterial vaginosis. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Nkamga, V.D.; Lakhe, N.A.; Régis, J.-M.; Dufour, H.; Fournier, P.-E.; Bechah, Y.; Scheld, W.M.; Raoult, D. Evidence of Archaeal Methanogens in Brain Abscess. Clin. Infect. Dis. 2017, 65, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Nkamga, V.; Lotte, R.; Chirio, D.; Lonjon, M.; Roger, P.-M.; Drancourt, M.; Ruimy, R. Methanobrevibacter oralis detected along with Aggregatibacter actinomycetemcomitans in a series of community-acquired brain abscesses. Clin. Microbiol. Infect. 2018, 24, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Sogodogo, E.; Fellag, M.; Loukil, A.; Nkamga, V.D.; Michel, J.; Dessi, P.; Fournier, P.-E.; Drancourt, M. Nine Cases of Methanogenic Archaea in Refractory Sinusitis, an Emerging Clinical Entity. Front. Public Health 2019, 7, 38. [Google Scholar] [CrossRef]
- Nkamga, V.D.; Lotte, R.; Roger, P.-M.; Drancourt, M.; Ruimy, R. Methanobrevibacter smithii and Bacteroides thetaiotaomicron cultivated from a chronic paravertebral muscle abscess. Clin. Microbiol. Infect. 2016, 22, 1008–1009. [Google Scholar] [CrossRef]
- Dubourg, G.; Abat, C.; Rolain, J.-M.; Raoult, D. Correlation between Sputum and Bronchoalveolar Lavage Fluid Cultures. J. Clin. Microbiol. 2014, 53, 994–996. [Google Scholar] [CrossRef]
- La Scola, B.; Khelaifia, S.; Lagier, J.-C.; Raoult, D. Aerobic culture of anaerobic bacteria using antioxidants: A preliminary report. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1781–1783. [Google Scholar] [CrossRef]
- Nkamga, V.D.; Huynh, H.T.T.; Aboudharam, G.; Ruimy, R.; Drancourt, M. Diversity of Human-Associated Methanobrevibacter smithii Isolates Revealed by Multispacer Sequence Typing. Curr. Microbiol. 2015, 70, 810–815. [Google Scholar] [CrossRef]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High Prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae Detected in the Human Gut Using an Improved DNA Detection Protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef]
- Balch, W.E.; Wolfe, R.S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl. Environ. Microbiol. 1976, 32, 781–791. [Google Scholar] [CrossRef]
- Hungate, R.E.; Macy, J. Roll tube method for cultivation of strict anaerobes. Bulletins from the Ecological Research Committee-NFR (Statens Naturvetenskapliga Forskningsrad). Bull. Ecol. Res. Comm. 1972, 17, 123–126. [Google Scholar]
- Khelaifia, S.; Raoult, D.; Drancourt, M. A Versatile Medium for Cultivating Methanogenic Archaea. PLoS ONE 2013, 8, e61563. [Google Scholar] [CrossRef] [PubMed]
- Khelaifia, S.; Lagier, J.-C.; Nkamga, V.D.; Guilhot, E.; Drancourt, M.; Raoult, D. Aerobic culture of methanogenic archaea without an external source of hydrogen. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Raskin, L.; Stromley, J.M.; Rittmann, B.E.; Stahl, D.A. Group-Specific 16S Ribosomal-Rna Hybridization Probes to Describe Natural Communities of Methanogens. Appl. Environ. Microbiol. 1994, 60, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Luton, P.E.; Wayne, J.M.; Sharp, R.J.; Riley, P.W. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill b bThe GenBank accession numbers for the mcrA sequences reported in this paper are AF414034–AF414051 (see Fig. 2) and AF414007–AF414033 (environmental isolates in Fig. 3). Microbiology 2002, 148, 3521–3530. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, K.; Pausan, M.R.; Perras, A.K.; Beck, M.; Bang, C.; Mora, M.; Schilhabel, A.; Schmitz, R.; Moissl-Eichinger, C. First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 2017, 8, e00824-17. [Google Scholar] [CrossRef] [PubMed]
- Guindo, C.O.; Terrer, E.; Chabrière, E.; Aboudharam, G.; Drancourt, M.; Grine, G. Culture of salivary methanogens assisted by chemically produced hydrogen. Anaerobe 2020, 61, 102128. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Mihajlovski, A.; Alric, M.; Brugère, J.-F. A putative new order of methanogenic Archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Res. Microbiol. 2008, 159, 516–521. [Google Scholar] [CrossRef]
- Togo, A.H.; Grine, G.; Khelaifia, S.; Robert, C.D.; Brevaut, V.; Caputo, A.; Baptiste, E.; Bonnet, M.; Levasseur, A.; Drancourt, M.; et al. Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Drancourt, M.; Djemai, K.; Gouriet, F.; Grine, G.; Loukil, A.; Bedotto, M.; Levasseur, A.; Lepidi, H.; Bou-Khalil, J.; Khelaifia, S.; et al. Methanobrevibacter smithii Archaemia in Febrile Patients With Bacteremia, Including Those With Endocarditis. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Grine, G.; Royer, A.; Terrer, E.; Diallo, O.O.; Drancourt, M.; Aboudharam, G. Tobacco Smoking Affects the Salivary Gram-Positive Bacterial Population. Front. Public Health 2019, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Lyes, M.; Sladewski, K.; Enany, S.; McEachern, E.; Mathew, D.P.; Das, S.; Moshensky, A.; Bapat, S.; Pride, D.T.; et al. Electronic cigarette inhalation alters innate immunity and airway cytokines while increasing the virulence of colonizing bacteria. J. Mol. Med. 2016, 94, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Bozier, J.; Chivers, E.K.; Chapman, D.G.; Larcombe, A.N.; Bastian, N.A.; Masso-Silva, J.A.; Byun, M.K.; McDonald, C.F.; Alexander, L.E.C.; Ween, M.P. The Evolving Landscape of e-Cigarettes. Chest 2020, 157, 1362–1390. [Google Scholar] [CrossRef]
- Hswen, Y.; Brownstein, J.S. Real-Time Digital Surveillance of Vaping-Induced Pulmonary Disease. N. Engl. J. Med. 2019, 381, 1778–1780. [Google Scholar] [CrossRef]

| Nature of Sample | Sputum | Bronchoalveolar Lavage | Bronchial Aspirates |
|---|---|---|---|
| Number of analyzed samples | 527 | 188 | 193 |
| Archaea 16S rRNA PCR | 21/527 (3.9%) | 2/188 (1.06%) | 0 |
| Sequencing | M. oralis (n = 19); M. smithii (n = 2) | M. oralis (n = 1); M. smithii (n = 1) | 0 |
| RT PCR (M. smithii) | n = 2 (with load of 1.32 × 104 ± 0.56 × 104/mL) | n = 1 (with load of 0.8 × 104/mL) | 0 |
| RT PCR (M. oralis) | n = 19 (with load of 2.61 × 105 ± 1.23 × 105/mL) | n = 1 (with load of 1.41 × 104/mL) | 0 |
| Methanogen culture | 0 | 0 | 0 |
| FISH | n = 3 | n = 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassani, Y.; Brégeon, F.; Aboudharam, G.; Drancourt, M.; Grine, G. Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota. Microorganisms 2020, 8, 1866. https://doi.org/10.3390/microorganisms8121866
Hassani Y, Brégeon F, Aboudharam G, Drancourt M, Grine G. Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota. Microorganisms. 2020; 8(12):1866. https://doi.org/10.3390/microorganisms8121866
Chicago/Turabian StyleHassani, Yasmine, Fabienne Brégeon, Gérard Aboudharam, Michel Drancourt, and Ghiles Grine. 2020. "Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota" Microorganisms 8, no. 12: 1866. https://doi.org/10.3390/microorganisms8121866
APA StyleHassani, Y., Brégeon, F., Aboudharam, G., Drancourt, M., & Grine, G. (2020). Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota. Microorganisms, 8(12), 1866. https://doi.org/10.3390/microorganisms8121866




