Correlation between Human Papillomavirus Codetection Profiles and Cervical Intraepithelial Neoplasia in Japanese Women
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Ethical Approval
2.3. HPV Genotyping
2.4. Statistical Analysis
3. Results
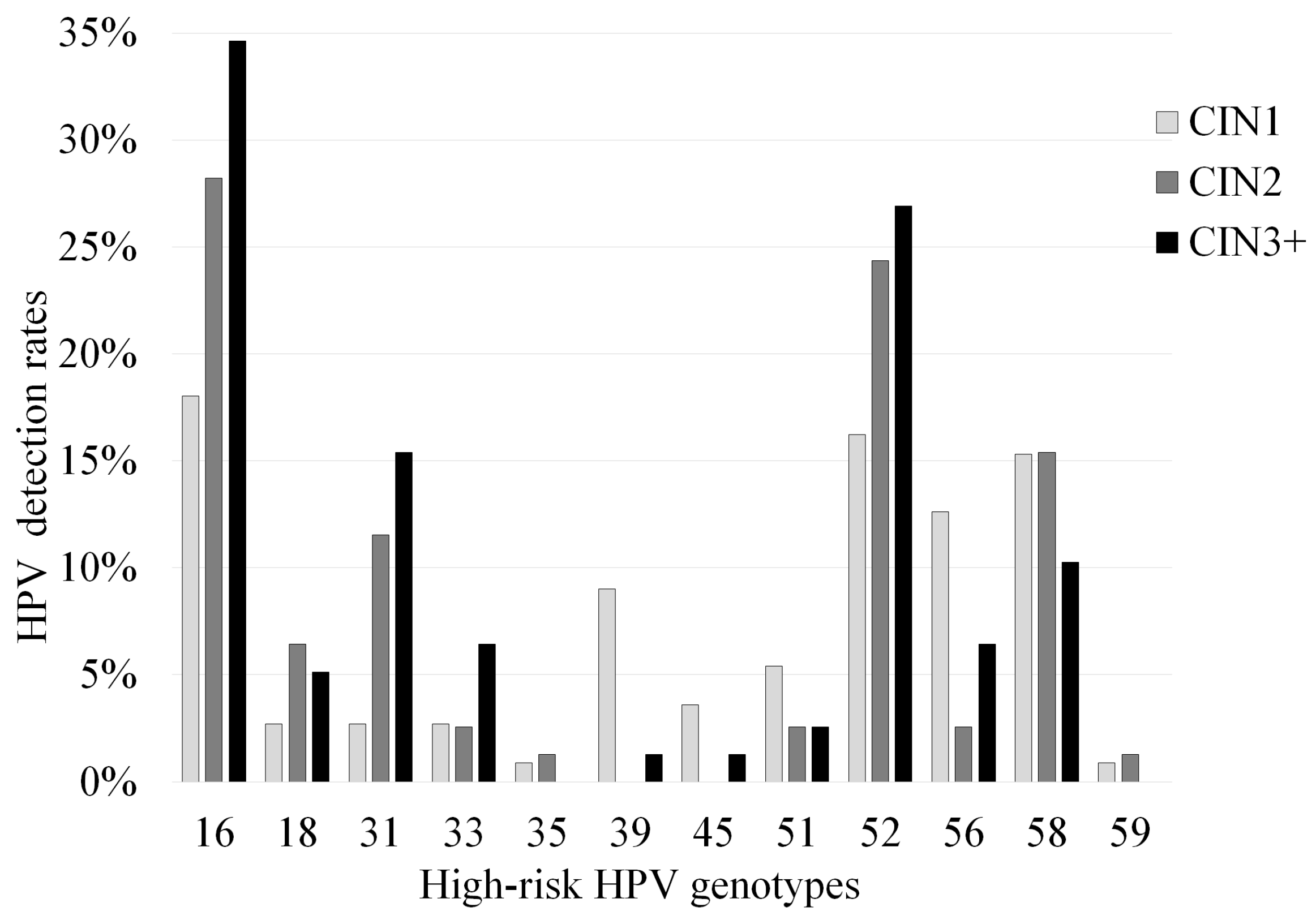
3.1. Correlation between Pathological Findings and HPV Detection Rates
3.2. Relationships among Detected HPV Genotypes, Codetection Rate, and CIN Grade
4. Discussions
Author Contributions
Funding
Conflicts of Interest
References
- Walboomers, J.M.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.F.; Peto, J.; Meijer, C.J.L.M.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Dunne, E.F.; Markowitz, L.E. Genital human papillomavirus infection. Clin. Infects Dis. 2006, 43, 624–629. [Google Scholar] [CrossRef]
- Cuschieri, K.S.; Cubie, H.A.; Whitley, M.W.; Gilkison, G.; Arends, M.J.; Graham, C.; McGoogan, E. Persistent high risk HPV infection associated with development of cervical neoplasia in a prospective population study. J. Clin. Pathol. 2005, 58, 946–950. [Google Scholar] [CrossRef]
- Brummer, O.; Hollwitz, B.; Böhmer, G.; Kühnle, H.; Petry, K.U. Human papillomavirus-type persistence patterns predict the clinical outcome of cervical intraepithelial neoplasia. Gynecol. Oncol. 2006, 102, 517–522. [Google Scholar] [CrossRef]
- Nobbenhuis, M.A.; Walboomers, J.M.; Helmerhorst, T.J.; Rozendaal, L.; Remmink, A.J.; Risse, E.K.; van der Linden, H.C.; Voorhorst, F.J.; Kenemans, P.; Meijer, C.J. Relation of human papilloma virus status to cervical lesions and consequences for cervical-cancer screening: A prospective study. Lancet 1991, 354, 20–25. [Google Scholar] [CrossRef]
- Bosch, F.X.; Lorincz, A.; Muñoz, N.; Meijer, C.J.L.M.; Shah, K.V. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 2002, 55, 244–265. [Google Scholar] [CrossRef]
- Pinto, A.P.; Crum, C.P. Natural history of cervical neoplasia: Defining progression and its consequence. Clin. Obstet. Gynecol. 2000, 43, 352–362. [Google Scholar] [CrossRef]
- Munoz, N.; Castellsagué, X.; de González, A.B.; Gissmann, L. HPV in the etiology of human cancer. Vaccine 2006, 24, S1–S10. [Google Scholar] [CrossRef]
- Castellsagué, X.; Díaz, M.; de Sanjosé, S.; Muñoz, N.; Herrero, R.; Franceschi, S.; Peeling, R.W.; Ashley, R.; Smith, J.S.; Snijders, P.J.F.; et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: Implications for screening and prevention. J. Natl. Cancer Inst. 2006, 98, 303–315. [Google Scholar] [CrossRef]
- Jacobs, M.V.; Snijders, P.J.; van den Brule, A.J.; Helmerhorst, T.J.; Meijer, C.J.; Walboomers, J.M. A general primer GP5+/GP6 (+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 1997, 35, 791–795. [Google Scholar] [CrossRef]
- An, H.J.; Cho, N.H.; Lee, S.Y.; Kim, I.H.; Lee, C.; Kim, S.J.; Mun, M.S.; Kim, S.H.; Jeong, J.K. Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with the HPV DNA chip microarray method. Cancer 2004, 97, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.D.; Jung, W.W.; Nam, J.H.; Choi, H.S.; Park, C.S. Detection of HPV genotypes in cervical lesions by the HPV DNA Chip and sequencing. Gynecol. Oncol. 2005, 98, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.Y.; Kim, Y.W.; Bae, S.M.; Kwon, E.H.; Chaturvedi, P.K.; Battogtokh, G.; Ahn, W.S. Development of a bead-based multiplex genotyping method for diagnostic characterization of HPV infection. PLoS ONE 2012, 7, e32259. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, Z.; Wang, Q.; Si, T.; Pei, S.; Wang, C.; Qu, H.; Zhong, J.; Ma, Y.; Nie, C.; et al. Human papillomavirus genotypes associated with cervical precancerous lesions and cancer in the highest area of cervical cancer mortality, Longnan, China. Infect. Agent Cancer 2017, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Onuki, M.; Matsumoto, K.; Satoh, T.; Oki, A.; Okada, S.; Minaguchi, T.; Ochi, H.; Nakao, S.; Someya, K.; Yamada, N.; et al. Human papillomavirus infections among Japanese women: Age-related prevalence and type-specific risk for cervical cancer. Cancer Sci. 2009, 100, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.X.; Zhou, J.S.; Yuan, S.H.; Yu, H.; Lou, H.M. Distribution of HPV genotype in invasive cervical carcinoma and cervical intraepithelial neoplasia in Zhejiang province, southeast China: Establishing the baseline for surveillance. Int. J. Environ. Res. Public Health 2015, 12, 10794–10805. [Google Scholar] [CrossRef]
- Geraets, D.; Alemany, L.; Guimera, N.; de Sanjose, S.; de Koning, M.; Molijn, A.; Jenkins, D.; Bosch, X.; Quint, W. Detection of rare and possibly carcinogenic human papillomavirus genotypes as single infections in invasive cervical cancer. J. Pathol. 2012, 228, 534–543. [Google Scholar] [CrossRef]
- Naryshkin, S.; Austin, R.M. Limitations of widely used high-risk human papillomavirus laboratory-developed testing in cervical cancer screening. Drug Healthc. Patient Saf. 2012, 4, 167. [Google Scholar] [CrossRef]
- Eltoum, I.A.; Chhieng, D.C.; Crowe, D.R.; Roberson, J.; Jin, G.; Broker, T.R. Significance and possible causes of false-negative results of reflex human papillomavirus infection testing. Cancer Cytopathol. 2007, 111, 154–159. [Google Scholar] [CrossRef]
- Okodo, M.; Okayama, K.; Teruya, K.; Sasagawa, T. Uniplex E6/E7 PCR method detecting E6 or E7 genes in 39 human papillomavirus types. J. Med. Virol. 2018, 90, 981–988. [Google Scholar] [CrossRef]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological Agents. Volume 100 B. A Review of Human Carcinogens; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Lyon, France, 2012; Available online: https://monographs.iarc.fr/wp-content/uploads/2018/06/mono100B.pdf (accessed on 10 May 2019).
- Greer, C.E.; Peterson, S.L.; Kiviat, N.B.; Manos, M.M. PCR amplification from paraffin-embedded tissues: Effects of fixative and fixation time. Am. J. Clin. Pathol. 1991, 95, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Wright, T.C.; Stoler, M.H.; Behrens, C.M.; Sharma, A.; Zhang, G.; Wright, T.L. Primary cervical cancer screening with human papillomavirus: End of study results from the ATHENA study using HPV as the first-line screening test. Gynecol. Oncol. 2015, 136, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.F.; Peng, Z.; Clark, K.M.; Adamson, C.S.; Ma, X.J.; Wu, X.; Wang, H.; Luo, Y.; Cooper, K. HPV E6/E7 RNA in situ hybridization signal patterns as biomarkers of three-tier cervical intraepithelial neoplasia grade. PLoS ONE 2014, 9, e91142. [Google Scholar] [CrossRef]
- Coutlee, F.; Ratnam, S.; Ramanakumar, A.V.; Insinga, R.R.; Bentley, J.; Escott, N.; Ghatage, P.; Koushik, A.; Ferenczy, A.; Franco, E.L. Distribution of human papillomavirus genotypes in cervical intraepithelial neoplasia and invasive cervical cancer in Canada. J. Med. Virol. 2011, 83, 1034–1041. [Google Scholar] [CrossRef]
- Xiao, M.; Xu, Q.; Li, H.; Gao, H.; Bie, Y.; Zhang, Z. Prevalence of human papillomavirus genotypes among women with high-grade cervical lesions in Beijing, China. Medicine 2016, 95, e2555. [Google Scholar] [CrossRef]
- Hou, R.; Xu, C.; Zhang, S.; Wu, M.; Zhang, W. Distribution of human papillomavirus genotype and cervical neoplasia among women with abnormal cytology in Beijing, China. Int. J. Gynaecol. Obstet. 2012, 119, 257–261. [Google Scholar] [CrossRef]
- Li, X.; Wan, X.; Zheng, F.; Zhu, H.; Zhu, X.; Yu, J. Comparison of human papillomavirus genotype distributions in cervical intraepithelial neoplasia and cervical cancer. Biomed. Res. 2017, 28, 2284–2289. [Google Scholar]
- De Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Bernard, E.; Pons-Salort, M.; Favre, M.; Heard, I.; Delarocque-Astagneau, E.; Guillemot, D.; Thiébaut, A.C.M. Comparing human papillomavirus prevalences in women with normal cytology or invasive cervical cancer to rank genotypes according to their oncogenic potential: A meta-analysis of observational studies. BMC Infect. Dis. 2013, 13, 373. [Google Scholar] [CrossRef]
- Jacobs, M.V.; Walboomers, J.M.; Snijders, P.J.; Voorhorst, F.J.; Verheijen, R.H.; Fransen-Daalmeijer, N.; Meijer, C.J. Distribution of 37 mucosotropic HPV types in women with cytologically normal cervical smears: The age-related patterns for high-risk and low-risk types. Int. J. Cancer. 2000, 87, 221–227. [Google Scholar] [CrossRef]
- Kurokawa, T.; Yoshida, Y.; Iwanari, O.; Oishi, T.; Kasai, T.; Hamada, M.; Fujita, H.; Fujiwara, H.; Yokoyama, M.; Sakuragi, N.; et al. Implementation of primary HPV testing in Japan. Mol. Clin. Oncol. 2020, 13, 22. [Google Scholar] [CrossRef] [PubMed]

| Single HPV Genotype Detected | Multiple HPV Genotypes Detected | ||||||
|---|---|---|---|---|---|---|---|
| High-Risk | Other | Total | High-Risk | High-Risk + Other | Other | Total | |
| Genotypes | Genotypes | Genotypes | Genotypes | Genotypes | |||
| CIN1 | 47.70% | 18.00% | 65.80% | 3.60% | 27.00% | 3.60% | 34.20% |
| N = 111 | (53/111) | (20/111) | (73/111) | (4/111) | (30/111) | (4/111) | (38/111) |
| CIN2 | 66.70% | 6.40% | 73.10% | 5.10% | 17.90% | 3.80% | 26.90% |
| N = 78 | (52/78) | (5/78) | (57/78) | (4/78) | (14/78) | (3/78) | (21/78) |
| CIN3 | 64.10% | 7.70% | 71.80% | 6.40% | 20.50% | 1.30% | 28.20% |
| N = 78 | (50/78) | (6/78) | (56/78) | (5/78) | (16/78) | (1/78) | (22/78) |
| Total | 58.10% | 11.60% | 69.70% | 4.90% | 22.50% | 3.00% | 30.30% |
| N = 267 | (155/267) | (31/267) | (186/267) | (13/267) | (60/267) | (8/267) | (81/267) |
| High-Risk Genotype | Single Detection | Multiple Detection (High-Risk Genotypes Alone) | Multiple Detection (High-Risk and Other Genotypes) | Total |
|---|---|---|---|---|
| 16 | 78.3% (54/69) | 5.8% (4/69) | 15.9% (11/69) | 26.8% (69/257) |
| 18 | 75.0% (9/12) | 16.7% (2/12) | 8.3% (1/12) | 4.7% (12/257) |
| 31 | 58.3% (14/24) | 16.7% (4/24) | 25.0% (6/24) | 9.3% (24/257) |
| 33 | 50.0% (5/10) | 0.0% (0/10) | 50.0% (5/10) | 3.9% (10/257) |
| 35 | 0.0% (0/2) | 0.0% (0/2) | 100% (2/2) | 0.8% (2/257) |
| 39 | 45.5% (5/11) | 9.1% (1/11) | 45.5% (5/11) | 4.3% (11/257) |
| 45 | 75.0% (3/4) | 0.0% (0/4) | 25.0% (1/4) | 1.6% (4/257) |
| 51 | 22.2% (2/9) | 0.0% (0/9) | 77.8% (7/9) | 3.5% (9/257) |
| 52 | 51.8% (29/56) | 14.3% (8/56) | 33.9% (19/56) | 21.8% (56/257) |
| 56 | 38.1% (8/21) | 14.3% (3/21) | 47.6% (10/21) | 8.2% (21/257) |
| 58 | 70.3% (26/37) | 10.8% (4/37) | 18.9% (7/37) | 14.4% (37/257) |
| 59 | 0.0% (0/2) | 50.0% (1/2) | 50.0% (1/2) | 0.8% (2/257) |
| Total | 60.3% (155/257) | 10.5% (27/257) | 29.2% (75/257) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okayama, K.; Kimura, H.; Teruya, K.; Ishii, Y.; Fujita, K.; Fujii, M.; Oda, M.; Sasagawa, T.; Okodo, M. Correlation between Human Papillomavirus Codetection Profiles and Cervical Intraepithelial Neoplasia in Japanese Women. Microorganisms 2020, 8, 1863. https://doi.org/10.3390/microorganisms8121863
Okayama K, Kimura H, Teruya K, Ishii Y, Fujita K, Fujii M, Oda M, Sasagawa T, Okodo M. Correlation between Human Papillomavirus Codetection Profiles and Cervical Intraepithelial Neoplasia in Japanese Women. Microorganisms. 2020; 8(12):1863. https://doi.org/10.3390/microorganisms8121863
Chicago/Turabian StyleOkayama, Kaori, Hirokazu Kimura, Koji Teruya, Yasuyoshi Ishii, Kiyotaka Fujita, Masahiko Fujii, Mizue Oda, Toshiyuki Sasagawa, and Mitsuaki Okodo. 2020. "Correlation between Human Papillomavirus Codetection Profiles and Cervical Intraepithelial Neoplasia in Japanese Women" Microorganisms 8, no. 12: 1863. https://doi.org/10.3390/microorganisms8121863
APA StyleOkayama, K., Kimura, H., Teruya, K., Ishii, Y., Fujita, K., Fujii, M., Oda, M., Sasagawa, T., & Okodo, M. (2020). Correlation between Human Papillomavirus Codetection Profiles and Cervical Intraepithelial Neoplasia in Japanese Women. Microorganisms, 8(12), 1863. https://doi.org/10.3390/microorganisms8121863






