Accelerated Electro-Fermentation of Acetoin in Escherichia coli by Identifying Physiological Limitations of the Electron Transfer Kinetics and the Central Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector and Strain Construction
2.2. Cell Cultivation
2.3. Adaptive Laboratory Evolution (ALE)
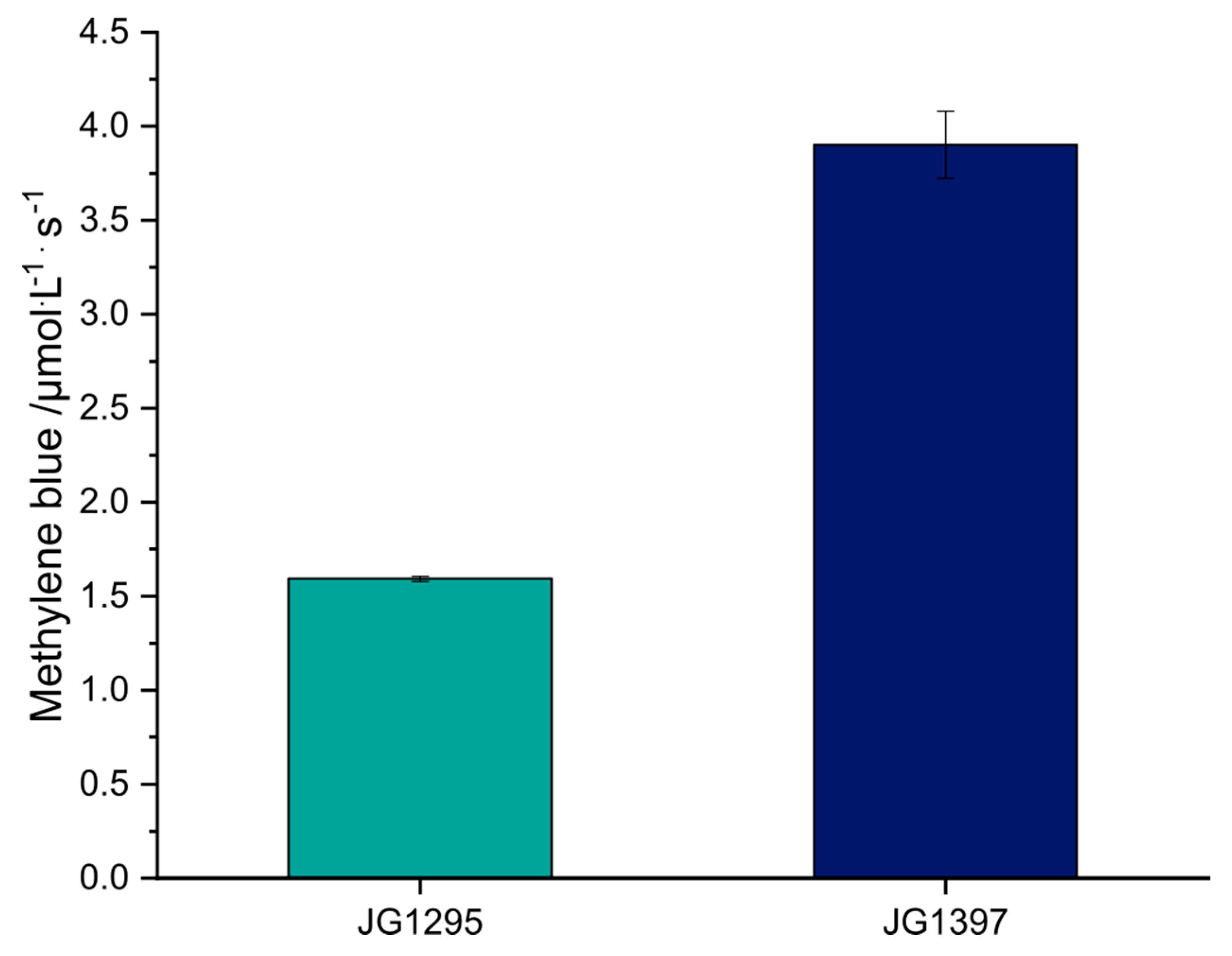
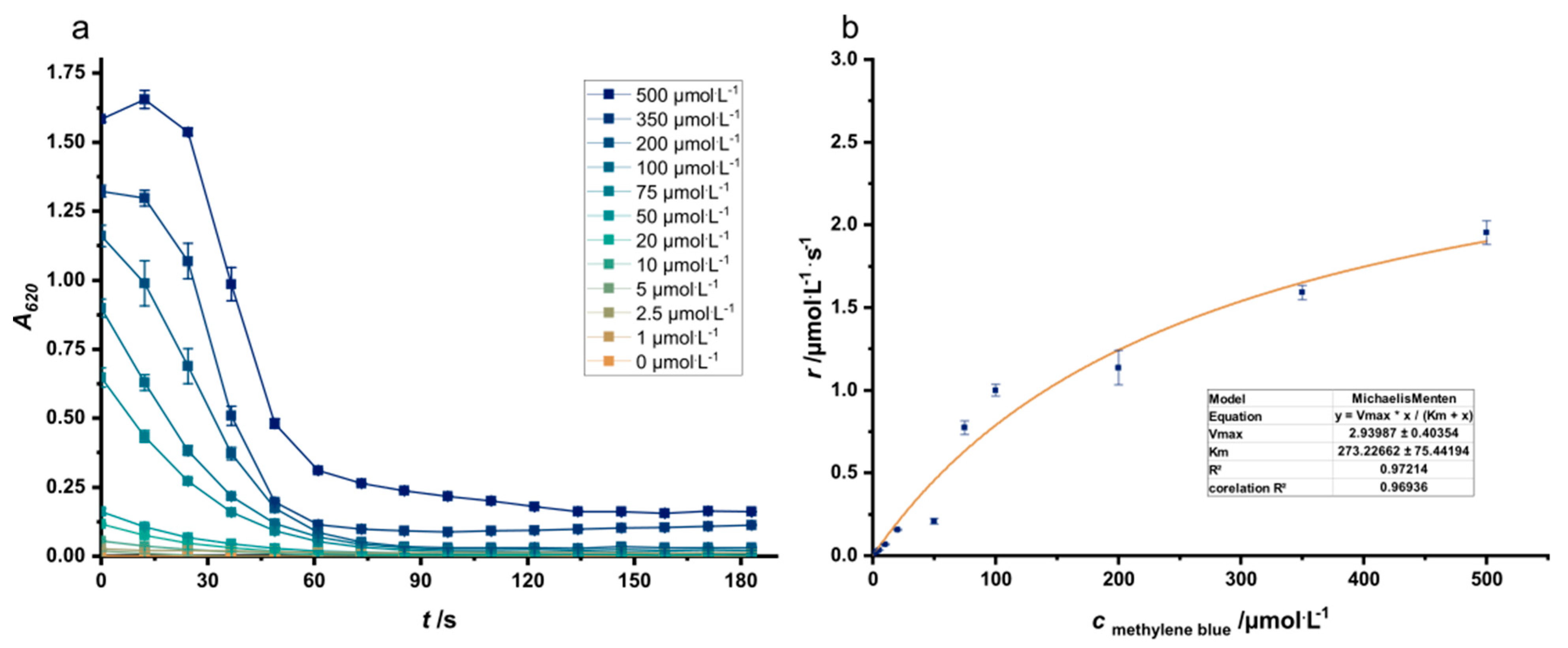
2.4. Methylene Blue Reduction Assay
2.5. Preparation of Nucleic Acids
2.6. Sequencing
2.7. Bioinformatic Analysis
2.8. Electro-Fermentation
2.9. Analytical Methods
2.10. Calculation of Evaluation Parameters
3. Results and Discussion
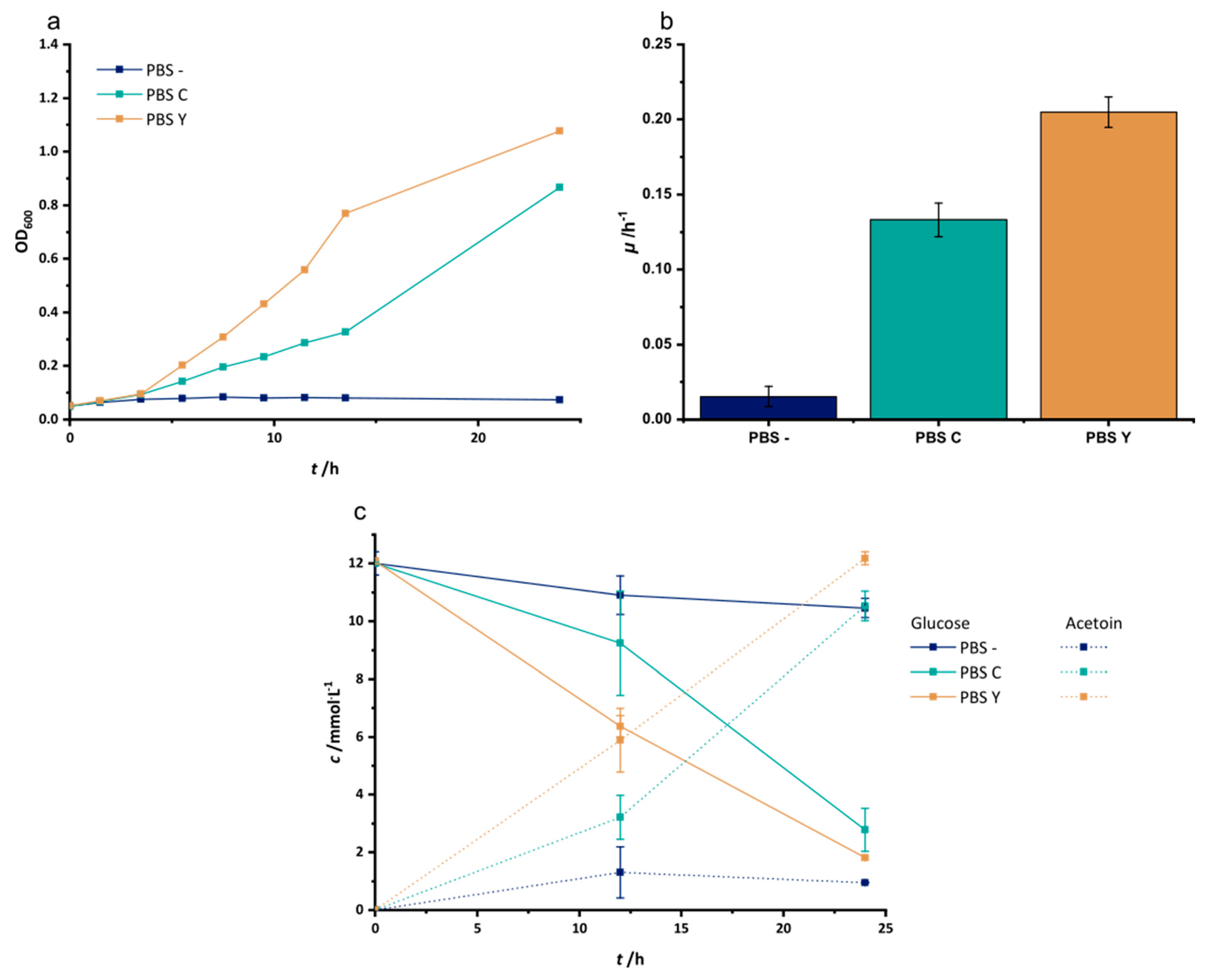
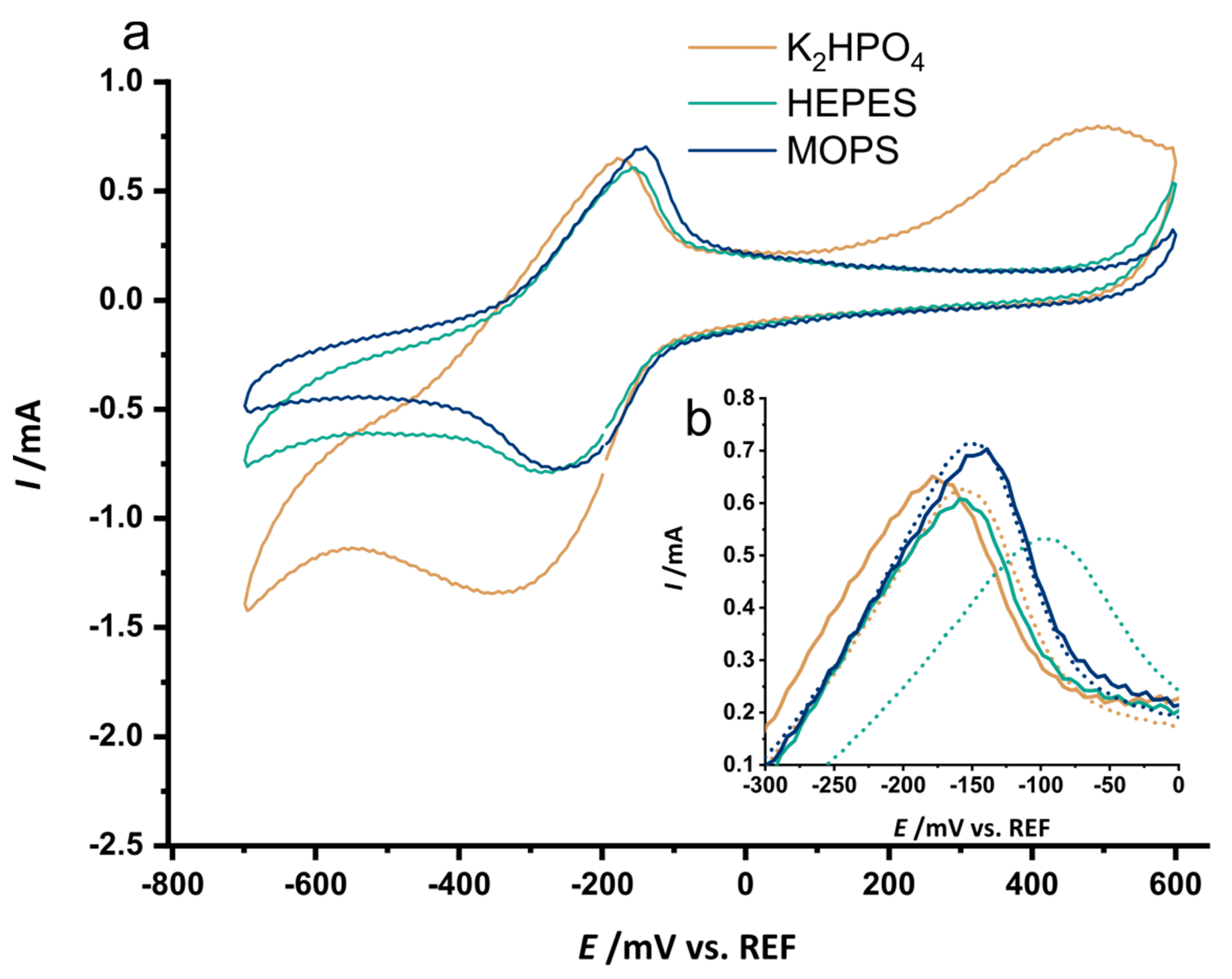
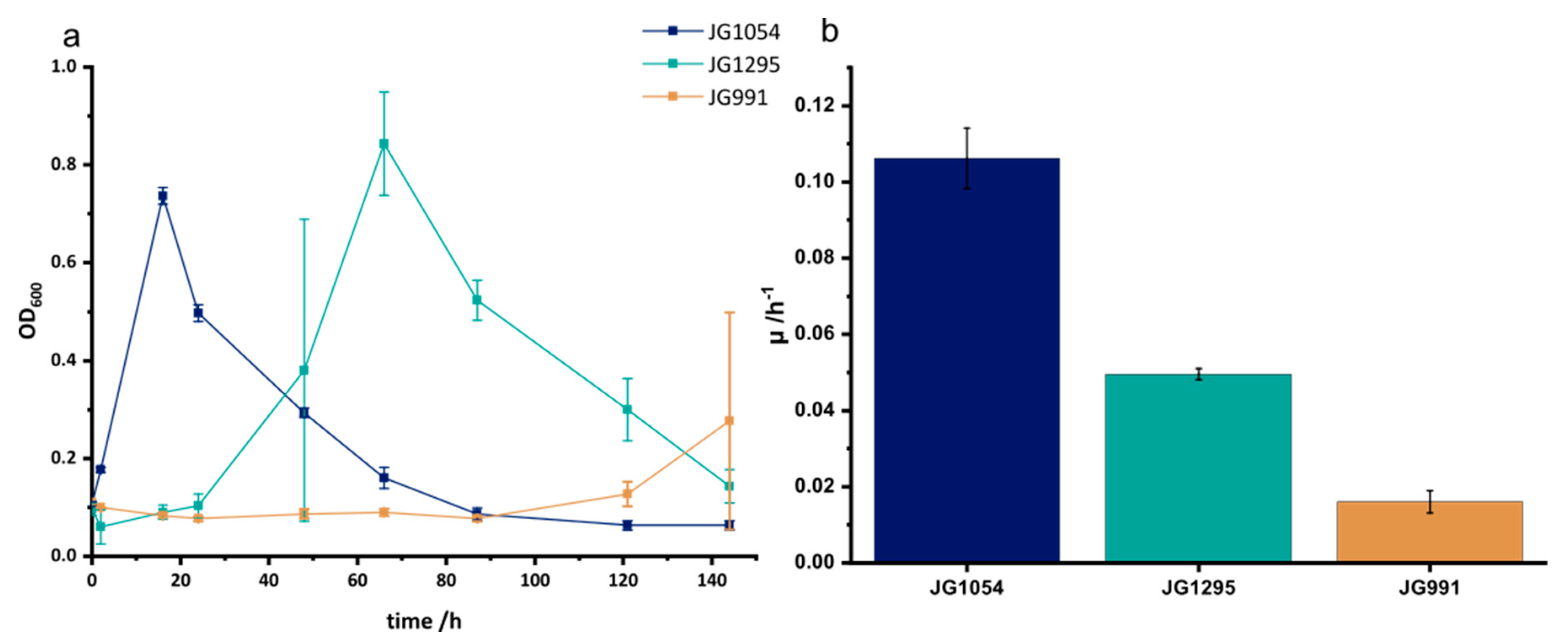
3.1. Adaptation to Improved Anoxic Respiration
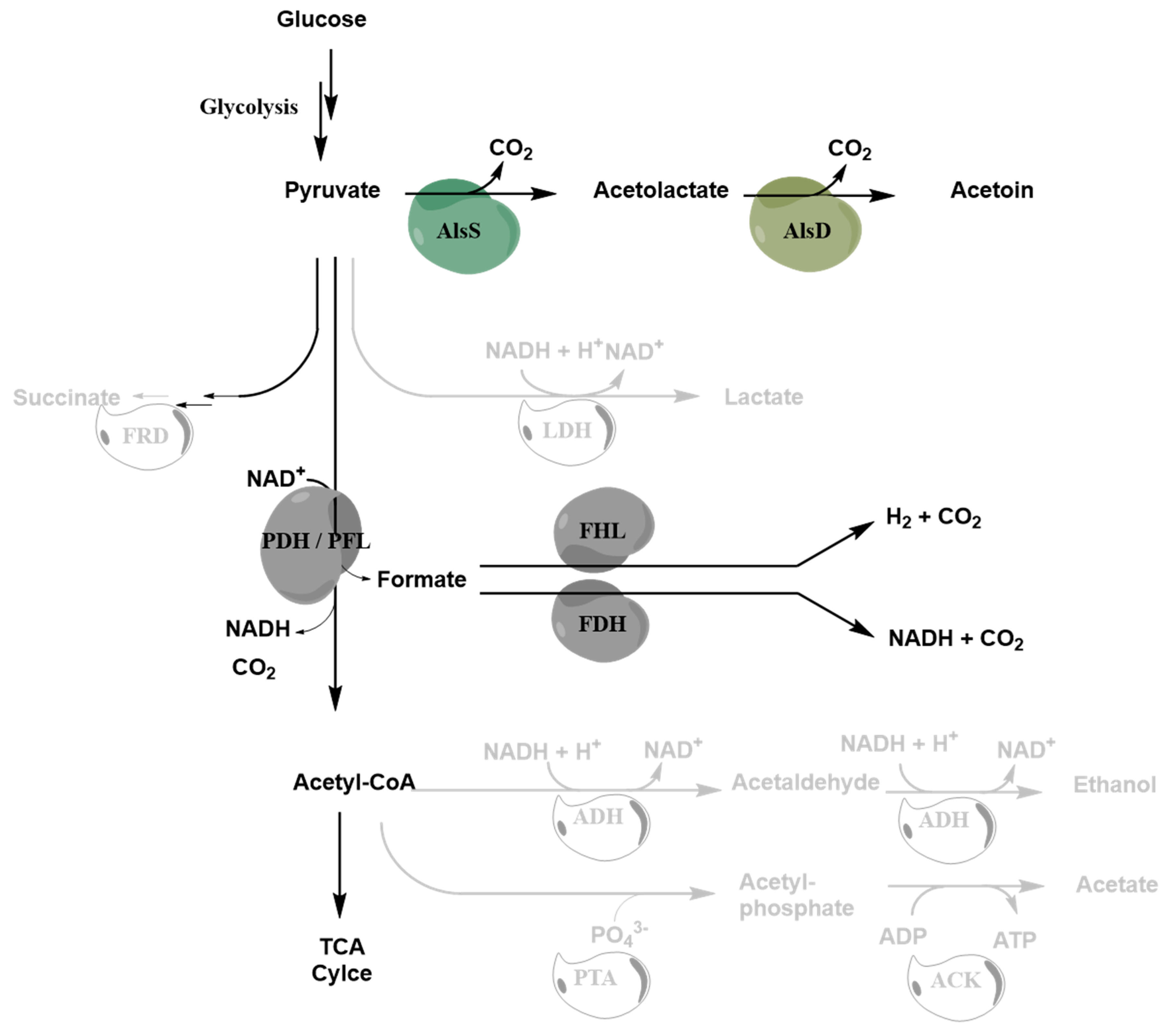
3.2. The Adaptation to Growth on DMSO Is Based on Three Point Mutations
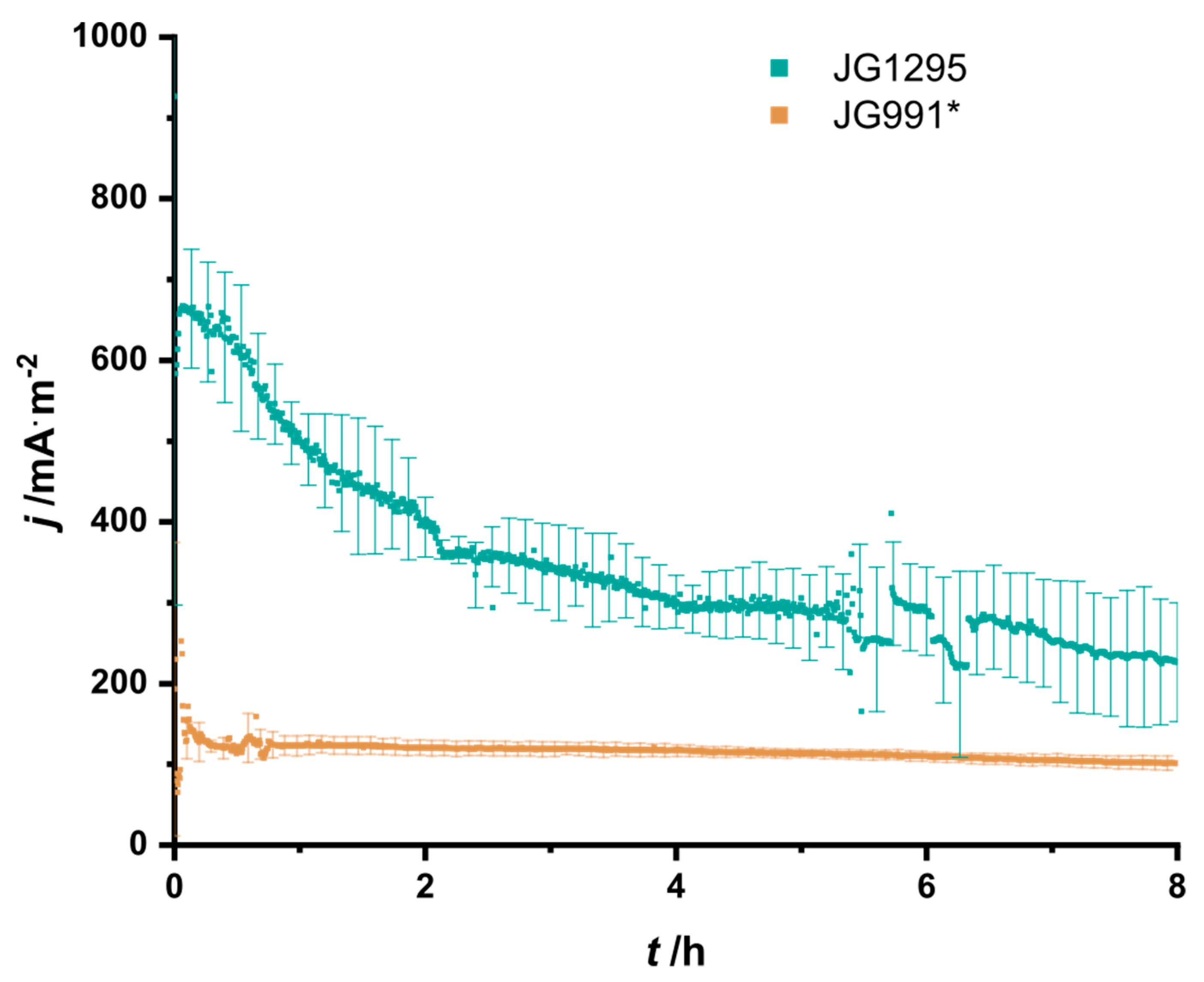
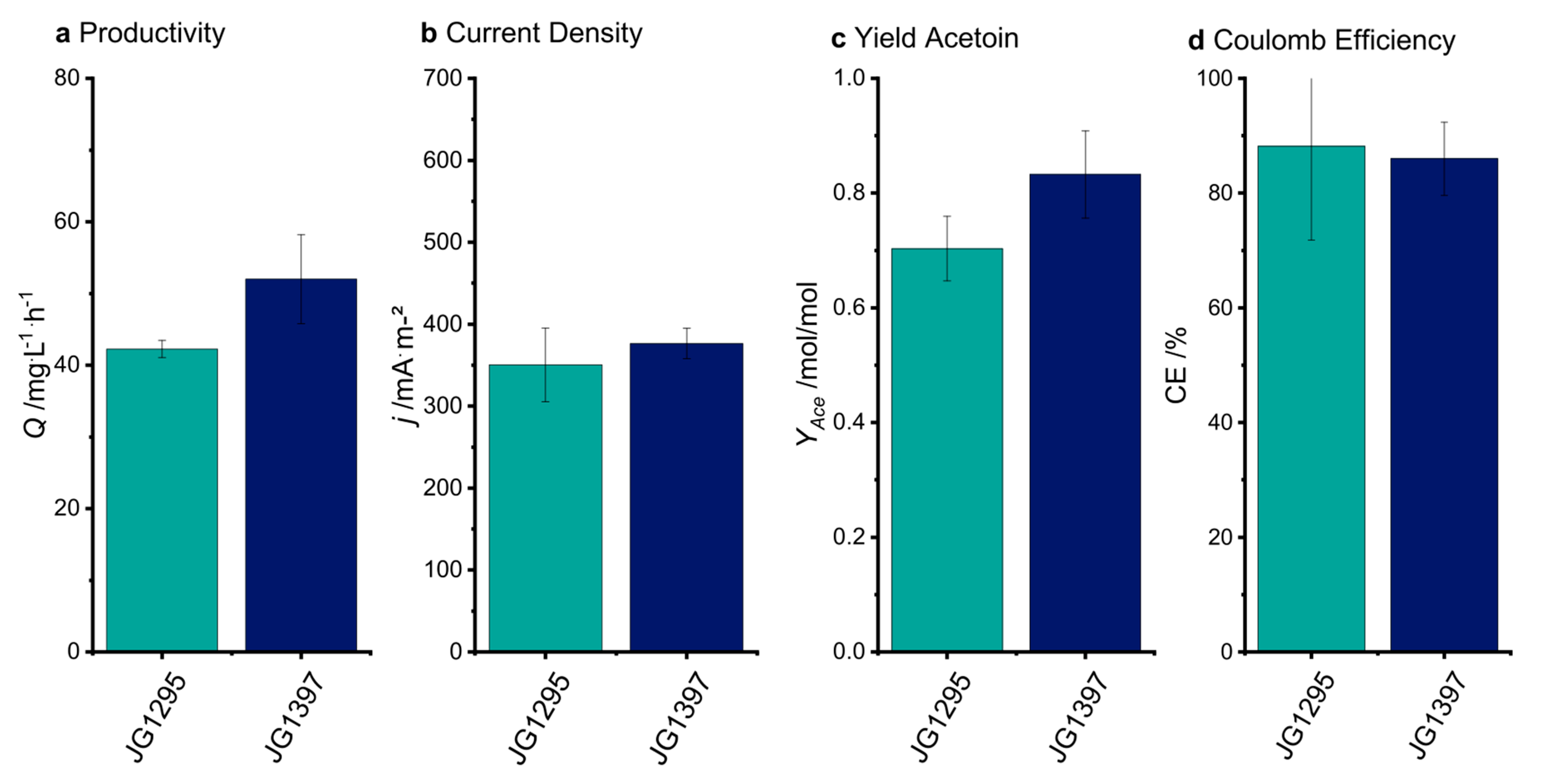
3.3. The Selected Mutations Increase Productivity and Current Production in Bioelectrochemical Systems
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Causey, T.B.; Zhou, S.; Shanmugam, K.T.; Ingram, L.O. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: Homoacetate production. Proc. Natl. Acad. Sci. USA 2003, 100, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; May, H.D.; Lu, L.; Liang, P.; Huang, X.; Ren, Z.J. Carbon dioxide and organic waste valorization by microbial electrosynthesis and electro-fermentation. Water Res. 2019, 149, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Bhagchandanii, D.D.; Babu, R.P.; Sonawane, J.M.; Khanna, N.; Pandit, S.; Jadhav, D.A.; Khilari, S.; Prasad, R. A Comprehensive Understanding of Electro-Fermentation. Fermentation 2020, 6, 92. [Google Scholar] [CrossRef]
- Philipp, L.-A.; Edel, M.; Johnston, C. Genetic engineering for enhanced productivity in bioelectrochemical systems. Adv. Virus Res. 2020, 111, 1–31. [Google Scholar] [CrossRef]
- Korth, B.; Harnisch, F. Spotlight on the Energy Harvest of Electroactive Microorganisms: The Impact of the Applied Anode Potential. Front. Microbiol. 2019, 10, 1352. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Fan, Y.; Liu, H. Hydrogen production in single-chamber tubular microbial electrolysis cells using non-precious-metal catalysts. Int. J. Hydrogen Energy 2009, 34, 8535–8542. [Google Scholar] [CrossRef]
- Cusick, R.D.; Bryan, B.; Parker, D.S.; Merrill, M.D.; Mehanna, M.; Kiely, P.D.; Liu, G.; Logan, B.E. Performance of a pilot-scale continuous flow microbial electrolysis cell fed winery wastewater. Appl. Microbiol. Biotechnol. 2011, 89, 2053–2063. [Google Scholar] [CrossRef]
- Edel, M.; Horn, H.; Johnston, C. Biofilm systems as tools in biotechnological production. Appl. Microbiol. Biotechnol. 2019, 103, 5095–5103. [Google Scholar] [CrossRef]
- McMillan, D.G.G.; Marritt, S.J.; Butt, J.N.; Jeuken, L.J.C. Menaquinone-7 Is Specific Cofactor in Tetraheme Quinol Dehydrogenase CymA. J. Biol. Chem. 2012, 287, 14215–14225. [Google Scholar] [CrossRef]
- Fonseca, B.M.; Paquete, C.M.; Neto, S.E.; Pacheco, I.; Soares, C.M.; Louro, R.O. Mind the gap: Cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem. J. 2012, 449, 101–108. [Google Scholar] [CrossRef]
- Sturm, G.; Richter, K.; Doetsch, A.; Heide, H.; Louro, R.O.; Johnston, C. A dynamic periplasmic electron transfer network enables respiratory flexibility beyond a thermodynamic regulatory regime. ISME J. 2015, 9, 1802–1811. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, B.; Wang, Z.; Elias, D.A.; Mayer, M.U.; Gorby, Y.A.; Ni, S.; Lower, B.H.; Kennedy, D.W.; Wunschel, D.S.; et al. Isolation of a High-Affinity Functional Protein Complex between OmcA and MtrC: Two Outer Membrane Decaheme c-Type Cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 2006, 188, 4705–4714. [Google Scholar] [CrossRef] [PubMed]
- Beblawy, S.; Bursac, T.; Paquete, C.; Louro, R.; Clarke, T.A.; Johnston, C. Extracellular reduction of solid electron acceptors by Shewanella oneidensis. Mol. Microbiol. 2018, 109, 571–583. [Google Scholar] [CrossRef]
- Costa, N.L.; Clarke, T.A.; Philipp, L.-A.; Gescher, J.; Louro, R.O.; Paquete, C.M. Electron transfer process in microbial electrochemical technologies: The role of cell-surface exposed conductive proteins. Bioresour. Technol. 2018, 255, 308–317. [Google Scholar] [CrossRef]
- Yu, S.; Lai, B.; Plan, M.R.; Hodson, M.P.; Lestari, E.A.; Song, H.; Krömer, J.O. Improved performance of Pseudomonas putida in a bioelectrochemical system through overexpression of periplasmic glucose dehydrogenase. Biotechnol. Bioeng. 2018, 115, 145–155. [Google Scholar] [CrossRef]
- Lai, B.; Yu, S.; Bernhardt, P.V.; Rabaey, K.; Virdis, B.; Krömer, J.O. Anoxic metabolism and biochemical production in Pseudomonas putida F1 driven by a bioelectrochemical system. Biotechnol. Biofuels 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Vassilev, I.; Gießelmann, G.; Schwechheimer, S.K.; Wittmann, C.; Virdis, B.; Krömer, J.O. Anodic electro-fermentation: Anaerobic production of L-Lysine by recombinant Corynebacterium glutamicum. Biotechnol. Bioeng. 2018, 115, 1499–1508. [Google Scholar] [CrossRef]
- Gescher, J.S.; Cordova, C.D.; Spormann, A.M. Dissimilatory iron reduction in Escherichia coli: Identification of CymA of Shewanella oneidensis and NapC of E. coli as ferric reductases. Mol. Microbiol. 2008, 68, 706–719. [Google Scholar] [CrossRef]
- Jensen, H.M.; Albers, A.E.; Malley, K.R.; Londer, Y.Y.; Cohen, B.E.; Helms, B.A.; Weigele, P.; Groves, J.T.; Ajo-Franklin, C.M. Engineering of a synthetic electron conduit in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 19213–19218. [Google Scholar] [CrossRef]
- Sturm-Richter, K.; Golitsch, F.; Sturm, G.; Kipf, E.; Dittrich, A.; Beblawy, S.; Kerzenmacher, S.; Johnston, C. Unbalanced fermentation of glycerol in Escherichia coli via heterologous production of an electron transport chain and electrode interaction in microbial electrochemical cells. Bioresour. Technol. 2015, 186, 89–96. [Google Scholar] [CrossRef]
- TerAvest, M.A.; Zajdel, T.J.; Ajo-Franklin, C.M. The Mtr Pathway of Shewanella oneidensis MR-1 Couples Substrate Utilization to Current Production in Escherichia coli. ChemElectroChem 2014, 1, 1874–1879. [Google Scholar] [CrossRef]
- Förster, A.H.; Beblawy, S.; Golitsch, F.; Johnston, C. Electrode-assisted acetoin production in a metabolically engineered Escherichia coli strain. Biotechnol. Biofuels 2017, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Lutz, R. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997, 25, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Arslan, E.; Schulz, H.; Zufferey, R.; Künzler, P.; Thöny-Meyer, L. Overproduction of the Bradyrhizobium japonicum c-Type Cytochrome Subunits of thecbb3Oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 1998, 251, 744–747. [Google Scholar] [CrossRef]
- Tuite, E.M.; Kelly, J.M. New trends in photobiology. Photochemical Interactions of Methylene Blue and Analogues with DNA and Other Biological Substrates. J. Photochem. Photobiol. B Biol. 1993, 21, 103–124. [Google Scholar] [CrossRef]
- Usacheva, M.N.; Ba, M.C.T.; Biel, M.A. Comparison of the methylene blue and toluidine blue photobactericidal efficacy against gram-positive and gram-negative microorganisms. Lasers Surg. Med. 2001, 29, 165–173. [Google Scholar] [CrossRef]
- Kim, Y.; Ingram, L.O.; Shanmugam, K.T. Construction of an Escherichia coli K-12 Mutant for Homoethanologenic Fermentation of Glucose or Xylose without Foreign Genes. Appl. Environ. Microbiol. 2007, 73, 1766–1771. [Google Scholar] [CrossRef]
- Bleile, D.M.; Munk, P.; Oliver, R.M.; Reed, L.J. Subunit structure of dihydrolipoyl transacetylase component of pyruvate dehydrogenase complex from Escherichia coli. Proc. Natl. Acad. Sci. USA 1979, 76, 4385–4389. [Google Scholar] [CrossRef]
- Stephens, P.E.; Darlison, M.G.; Lewis, H.M.; Guest, J.R. The Pyruvate Dehydrogenase Complex of Escherichia coli K12. Nucleotide Sequence Encoding the Dihydrolipoamide Acetyltransferase Component. JBIC J. Biol. Inorg. Chem. 1983, 133, 481–489. [Google Scholar] [CrossRef]
- Kim, Y.; Ingram, L.O.; Shanmugam, K.T. Dihydrolipoamide Dehydrogenase Mutation Alters the NADH Sensitivity of Pyruvate Dehydrogenase Complex of Escherichia coli K-12. J. Bacteriol. 2008, 190, 3851–3858. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.; Henning, U. Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochim. Biophys. Acta (BBA) Enzym. Biol. Oxid. 1966, 122, 355–358. [Google Scholar] [CrossRef]
- Schmincke-Ott, E.; Bisswanger, H. Dihydrolipoamide Dehydrogenase Component of the Pyruvate Dehydrogenase Complex from Escherichia coli K12. Comparative Characterization of the Free and the Complex-Bound Component. JBIC J. Biol. Inorg. Chem. 1981, 114, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.C.; Atkinson, D.E. Regulation of pyruvate dehydrogenase from Escherichia coli. Interactions of adenylate energy charge and other regulatory parameters. J. Biol. Chem. 1970, 245, 5974–5978. [Google Scholar] [PubMed]
- Brass, E.P. Overview of coenzyme A metabolism and its role in cellular toxicity. Chem. Interact. 1994, 90, 203–214. [Google Scholar] [CrossRef]
- Gokarn, R.R.; Eiteman, M.A.; Altman, E. Metabolic Analysis of Escherichia coli in the Presence and Absence of the Carboxylating Enzymes Phosphoenolpyruvate Carboxylase and Pyruvate Carboxylase. Appl. Environ. Microbiol. 2000, 66, 1844–1850. [Google Scholar] [CrossRef]
- Sawers, R. The aerobic/anaerobic interface. Curr. Opin. Microbiol. 1999, 2, 181–187. [Google Scholar] [CrossRef]
- Sawers, G.; Böck, A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J. Bacteriol. 1988, 170, 5330–5336. [Google Scholar] [CrossRef]
- Murphy, H.; Cashel, M. Isolation of RNA Polymerase Suppressors of a (p)ppGpp Deficiency. Methods Enzym. 2003, 371, 596–601. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D.; Santos, C.N.S.; Yu, H.; Stephanopoulos, G. Mutagenesis of the Bacterial RNA Polymerase Alpha Subunit for Improvement of Complex Phenotypes. Appl. Environ. Microbiol. 2009, 75, 2705–2711. [Google Scholar] [CrossRef]
- Goldbeck, C.P.; Jensen, H.M.; TerAvest, M.A.; Beedle, N.; Appling, Y.; Hepler, M.; Cambray, G.; Mutalik, V.; Angenent, L.T.; Ajo-Franklin, C.M. Tuning Promoter Strengths for Improved Synthesis and Function of Electron Conduits in Escherichia coli. ACS Synth. Biol. 2013, 2, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Vellingiri, A.; Song, Y.E.; Munussami, G.; Kim, C.; Park, C.; Jeon, B.H.; Lee, S.-G.; Kim, J.R. Overexpression of c-type cytochrome, CymA in Shewanella oneidensis MR-1 for enhanced bioelectricity generation and cell growth in a microbial fuel cell. J. Chem. Technol. Biotechnol. 2019, 94, 2115–2122. [Google Scholar] [CrossRef]
- Sugino, Y. Mutants of Escherichia coli sensitive to methylene blue and acridines. Genet. Res. 1966, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.M.; Walker, J.R. Genetic analysis of acrA and lir mutations of Escherichia coli. J. Bacteriol. 1982, 152, 1301–1302. [Google Scholar]

| No. | Based on | Relevant Genotype | Reference |
|---|---|---|---|
| JG22 | E. coli DH5αZ1 | aciq, PN25-tetR, SpR, deoR, supE44, Δ(lacZYA argFV169), Phi80 lacZΔM15 | [24] |
| JG1054 | JG22 | pMAL alsSD | this study |
| JG991 | JG22 | Δ(napC-F), ΔfrdA-D::(Ptet cymA mtrA, mtrB), ΔgalK, ΔadhE, idh::PAra stc, ΔldhA::cscRAKB, Δpta ackA::galK pMAL alsSD pEC86 | [22] |
| JG1295 | JG991 | pfl−, aceF*, rpoC− | this study |
| JG1352 | JG1295 | Δ(napC-F), ΔfrdA-D::(Ptet cymA mtrA, mtrB), ΔgalK, ΔadhE, idh::PAra stc,
pfl−, aceF*, rpoC− ΔldhA::cscRAKB, Δpta ackA::galK | this study |
| JG1397 | JG1352 | pMAL alsSD_cymA_stc pEC86 | this study |
| Name | Relevant Genotype | Reference |
|---|---|---|
| pMAL alsSD | bla, PTacalsSD | [22] |
| pMAL alsSD_cymA_stc | bla, PTacalsSD, cymA, stc | this study |
| pEC86 | cat, PTet ccmABCDEFGH | [25] |
| No. | Sequence |
|---|---|
| 1 | CCCTTTTAGCAGGGCTTTCTGGAAGGAGATATACATACCATGAACTG |
| 2 | AAACGACGGCCAGTGCCAAGCTTGCCTGCAGGTTATTTTTTCAGAACAGATGCGC |
| Base | Gene | Type | Mutation | Effect | Fidelity |
|---|---|---|---|---|---|
| 4320496 | pflB | SNV | G → A | Gln to Stop (AA431) | 112/112 (100%) |
| 3009449 | rpoC | SNV | T → C | Pro to Leu (AA359) | 155/156 (99.4%) |
| 3593620 | aceF | SNV | A → G | Asp to Gly (AA430) | 136/138 (98.6%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beblawy, S.; Philipp, L.-A.; Gescher, J. Accelerated Electro-Fermentation of Acetoin in Escherichia coli by Identifying Physiological Limitations of the Electron Transfer Kinetics and the Central Metabolism. Microorganisms 2020, 8, 1843. https://doi.org/10.3390/microorganisms8111843
Beblawy S, Philipp L-A, Gescher J. Accelerated Electro-Fermentation of Acetoin in Escherichia coli by Identifying Physiological Limitations of the Electron Transfer Kinetics and the Central Metabolism. Microorganisms. 2020; 8(11):1843. https://doi.org/10.3390/microorganisms8111843
Chicago/Turabian StyleBeblawy, Sebastian, Laura-Alina Philipp, and Johannes Gescher. 2020. "Accelerated Electro-Fermentation of Acetoin in Escherichia coli by Identifying Physiological Limitations of the Electron Transfer Kinetics and the Central Metabolism" Microorganisms 8, no. 11: 1843. https://doi.org/10.3390/microorganisms8111843
APA StyleBeblawy, S., Philipp, L.-A., & Gescher, J. (2020). Accelerated Electro-Fermentation of Acetoin in Escherichia coli by Identifying Physiological Limitations of the Electron Transfer Kinetics and the Central Metabolism. Microorganisms, 8(11), 1843. https://doi.org/10.3390/microorganisms8111843




