E6/E7 Sequence Diversity of High-Risk Human Papillomaviruses in Two Geographically Isolated Populations of French Guiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Conduct
2.2. Sample Collection
2.3. Type-Specific E6 and E7 Gene Amplification
2.4. Phylogenetic Analysis
2.5. Data Collection and Analysis
2.6. Ethical and Regulatory Aspects
3. Results
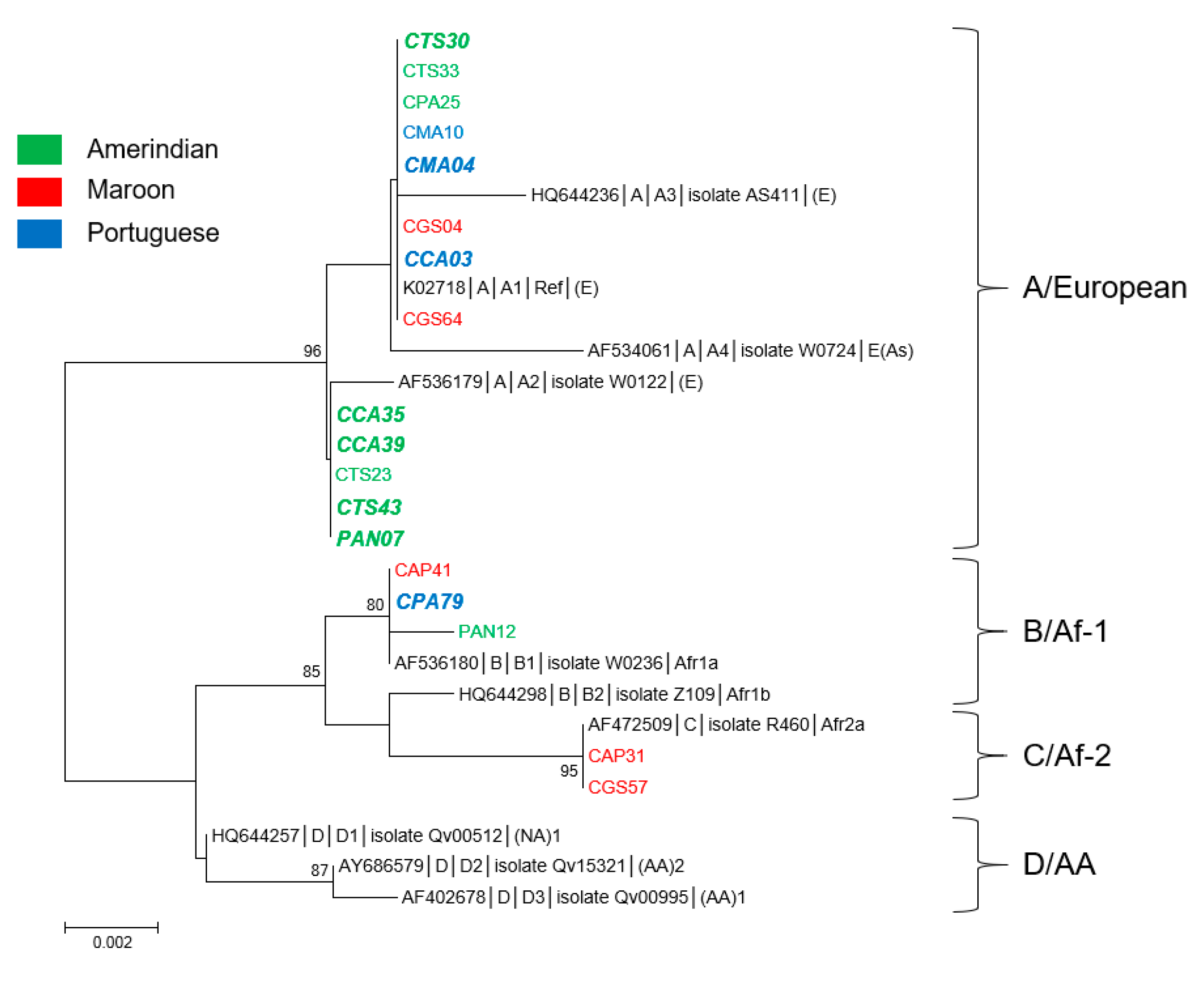
3.1. Genetic Diversity and Phylogeny of HPV16 Sequences
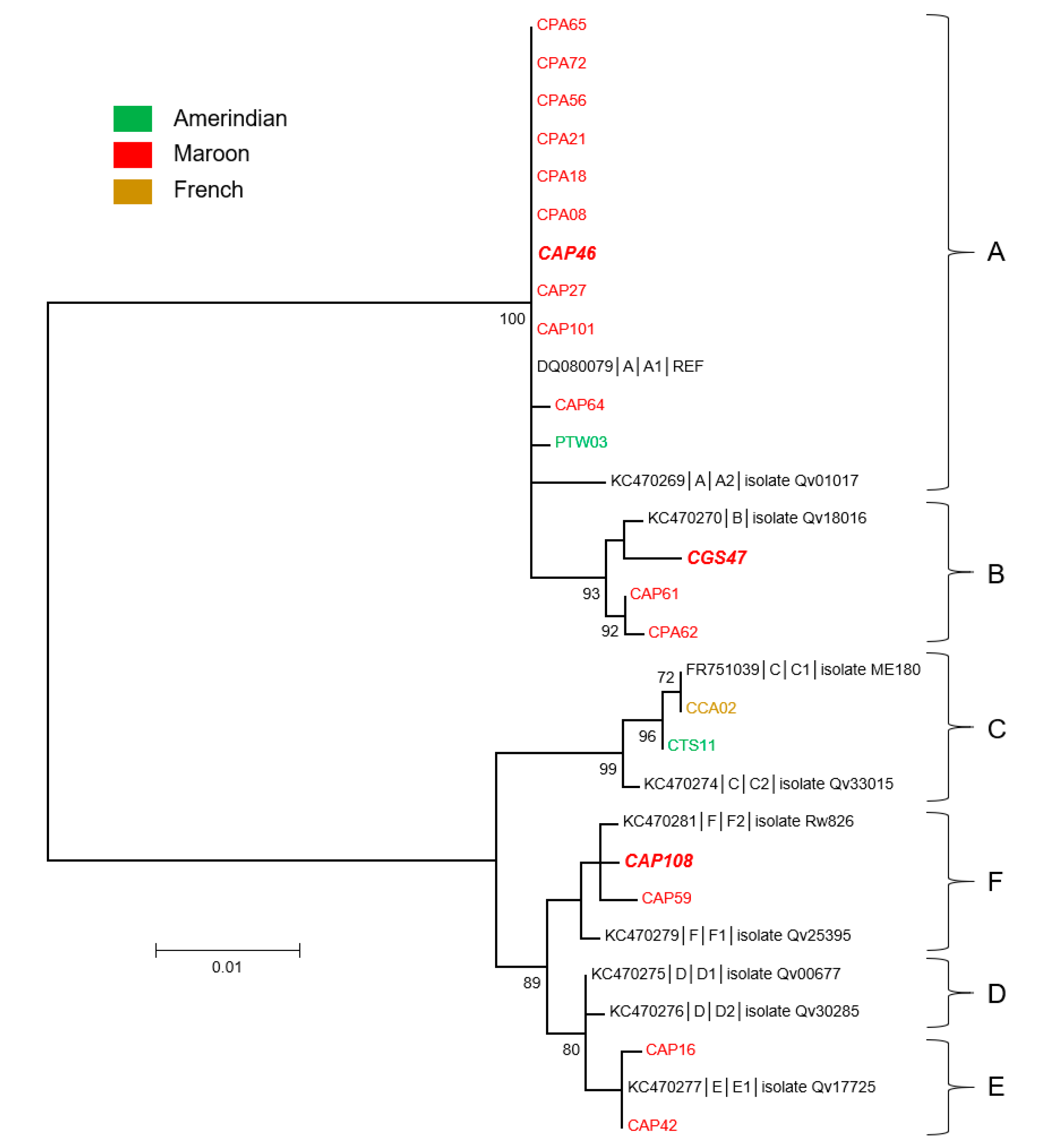
3.2. Genetic Diversity and Phylogeny of HPV18 Sequences
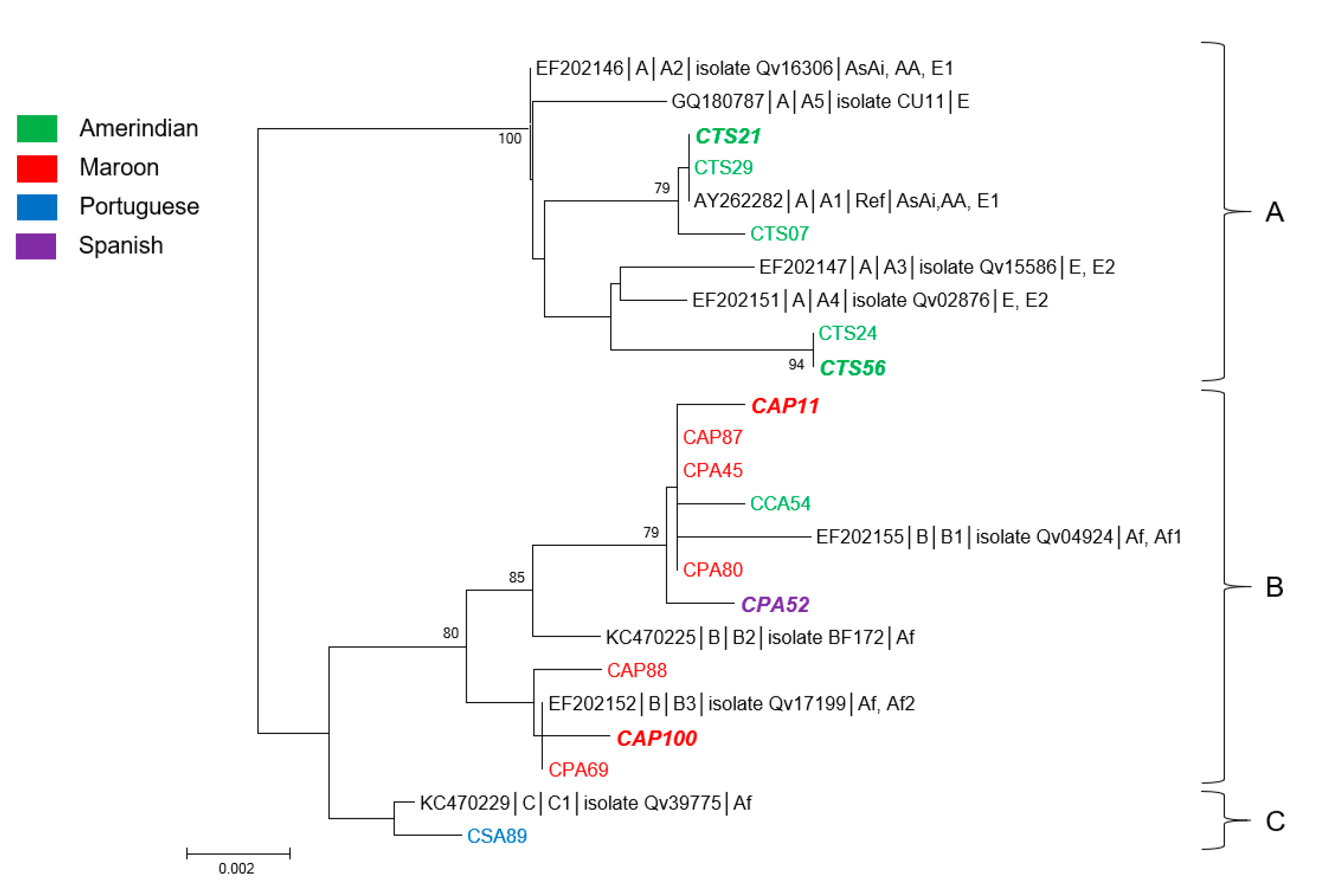
3.3. Genetic Diversity and Phylogeny of HPV31 Sequences
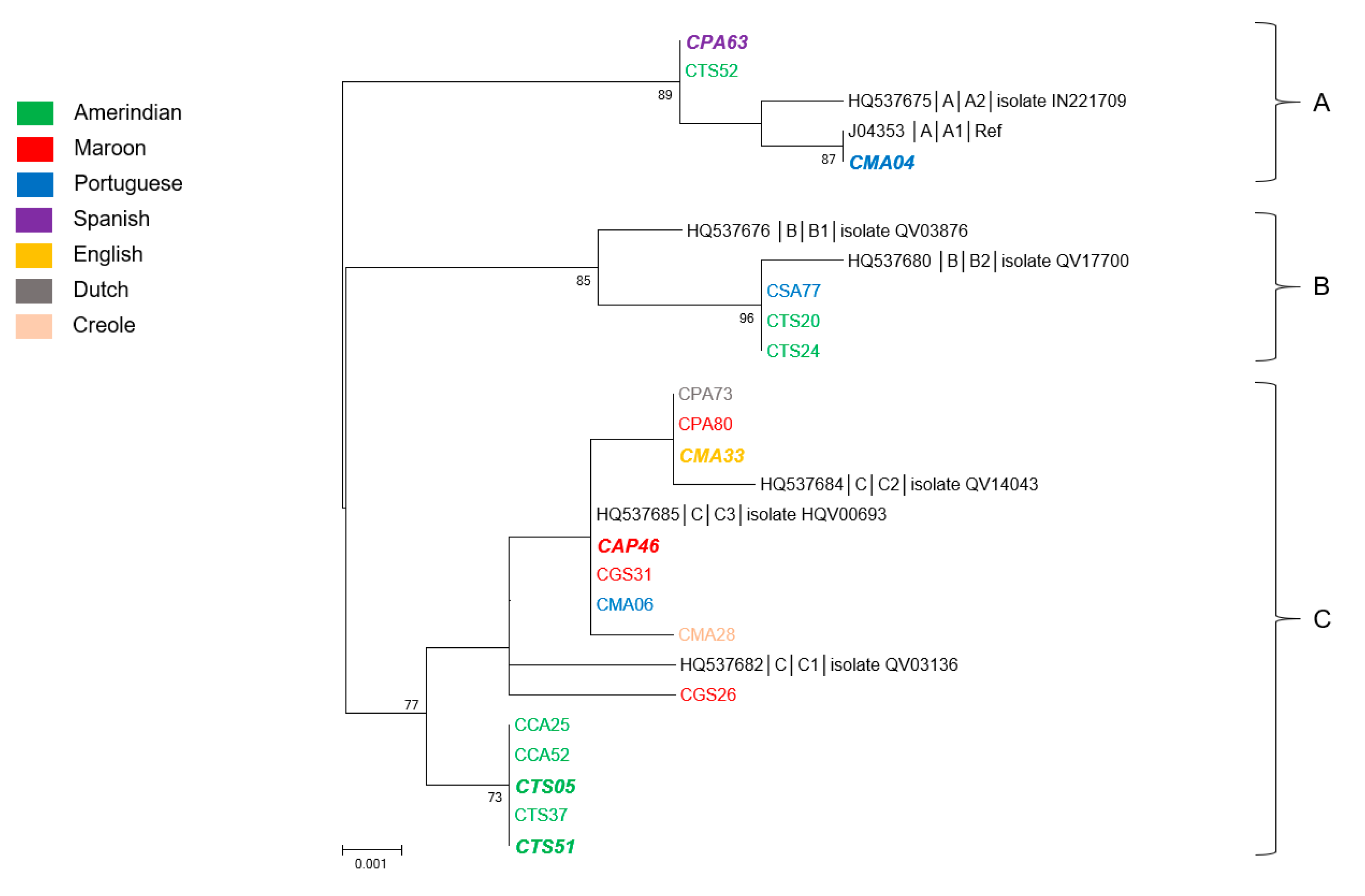
3.4. Genetic Diversity and Phylogeny of HPV52 Sequences
3.5. Genetic Diversity and Phylogeny of HPV58 Sequences
3.6. Genetic Diversity and Phylogeny of HPV68 Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Roue, T.; Nacher, M.; Fior, A.; Plenet, J.; Belliardo, S.; Gandolfo, N.; Deshayes, J.-L.; Laborde, O.; Carles, G.; Thomas, N.; et al. Cervical cancer incidence in French Guiana. Int. J. Gynecol. Cancer 2012, 22, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Douine, M.; Roué, T.; Fior, A.; Adenis, A.; Thomas, N.; Nacher, M. Survival of patients with invasive cervical cancer in French Guiana, 2003–2008. Int. J. Gynecol. Obstet. 2014, 125, 166–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Adenis, A.; Dufit, V.; Douine, M.; Corlin, F.; Ayhan, G.; Najioullah, F.; Molinie, V.; Brousse, P.; Carles, G.; Lacoste, V.; et al. High prevalence of HPV infection in the remote villages of French Guiana: an epidemiological study. Epidemiol. Infect. 2017, 145, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Van Melle, A.; Parriault, M.-C.; Basurko, C.; Jolivet, A.; Flamand, C.; Pigeon, P.; Caudal, J.; Lydié, N.; Halfen, S.; Goerger-Sow, M.-T.; et al. Knowledge, attitudes, behaviors, and practices differences regarding HIV in populations living along the Maroni river: Particularities of operational interest for Amerindian and Maroon populations. AIDS Care 2015, 27, 1112–1117. [Google Scholar] [CrossRef] [PubMed]
- Nacher, M.; Vantilcke, V.; Parriault, M.C.; Van Melle, A.; Hanf, M.; Labadie, G.; Romeo, M.; Adriouch, L.; Carles, G.; Couppié, P. What is driving the HIV epidemic in French Guiana? Int. J. STD AIDS 2010, 21, 359–361. [Google Scholar] [CrossRef]
- Adenis, A.; Dufit, V.; Douine, M.; Najioullah, F.; Molinie, V.; Catherine, D.; Kilié, O.; Thomas, N.; Deshayes, J.L.; Brousse, P.; et al. The singular epidemiology of HPV infection among French Guianese women with normal cytology. BMC Public Heal. 2017, 17, 279. [Google Scholar] [CrossRef]
- Adenis, A.; Dufit, V.; Douine, M.; Ponty, J.; Bianco, L.; Najioullah, F.; Kilié, O.; Catherine, D.; Thomas, N.; Deshayes, J.L.; et al. Predictors of abnormal cytology among HPV-infected women in remote territories of French Guiana. BMC Women’s Heal. 2018, 18, 25. [Google Scholar] [CrossRef]
- Da Fonseca, A.J.; Taeko, D.; Chaves, T.A.; Amorim, L.D.D.C.; Murari, R.S.W.; Miranda, A.E.; Chen, Z.; Burk, R.D.; Ferreira, L.C.L. HPV infection and cervical screening in socially isolated indigenous women inhabitants of the Amazonian rainforest. PLoS ONE 2015, 10, e0133635. [Google Scholar] [CrossRef]
- Bergeron, C.; Cartier, I.; Guldner, L.; Lassalle, M.; Savignoni, A.; Asselain, B. Lésions précancéreuses et cancers du col de l’utérus diagnostiqués par le frottis cervical, Ile-de-France, enquête Crisap, 2002. Bull Epidémiolog. Hebd 2005, 2, 5–6. [Google Scholar]
- Metcalfe, S.; Roger, M.; Faucher, M.-C.; Coutlée, F.; Franco, E.L.; Brassard, P. The association between human leukocyte antigen (HLA)-G polymorphisms and human papillomavirus (HPV) infection in Inuit women of northern Quebec. Hum. Immunol. 2013, 74, 1610–1615. [Google Scholar] [CrossRef]
- Berumen, J.; OrdoñezR, M.; Lazcano, E.; Salmeron, J.; Galvan, S.C.; Estrada, R.A.; Yunes, E.; Garcia-Carranca, A.; Gonzalez-Lira, G.; La Campa, A.M.-D. Asian-American variants of human Papillomavirus 16 and risk for cervical cancer: A case-control study. J. Natl. Cancer Inst. 2001, 93, 1325–1330. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.D.; Terai, M.; Gravitt, P.E.; Brinton, L.A.; Kurman, R.J.; Barnes, W.A.; Greenberg, M.D.; Hadjimichael, O.C.; Fu, L.; McGowan, L.; et al. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res. 2003, 63, 7215–7220. [Google Scholar] [PubMed]
- Castro, M.; Farias, I.; Borborema-Santos, C.; Correia, G.; Astolfi-Filho, S. Prevalence of human papillomavirus (HPV) type 16 variants and rare HPV types in the central Amazon region. Genet. Mol. Res. 2011, 10, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.R.; Cerqueira, D.M.; Cruz, W.B.; Camara, G.N.L.; Brígido, M.M.; Silva, E.O.; Carvalho, L.G.S.; Martins, C. Prevalence of human papillomavirus type 16 variants in the Federal District, Central Brazil. Mem. Instit. Oswaldo Cruz 2004, 99, 281–282. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, J.-W.; Fang, F.; Guo, Y.; Zhu, T.-L.; Yu, Y.-Y.; Kong, F.-F.; Han, L.-F.; Chen, D.-S.; Li, F. HPV16 integration probably contributes to cervical oncogenesis through interrupting tumor suppressor genes and inducing chromosome instability. J. Exp. Clin. Cancer Res. 2016, 35, 1–14. [Google Scholar] [CrossRef]
- Ganguly, N.; Parihar, S.P. Human papillomavirus E6 and E7 oncoproteins as risk factors for tumorigenesis. J. Biosci. 2009, 34, 113–123. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Burk, R.D.; Harari, A.; Chen, Z. Human papillomavirus genome variants. Virology 2013, 445, 232–243. [Google Scholar] [CrossRef]
- Chen, Z.; Schiffman, M.; Herrero, R.; DeSalle, R.; Anastos, K.; Segondy, M.; Sahasrabuddhe, V.V.; Gravitt, P.E.; Hsing, A.W.; Burk, R.D. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS ONE 2013, 8, e72565. [Google Scholar] [CrossRef]
- Salgado, A.H.; Martín-Gámez, D.C.; Moreno, P.; Murillo, R.; Bravo, M.M.; Villa, L.; Molano, M. E6 molecular variants of human papillomavirus (HPV) type 16: An updated and unified criterion for clustering and nomenclature. Virology 2011, 410, 201–215. [Google Scholar] [CrossRef]
- Junes-Gill, K.; Sichero, L.; Maciag, P.C.; Mello, W.; Noronha, V.; Villa, L.L. Human papillomavirus type 16 variants in cervical cancer from an admixtured population in Brazil. J. Med Virol. 2008, 80, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Schiffman, M.; Herrero, R.; DeSalle, R.; Anastos, K.; Segondy, M.; Sahasrabuddhe, V.V.; Gravitt, P.E.; Hsing, A.W.; Burk, R.D. Evolution and taxonomic classification of human Papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS ONE 2011, 6, e20183. [Google Scholar] [CrossRef] [PubMed]
- Douine, M.; Mosnier, E.; Le Hingrat, Q.; Charpentier, C.; Corlin, F.; Hureau, L.; Adenis, A.; Lazrek, Y.; Niemetsky, F.; Aucouturier, A.-L.; et al. Illegal gold miners in French Guiana: a neglected population with poor health. BMC Public Heal. 2017, 18, 23. [Google Scholar] [CrossRef]
- Gaubert-Maréchal, E.; Jolivet, A.; Van-Melle, A.; Parriault, M.-C.; Basurko, C.; Adenis, A.; Hanf, M.; Vantilcke, V.; Halfen, S.; Couppie, P.; et al. Knowledge, attitudes, beliefs and practices on HIV/AIDS among boatmen on the Maroni river: A neglected bridging group? J. AIDS Clin. Res. 2012, 3, 1000181. [Google Scholar]
- Parriault, M.C.; Basurko, C.; Van Melle, A.; Gaubert-Maréchal, E.; Rogier, S.; Couppié, P.; Nacher, M. Predictive factors of unprotected sex for female sex workers: first study in French Guiana, the French territory with the highest HIV prevalence. Int. J. STD AIDS 2014, 26, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Parriault, M.-C.; Van Melle, A.; Basurko, C.; Gaubert-Marechal, E.; Macena, R.H.M.; Rogier, S.; Kerr, L.; Nacher, M. HIV-testing among female sex workers on the border between Brazil and French Guiana: The need for targeted interventions. Cad. Saúde Pública 2015, 31, 1615–1622. [Google Scholar] [CrossRef]


| Genotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Age | Country of Birth | Monther Tongue | Cytology | 16 | 18 | 31 | 52 | 58 | 68 | |
| Saint Georges (8 seqs) | CSA89 | 43 | Brazil | Portuguese | ⚫ | ||||||
| CSA77 | 32 | Brazil | Portuguese | ⚫ | |||||||
| CSA06 | 26 | Brazil | Portuguese | ⚫ | |||||||
| CSA17 | 26 | French Guiana | Portuguese | ⚫ | |||||||
| CSA51 | 30 | Brazil | Portuguese | ASCUS | ⚫ | ||||||
| CSA67 | 42 | French Guiana | Amerindian | HSIL | ⚫ | ||||||
| CSA84 | 29 | French Guiana | Amerindian | ⚫ | |||||||
| CSA13 | 23 | Brazil | Portuguese | ⚫ | |||||||
| Camopi (9 seqs) | CCA03 | 48 | Brazil | Portuguese | ASCUS | ⚫ | |||||
| CCA35 | 28 | French Guiana | Amerindian | LSIL | ⚫ | ||||||
| CCA39 | 35 | French Guiana | Amerindian | ASCUS | ⚫ | ||||||
| CCA54 | 31 | French Guiana | Amerindian | ⚫ | |||||||
| CCA25 | 32 | French Guiana | Amerindian | ⚫ | |||||||
| CCA52 | 56 | French Guiana | Amerindian | ⚫ | |||||||
| CCA11 | 42 | French Guiana | Amerindian | ⚫ | |||||||
| CCA32 | 41 | French Guiana | Amerindian | LSIL | ⚫ | ||||||
| CCA02 | 22 | French Guiana | French | ⚫ | |||||||
| Trois Sauts (30 seqs) | CTS23 | 25 | French Guiana | Amerindian | ⚫ | ||||||
| CTS30 | 26 | French Guiana | Amerindian | LSIL | ⚫ | ⚫ | |||||
| CTS33 | 26 | French Guiana | Amerindian | ⚫ | |||||||
| CTS43 | 39 | French Guiana | Amerindian | ASC-H | ⚫ | ||||||
| CTS07 | 30 | French Guiana | Amerindian | ⚫ | |||||||
| CTS21 | 33 | French Guiana | Amerindian | ASCUS | ⚫ | ⚫ | |||||
| CTS24 | 31 | French Guiana | Amerindian | ⚫ | ⚫ | ||||||
| CTS29 | 33 | French Guiana | Amerindian | ⚫ | |||||||
| CTS56 | 37 | French Guiana | Amerindian | LSIL | ⚫ | ⚫ | |||||
| CTS05 | 28 | French Guiana | Amerindian | HSIL | ⚫ | ||||||
| CTS20 | 52 | French Guiana | Amerindian | ⚫ | |||||||
| CTS37 | 29 | French Guiana | Amerindian | ⚫ | |||||||
| CTS51 | 38 | French Guiana | Amerindian | ASC-H | ⚫ | ⚫ | |||||
| CTS52 | 29 | French Guiana | Amerindian | ⚫ | |||||||
| CTS08 | 22 | French Guiana | Amerindian | ⚫ | |||||||
| CTS14 | 33 | French Guiana | Amerindian | LSIL | ⚫ | ||||||
| CTS35 | 34 | French Guiana | Amerindian | ⚫ | |||||||
| CTS36 | 53 | French Guiana | Amerindian | ⚫ | |||||||
| CTS01 | 35 | French Guiana | Amerindian | ASCUS | ⚫ | ||||||
| CTS16 | 35 | French Guiana | Amerindian | LSIL | ⚫ | ||||||
| CTS42 | 34 | French Guiana | Amerindian | ⚫ | |||||||
| CTS60 | 22 | French Guiana | Amerindian | ASCUS | ⚫ | ||||||
| CTS63 | 32 | French Guiana | Amerindian | ASCUS | ⚫ | ||||||
| CTS78 | 29 | French Guiana | Amerindian | ASC-H | ⚫ | ||||||
| CTS61 | 20 | French Guiana | Amerindian | ||||||||
| CTS11 | 25 | French Guiana | Amerindian | ⚫ | |||||||
| Total/genotype | 7 | 7 | 9 | 14 | 8 | 2 | |||||
| Genotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Age | Country of Birth | Monther Tongue | Cytology | 16 | 18 | 31 | 52 | 58 | 68 | |
| Apatou (27 seqs) | CAP31 | 34 | Suriname | Bushinengue | ⚫ | ||||||
| CAP41 | 37 | Suriname | Bushinengue | ⚫ | |||||||
| CAP11 | 46 | Suriname | Bushinengue | HSIL | ⚫ | ||||||
| CAP87 | 29 | Suriname | Bushinengue | ⚫ | ⚫ | ||||||
| CAP88 | 50 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP100 | 26 | French Guiana | Bushinengue | LSIL | ⚫ | ||||||
| CAP46 | 20 | French Guiana | Bushinengue | ASCUS | ⚫ | ⚫ | ⚫ | ||||
| CAP10 | 49 | Suriname | Bushinengue | ⚫ | |||||||
| CAP47 | 55 | Suriname | Bushinengue | HSIL & GA | ⚫ | ⚫ | |||||
| CAP58 | 60 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP78 | 52 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP96 | 22 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP113 | 26 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP63 | 38 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP110 | 58 | Suriname | Bushinengue | ⚫ | |||||||
| CAP57 | 22 | French Guiana | Bushinengue | ||||||||
| CAP16 | 51 | Suriname | Bushinengue | ⚫ | |||||||
| CAP27 | 64 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP42 | 54 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP59 | 60 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP61 | 48 | French Guiana | Bushinengue | ⚫ | |||||||
| CAP64 | 39 | Suriname | Bushinengue | ⚫ | |||||||
| CAP101 | 26 | Suriname | Bushinengue | ⚫ | |||||||
| CAP108 | 22 | French Guiana | Bushinengue | ASCUS | ⚫ | ||||||
| Grand Santi (8 seqs) | CGS04 | 37 | Suriname | Bushinengue | ⚫ | ||||||
| CGS57 | 23 | French Guiana | Bushinengue | ⚫ | |||||||
| CGS64 | 34 | French Guiana | Bushinengue | ⚫ | |||||||
| CGS26 | 24 | Suriname | Bushinengue | ⚫ | |||||||
| CGS31 | 56 | Suriname | Bushinengue | ⚫ | |||||||
| CGS14 | 26 | Suriname | Bushinengue | ASCUS | ⚫ | ||||||
| CGS69 | 43 | Suriname | Bushinengue | ASC-H | ⚫ | ||||||
| CGS47 | 28 | Suriname | Bushinengue | HSIL | ⚫ | ||||||
| Papaïchton (19 seqs) | CPA25 | 38 | French Guiana | Amerindian | ⚫ | ||||||
| CPA79 | 28 | Brazil | Portuguese | ASCUS | ⚫ | ||||||
| CPA45 | 55 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA52 | 26 | Dominican Rep. | Spanish | ASCUS | ⚫ | ||||||
| CPA69 | 45 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA80 | 50 | French Guiana | Bushinengue | ⚫ | ⚫ | ||||||
| CPA63 | 39 | Dominican Rep. | Spanish | LSIL | ⚫ | ||||||
| CPA73 | 47 | Suriname | Dutch | ⚫ | ⚫ | ||||||
| CPA19 | 44 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA46 | 58 | French Guiana | Bushinengue | ASCUS | ⚫ | ||||||
| CPA08 | 42 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA18 | 40 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA21 | 30 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA56 | 61 | Suriname | Bushinengue | ⚫ | |||||||
| CPA62 | 37 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA65 | 49 | Suriname | Bushinengue | ⚫ | |||||||
| CPA72 | 64 | French Guiana | Bushinengue | ⚫ | |||||||
| CPA01 | 33 | French Guiana | Bushinengue | ||||||||
| Maripasoula (8 seqs) | CMA04 | 26 | Brazil | Portuguese | ASC-H | ⚫ | ⚫ | ⚫ | |||
| CMA10 | 29 | Brazil | Portuguese | ⚫ | |||||||
| CMA52 | 44 | Brazil | Portuguese | ASC-H | |||||||
| CMA06 | 23 | Brazil | Portuguese | ⚫ | |||||||
| CMA28 | 29 | French Guiana | Guianese Creole | ⚫ | |||||||
| CMA33 | 32 | Guyana | English | LSIL | ⚫ | ||||||
| CMA37 | 39 | Brazil | Portuguese | ASC-H | ⚫ | ||||||
| Antecum Pata (3 seqs) | PAN07 | 55 | Brazil | Amerindian | LSIL | ⚫ | |||||
| PAN12 | 64 | French Guiana | Amerindian | ⚫ | |||||||
| CAN34 | 35 | French Guiana | Amerindian | ⚫ | |||||||
| Twenke-Taluen (2 seqs) | PTW01 | 30 | Suriname | Amerindian | HSIL | ⚫ | |||||
| CTW27 | 29 | Suriname | Amerindian | ||||||||
| PTW03 | 40 | Suriname | Amerindian | ⚫ | |||||||
| Total/genotype | 11 | 8 | 10 | 13 | 7 | 18 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nacher, M.; Godefroy, G.; Dufit, V.; Douine, M.; Najioullah, F.; Césaire, R.; Thomas, N.; Drak Alsibai, K.; Adenis, A.; Lacoste, V. E6/E7 Sequence Diversity of High-Risk Human Papillomaviruses in Two Geographically Isolated Populations of French Guiana. Microorganisms 2020, 8, 1842. https://doi.org/10.3390/microorganisms8111842
Nacher M, Godefroy G, Dufit V, Douine M, Najioullah F, Césaire R, Thomas N, Drak Alsibai K, Adenis A, Lacoste V. E6/E7 Sequence Diversity of High-Risk Human Papillomaviruses in Two Geographically Isolated Populations of French Guiana. Microorganisms. 2020; 8(11):1842. https://doi.org/10.3390/microorganisms8111842
Chicago/Turabian StyleNacher, Mathieu, Gersande Godefroy, Valentin Dufit, Maylis Douine, Fatiha Najioullah, Raymond Césaire, Nadia Thomas, Kinan Drak Alsibai, Antoine Adenis, and Vincent Lacoste. 2020. "E6/E7 Sequence Diversity of High-Risk Human Papillomaviruses in Two Geographically Isolated Populations of French Guiana" Microorganisms 8, no. 11: 1842. https://doi.org/10.3390/microorganisms8111842
APA StyleNacher, M., Godefroy, G., Dufit, V., Douine, M., Najioullah, F., Césaire, R., Thomas, N., Drak Alsibai, K., Adenis, A., & Lacoste, V. (2020). E6/E7 Sequence Diversity of High-Risk Human Papillomaviruses in Two Geographically Isolated Populations of French Guiana. Microorganisms, 8(11), 1842. https://doi.org/10.3390/microorganisms8111842







