Abstract
The relationship among bats, ectoparasites and associated microorganisms is important to investigate how humans can become exposed to zoonotic agents. Even though the diversity of Bartonella spp. in bats and ectoparasites has been previously reported, the occurrence of gltA genotypes within hosts has not been assessed so far. We aimed to investigate the genetic diversity of Bartonella spp. in non-hematophagous bats and associated ectoparasites by assessing cloned gltA Bartonella genotypes in intra- and inter-hosts levels, as well as by using three additional molecular markers. Overall, 13.5% (18/133) bat blood samples, 17.18% bat flies (11/64) and 23.8% (5/21) Macronyssidae mite pools showed to be positive for Bartonella spp. Seventeen positive samples were submitted to gltA-cloning and three clones were sequenced for each sample. We also obtained 11, seven and three sequences for nuoG, rpoB and ftsZ genes, respectively. None were positive for the other target genes. We found at least two genotypes among the three gltA-cloned sequences from each sample, and 13 between all the 51 sequences. Among the nuoG, rpoB and ftsZ sequences we found eight, five and three genotypes, respectively. In the phylogenetic analysis, the sequences were positioned mainly in groups related to Bartonella identified in rodents, bats and bat flies. Herein, we showed the genetic diversity of Bartonella in bat’s blood and associated ectoparasites samples at both intra- and inter-host levels.
1. Introduction
The interest in emerging and reemerging diseases has increased since the late 20th century. It is estimated that 75% of the emerging infectious diseases are zoonoses, which are mainly transmitted by vectors. The main cause of zoonoses’ emergence is the human encroachment on natural habitats changing parasite-host relationships due to its fragmentation and their close contact with wildlife, domestic animals and humans [1].
Bartonella spp. are gram-negative, fastidious and facultative intracellular bacteria that can be transmitted by arthropod vectors. These bacteria primarily parasitize erythrocytes and endothelial cells, but can also colonize dendritic cells, macrophages and monocytes from different mammals and humans [2,3].
The order Chiroptera represents the second largest group of mammals in the world [4]. In Brazil, 9 families, 69 genera and 182 species of bats have been described so far in all morphoclimatic regions [5]. Due to their high mobility, wide distribution, social behavior, longevity [6] and unique immunological system [7], these animals are frequently associated with several emerging infectious diseases and pointed out as reservoir hosts and the source of infection for potential zoonotic agents [8,9]. Indeed, these animals have already been incriminated as possible reservoirs of a diversity of important bacteria, such as Rickettsia spp. [10], Leptospira spp. [11], hemotropic mycoplasma [12,13] and Coxiella burnettii [14]; protozoans, such as Toxoplasma gondii [15,16], Leishmania spp. [17,18,19,20,21] and Trypanosoma spp. [17,18,22,23,24,25,26,27]; and different fungi and viruses such as rabies [28]. Furthermore, it has been suggested that bats are the most probable ancestral hosts for eubartonellae and that their ability to fly may have contributed to making these mammals ideal hosts for the spread of infectious agents, such as those caused by arthropod-borne bartonellae [29].
Due to the high abundance of bat ectoparasites in Brazil, which comprises so far 96 Streblidae species [30], 26 Nycteribiidae species [31] and 32 species of mites (Acari) [32], it is important to achieve a better understanding of the relationship among bats, ectoparasites and associated microorganisms in order to understand the epizooetiology of such agents [33].
Genotypic diversity of Bartonella spp. in bats and associated ectoparasites has been widely reported [6,34,35,36,37,38,39]. Even though bartonellae genotypes have been detected in bats [12,40] and ectoparasites [41], in Brazil, the occurrence of multiple citrate synthase gltA genotypes in both vertebrate and invertebrate hosts infected by Bartonella spp. has not been assessed so far, since most studies in the literature have focused on the analysis of genetic diversity by targeting different Bartonella housekeeping genes, as previously proposed [42].
Therefore, this work aimed to investigate the genetic diversity of Bartonella spp. in non-hematophagous bats and associated ectoparasites from central-western Brazil. For this purpose, two different strategies were used: while the first one was based on the assessment of the genetic diversity of cloned gltA Bartonella genotypes at both intra- and inter-hosts levels, the second one investigated such diversity using a set of three genes.
2. Materials and Methods
All experimental procedures were approved by the “Instituto Chico Mendes de Biodiversidade” (ICMBio) (SISBIO 57450-1) and the Ethics Committee on Animal Use of the School of Agricultural and Veterinary Sciences, UNESP (CEUA FCAV/UNESP 010050/17).
Between June 2017 and March 2018, bats samplings were performed quarterly in two distinct places in the periurban region of Campo Grande city, Mato Grosso do Sul state, central-western Brazil, namely “Centro de Educação Ambiental Polonês” (CEA Pol.; 54°34′49.816″ W and 20°26′50.673″ S) and “Florestinha” (CEA Flor.; 54°33′43.352″ W and 20°24′11.614″ S). Lactating and/or pregnant females were released after field screening, as well as individuals who reached the maximum number of species collected per location as per license permits. The animals and their ectoparasites were identified using previously described identification keys for bats [43,44] and ectoparasites [45,46,47] (Table S1). The bats were then taken to the Laboratory of Parasitc Biology of the Catholic University Dom Bosco. Bats were euthanized using 10% ketamine and 1% acepromazine association at a 9:1 ratio and 1 mL/kg injected intramuscularly or intraperitoneally. From each bat specimen, 0.2–2 mL blood samples (according to the animal size) and spleen fragments were collected by intracardiac puncture and necropsy in EDTA and absolute ethanol (Merck™, Darmstadt, Germany) containing RNAse- and DNAse-free microtubes (Kasvi®, São José dos Pinhais, Brazil) and maintained at −80 °C and −20 °C, respectively, until their use in molecular assays. All samples were collected in laminar flow, using appropriate and sterilized instruments to avoid environmental contamination. A total of 418 biological samples represented by 135 spleen tissues, 133 whole blood and 150 ectoparasites (comprising 64 bat flies, 67 mite pools, and 19 ticks) were collected from 135 individuals and used in the present study to investigate the genetic diversity of Bartonella spp. The small body size of one animal and the difficulty of getting enough samples from another one (represented by specimens of Myotis nigricans and Artibeus lituratus, respectively) precluded DNA extraction from those blood samples.
For DNA extraction, 64 bat flies and 19 ticks were individually processed. On the other hand, Spinturnicidae (specimens belonging to the same species) and Macronyssidae (specimens belonging to the same genus) mites collected from the same individual were pooled in 46 and 21 samples, respectively. Each pool sample contained up to 50 mites. All ectoparasites and spleen samples were purified using Illustra Tissue and Cells Genomic Prep Mini Spin Kit (GE Healthcare™, Chicago, IL, USA), whereas bat blood samples were purified using the lllustra Blood Mini Spin Kit (GE Healthcare™, Chicago, IL, USA), following the manufacturer’s recommendations. DNA samples were evaluated by optical spectrophotometry (Nanodrop, Thermo Fisher Scientific™, Waltham, MA, USA) to assess the concentration (µg/µL) and 260/280—260/230 ratios.
The DNA samples were firstly tested by conventional (c) PCR assays based on endogenous genes to verify the absence of PCR inhibitors. In this sense, for the DNA samples obtained from blood and spleen tissue, the cPCR protocol based on the mammals-glyceraldehyde 3-phosphate dehydrogenase (gapdh) gene was performed as previously described [48]. For the ectoparasites collected from the animals, two different cPCR protocols were used. While a PCR protocol based on cytochrome c oxidase subunit I (cox-1) gene [49] was used for DNA samples obtained from bat flies and mites, a protocol based on the 16S rRNA gene [50] was used for DNA samples obtained from tick larvae. All the obtained 16S rRNA tick larvae amplicons were submitted to dideoxynucleotide chain termination [51] sequencing to check the molecular identity by nBLAST analysis.
The positive samples for the endogenous genes protocols were then submitted to a screening quantitative (q) PCR for Bartonella spp. based on the NADH dehydrogenase gamma subunit (nuoG) gene, as previously described [52]. All positive samples on the qPCR assay were tested by cPCR assays based on the following genes: nuoG [53], riboflavin synthase (ribC) [54], citrate synthase (gltA) [55], β-subunit of RNA polymerase (rpoB) [56], hemin binding protein (pap-31) [57], heat-shock chaperonin protein (groEL) [56,58], cell division protein (fstZ) [56], 16S rRNA [56] and intergenic region 16S–23S rRNA (ITS) [59], following the protocols previously described.
Products amplified in the cPCR assays were submitted to horizontal electrophoresis on ethidium bromide stained-agarose gels (1.0%) at 100 V/150 mA for 50 min.
Bartonella-gltA amplicons were submitted to the cloning protocol using the vector pGEM-T Easy (Promega™, Madison, WI, USA) following the manufacturer’s recommendations. The ligation products were inserted into competent Escherichia coli One Shot Match 1T1R Chemically Competent Cells (Invitrogen™, Carlsbad, CA, USA - Cat # C8620-03) (109–1010 cfu/ng of DNA) by thermal shock method. After seeding on a solid Luria Bertani (LB) medium prepared with 100 μg/mL ampicillin, 40 μL of X-gal (5-bromo-4-chloro-3-indolyl-β- D-galactoside) (Sigma-Aldrich™, St. Louis, MO, USA) and 20 μL IPTG (isopropylthio-β-galactoside) (Sigma-Aldrich™, St. Louis, MO, USA), the plates were incubated at 37 °C overnight. Three individual white colonies of bacteria were chosen for each sample and transferred into tubes containing LB liquid medium and 100 μg/mL ampicillin. After incubation at 37 °C for up to 24 h, plasmid DNA was extracted from the transforming clones by the alkaline lysis method [60].
The obtained amplicons were purified using the Silica Bead DNA gel extraction kit (Thermo Fisher Scientific™, Waltham, MA, USA) and submitted to sequencing by dideoxynucleotide chain termination [51], performed in ABI PRISM 3700 DNA Analyzer sequencer (Applied Biosystems™, Foster City, CA, USA) at the “Centro de Recursos Biológicos e Biologia Genômica (CREBIO-FCAV—UNESP)”. All the sequences obtained were evaluated and a consensus sequence was generated using the Phred-Phrap software version 23 (Genome Sciences Department, University of Washington, Seattle, WA, USA) [61,62]. Then, all obtained consensus sequences were compared with sequences previously deposited in the GenBank (National Center for Biotechnology Information, U.S. National Library of Medicine, Bethesda, MD, USA) database by nBLAST (National Center for Biotechnology Information, U.S. National Library of Medicine, Bethesda, MD, USA) [63].
The cloned sequences were aligned to evaluate the nucleotide polymorphism within each bat blood/spleen and ectoparasite sample and among all the cloned samples. The number of genotypes (h), genotype diversity (Hd) and nucleotide diversity (π) were determined using the DnaSP 5 program, version 5.10.01 (Universitat de Barcelona, Barcelona, Espanha) [64]. The genotype networks were plotted using PopART (New Zealand) (http://popart.otago.ac.nz) with TCS networks [65]. Additionally, for the gltA gene, a genealogy network was created using the software Splitstree v4.11.3 (Eberhard Karls Universität Tübingen, Tübingen, Germany) [66] using Neighbor-net method and bootstrap of 1000 replicates.
Phylogenetic inferences were performed for all four molecular markers from which sequences were obtained. The sequences were aligned with other sequences from the GenBank database through BioEdit [67]. The best evolutionary model was chosen by the jModelTest2 program (version 2.1.6) [68], under the Bayesian Information Criterion (BIC) [69]. The Bayesian inference (BI) analysis was performed with MrBayes 3.1.2 [70] using the CIPRES Science Gateway on XSEDE (The Extreme Science and Engineering Discovery Environment, USA) [71]. The phylogenetic tree edition and rooting (outgroup) were performed using the Treegraph 2.0 beta software [72]. The evolutionary models used for Bayesian Inferences were TVM, TIM2ef, TIM1 and TrN for gltA, nuoG, ftsZ, and rpoB genes respectively. All were settled with proportion of invariable sites and gamma rates.
The sequences were deposited in the GenBank database under accession numbers MT874166-MT874216 for gltA sequences, MT874217-MT874227 for nuoG sequences, MT874228-MT874234 for rpoB sequences and MT831908-MT831910 for ftsZ sequences.
3. Results
3.1. PCR Assays and Sequencing
All DNA samples were positive in the cPCR assay based on the endogenous genes (gapdh for the blood and spleen samples, 16S rRNA to the ticks samples, and cox-1 to mite and fly samples). In qPCR assays for Bartonella spp. based on the nuoG gene, 34/418 (8.1%) samples showed positive results (efficiency ranging from 100.5% to 104.7%; slope from −3.311 to −3215; R2 from 0.969 to 0.997; y-intercept from 35.955 to 37.425). Overall, 18/34 (52.9%) were originated from bat blood samples and 16/34 (47.1%) were from ectoparasites samples. The bat blood positive samples [13.53% (18/133)] were represented by Platyrrhinus lineatus n = 12/23; Artibeus planirostris n = 3/33; Artibeus lituratus n = 2/37; and Carollia perspicillata n = 1/5. The positive bat flies [17.18% (11/64)] belonged to the following species: Trichobius dugesii complex n = 6/10; Trichobius costalimai n = 4/29; Megistopoda aranea n = 1/18. Bartonella-positive Macronyssidae pool mites [23.8% (5/21)] were represented by Steatonyssus spp. N = 5/21. One P. lineatus (#79) and one C. perspicillata (#42) each had the blood sample and associated ectoparasites (T. dugesii complex bat flies—one from animal #79 and two from the animal #42) positive in qPCR assay for Bartonella spp. No spleen samples, individual ticks and Spinturnicidae pool mites were positive for Bartonella spp. All the 34 positive samples were tested by cPCR to Bartonella spp. based on gltA, nuoG, rpoB and ftsZ genes. Due to the low volume of eluted DNA obtained in the nucleic acids extraction procedure from ectoparasites, they were not submitted to the cPCR protocols for ribC, groEL, pap-31 and 16S rRNA genes and ITS region.
Even though 23 samples were positive in the cPCR assay based on gltA gene, 17 (14 from bats and 3 from flies) were submitted to cloning due to the quality of the amplicons observed in agarose gel electrophoresis. For this specific gene, three clones were sequenced from each sample, totaling 51 sequences that were used to determine the genotypic diversity among the different species of bats and flies studied as well as within each individual host. Similarly, although 13 and 15 samples were positive in the cPCR assays for the rpoB and ftsZ genes, seven (5 from bats and 2 from flies) and three sequences (2 from bats and 1 from flies) were obtained from each gene, respectively. All the samples submitted to the nuoG cPCR protocol were positive, except for the DNA samples from mites (Macronyssidae). Only those samples that did not show positive results for any other gene but yielded nuoG amplicons were submitted to sequencing, totaling 11 sequences (3 from bats and 8 from flies). None of the 18 Bartonella nuoG qPCR-positive bat blood samples were positive in the cPCR protocols based ribC, groEL, pap-31 and 16S rRNA genes and ITS region (Table S2).
3.2. Sequences Homology Analysis
In the nBLAST analysis, three out of 51 gltA cloned showed 88.20%–88.55% identity (100% of query coverage) with Bartonella vinsonii strain NCTC12905 (GenBank Access Number LR134529). The 48 remaining gltA cloned sequences showed 91.29%–100% identity (query coverage value of 90%–100%) to Bartonella spp. sequences previously detected in bats and their ectoparasites sampled in Costa Rica (17/51 sequences showed identity to KJ816676—Myotis keaysi; 14/51 similar to MH234380—Glossophaga soricina; 5/51 were similar to KJ816682—Artibeus jamaicensis–Aspidoptera phyllostomatis; 3/51 sequences similar to MH234319—A. lituratus; 3/51 similar to MH234346—C. perspicillata; and 1/51 were similar to MH234347—C. perspicillata; 1/51 to MH234333—Artibeus glaucus watsoni; 1/51 to MH234322—A. lituratus; 1/51 to KJ816687—A. lituratus–Paratrichobius longicrus; 1/51 to KJ816666—Anoura geoffroyi; and 1/51 to KJ816673—Sturnira lilium–Aspidoptera delatorrei) (Table S3).
Regarding the nuoG gene, nine sequences showed 92.7%–98.98% identity (query coverage of 86%–100%) to Bartonella spp. previously detected in blood from vampire bats Desmodus rotundus (GenBank Access Number: MK314980/MK314982) and Diphylla ecaudata (GenBank Access Number: MK314979), and non-hematophagous bats C. perspicillata (GenBank Access Number: KY356757) and S. lillium (GenBank Access Number: KY356755) from Brazil. The remaining two sequences showed 91.72%–91.81% identity (query coverage of 99%–100%) to Bartonella spp. detected in Cervus nippon from Japan (GenBank Access Number: CP019781).
The five rpoB sequences obtained showed 85.66%–90.08% identity (query coverage of 99%–100%) to Bartonella spp. detected in a blood sample from Eidolon helvum bat (GenBank Access Number: KM215201) and to Bartonella taylorii detected in Apodemus flavicollis from Lithuania (GenBank Access Number: MH547315).
Finally, the three ftsZ sequences obtained showed 91.42% to 92.48% identity (query coverage of 100%) to Bartonella spp. detected in rodents (Meriones libycus) and bats (Rhinolophus euryale) from Georgia (GenBank Access Numbers: KT327035 e KX300105) and in the bat E. helvum from Tanzania (GenBank Access Number: KJ999693).
3.3. Genotype Analysis
The results found for the 51 gltA-cloned sequences, which were obtained from 17 Bartonella-positive samples, showed the presence of at least two different genotypes among the three cloned sequences from each bat fly or bat blood sample (Table 1). However, the number of variable sites found among the genotypes was high (at least 20 variable sites) in only 6/17 cloned samples (blood samples), while the others (8 from blood samples and 3 from bat flies) showed 1 to 9 variable sites. Thus, even with more than one genotype per sample, the sequences obtained from the blood of animals #05, #20, #24, #32, #81 and #114 (all from the same bat species, namely P. lineatus) showed a greater number of nucleotide differences among them. When all the 51 cloned gltA sequences obtained were analyzed together, 13 different genotypes were observed (Figure 1a). In the genotype network generated for this gene fragment, a higher diversity was observed among the sequences obtained from bat blood samples when compared to those obtained from bat flies. Among the nine sequences obtained from three bat flies (T. dugesii complex), two different genotypes (1 and 2) were found. Only one ectoparasite had the cloned sequences separate in the two genotypes. These two abovementioned genotypes were also represented by sequences obtained from bat blood samples. The genotype 1 was found in blood samples from bats #05 with two clones, #20 with one clone, and #32 with one clone, as well as in bat flies #79 and #83, both with the three clones. The genotype 2 was represented by sequences found in blood samples from bats #18 with three clones, #114 with one clone, and in bat fly #30 with one clone.

Table 1.
Polymorphism found among the three Bartonella gltA cloned sequences obtained from each bat blood and fly sample.
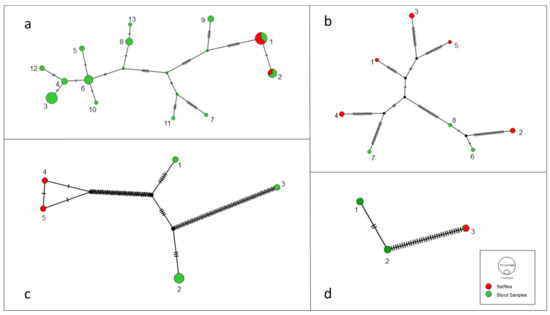
Figure 1.
Genotype network with gltA (a), nuoG (b), rpoB (c) and ftsZ (d) sequences obtained from bat blood and ectoparasite samples obtained in the present study.
In addition, polymorphism divergence analysis was performed between the group of sequences obtained from ectoparasites and bat blood samples and no significant difference between the two groups was found (Table 2).

Table 2.
Polymorphism found within each sample population (ectoparasite and bat blood samples) for gltA Bartonella sequences.
The genotype diversity analysis of the nuoG (Figure 1b), rpoB (Figure 1c) and ftsZ (Figure 1c) sequences demonstrated the presence of eight, five and three distinct genotypes, respectively, among 11 sequences for the nuoG gene, seven for the rpoB gene and three for the ftsZ gene. In the genotype network generated for these gene fragments, the sequences obtained from bat flies were grouped in different groups from those obtained from bat blood samples as previously observed among gltA sequences.
All the results from the genotype analyses are shown in Table 3.

Table 3.
Polymorphism found among the sequences obtained from ectoparasite and bat blood samples for each gene analyzed in the present study.
3.4. Phylogenetic Analysis
For the gltA gene fragment (790 bp), the Bartonella sequences were allocated in four groups into two large clades (Figure 2). The first one, containing only Bartonella sequences obtained from bats and including nine genotypes found in the present work, comprised two paraphyletic groups: one containing eight genotypes (3, 4, 5, 6, 8, 10, 11 and 13) closely related to each other, whereas the other one contained the genotype 7 and represented by only one clone from the bat blood sample #24. The other sequences obtained from this same bat specimen (blood sample #24) were positioned in the first paraphyletic group and represented by genotype 6, thus showing that the same bat blood sample contained more than one Bartonella genotype.

Figure 2.
Phylogenetic tree based on an alignment of 790 bp-length of Bartonella spp. gltA sequences, using the Bayesian Inference method and TVM+I+G as the evolutionary model. Numbers at nodes correspond to the posterior probability value support. Each sequence-clone or group of sequences-clone are highlighted according to their respective genotype. The colored bats are positioned according to the groups found in the genealogy network. Each sample is designated with the type of sample (bb = bat blood and bf = bat fly) followed by the referred animal and clone numbers.
The second large clade showed that the four remaining genotypes (1, 2, 9 and 12) were phylogenetically related to Bartonella sequences previously identified in rodents and other bats. In this clade, one genotype (9), which was represented by three clones from the same animal (bat blood #42), was positioned together with sequences previously detected in other bats from Peru and Brazil. The other three genotypes (1, 2 and 12), which comprised all the Bartonella gltA sequences obtained from ectoparasites as well as five sequences detected in bat blood samples, showed to be closely related to a Bartonella sequence obtained from Strebla guajiro in Brazil and other bat species from Brazil and Guatemala. Moreover, the 51 gltA-cloned sequences were submitted to a genealogy network analysis (Figure 3) and were distributed following the same pattern observed in the phylogenetic tree, where the bat figures represent each group found by different colors.
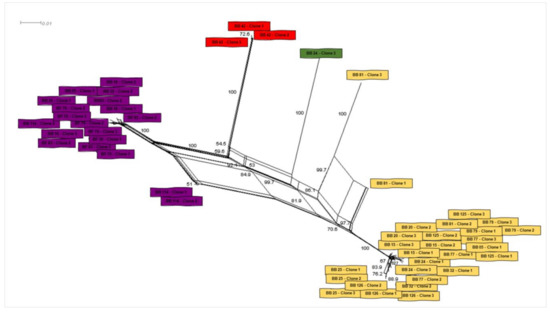
Figure 3.
Genealogy network analysis of Bartonella spp. gltA sequences-clones obtained from bat blood and flies sampled in the present study. The analysis was performed with Splitstree software using the parameters “Neighbor-Net” with bootstrap of 1000 replicates.
For the nuoG gene (368 pb) (Figure 4), seven different clusters were observed: the first and second ones are monophyletic and comprise three genotypes, genotype 2 with two sequences obtained from T. dugesii complex (#42M1 and #42M2) collected from the same animal (#42), and the other two genotypes (6 and 8) with sequences obtained from blood samples from the same animal species (A. planirostris) but different individuals (#16 and #44) that were positioned as monophyletic groups with 86% of posterior probability. Two nuoG Bartonella sequences from C. perspicillata and S. lilium previously sampled in Brazil were also positioned in the same branch. Two other genotypes (4 and 7), which comprised a sequence detected in P. lineatus (#34) and two sequences from T. costalimai (#59 and #56F1), were positioned as polytomic branches related to Bartonella clarridgeiae, B. rochalimae, B. bacilliformis, ruminant-related Bartonella and sequences previously detected in bats from Brazil and Ghana. The genotypes 1 and 5 were positioned as basal groups and comprised one sequence from T. dugesii complex (#23) and one from M. aranea (#120) that appeared to be closely related to Bartonella identified in vampire bats from Brazil. The last genotype comprising two sequences from T. costalimai flies (#55 and #57M) were positioned as one group basal from the others.
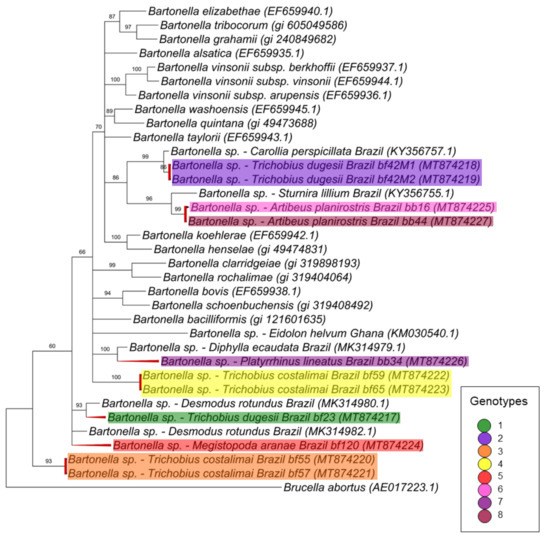
Figure 4.
Phylogenetic tree based on an alignment of 368 bp-length of Bartonella spp. nuoG sequences, using the Bayesian Inference method and TIM2ef+I+G as the evolutionary model. Numbers at nodes correspond to the posterior probability value support. Each sequence is highlighted according to its respective genotype. Each sample is designated with the type of sample (bb = bat blood and bf = bat fly) followed by the referred animal number.
In the phylogenetic analysis based on the rpoB gene (757 pb), the sequences were positioned in three separate clades (Figure 5). The first clade comprises two genotypes (4 and 5) with one sequence each and obtained from T. dugesii flies (#79 and #83) that were closely related to Bartonella genotypes previously detected in a bat and in a bat fly from Brazil, supported by 100% of posterior probability. The second clade, which was supported by 100% of posterior probability, was composed by four sequences detected in blood samples from P. lineatus (#05, #24, #25 and #81) and represented by two different genotypes (1 and 2). The most closely related sequences in this clade were represented by sequences obtained from bats from French Guiana and Brazil and Bartonella chomellii. The last sequence represented by genotype 3 and obtained from the blood sample of an A. lituratus individual appeared in a polytomic branch with Bartonella spp. detected in other bat species from Africa and Asia.
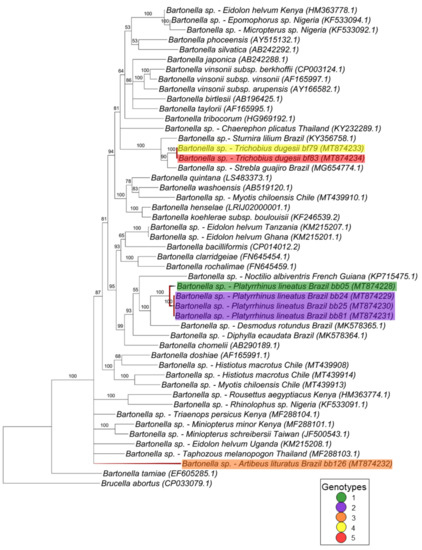
Figure 5.
Phylogenetic tree based on an alignment of 757 bp-length of Bartonella spp. rpoB sequences, using the Bayesian Inference method and TrN+I+G as the evolutionary model. Numbers at nodes correspond to the posterior probability value support. Each sequence is highlighted according to its respective genotype. Each sample is designated with the type of sample (bb = bat blood and bf = bat fly) followed by the referred animal number.
Finally, for the ftsZ gene fragment (521 bp), the three sequences corresponding to one genotype each were positioned in two different branches (Figure 6). The first branch comprised the genotype 1 (represented by a sequence detected in a bat blood sample [C. perspicillata #42]) that clustered together with Bartonella spp. detected in a rodent from Brazil and in a bat from Chile with 76% of posterior probability. The other two sequences, which were represented by genotypes 2 (represented by a sequence detected in a blood sample from P. lineatus #77) and 3 (represented by a sequence detected in M. aranea sample #120), clustered together in a polytomic branch—supported by 96% of posterior probability—with Bartonella bacilliformis, Bartonella naantalensis and Bartonella spp. detected in Myotis chiloensis from Chile.
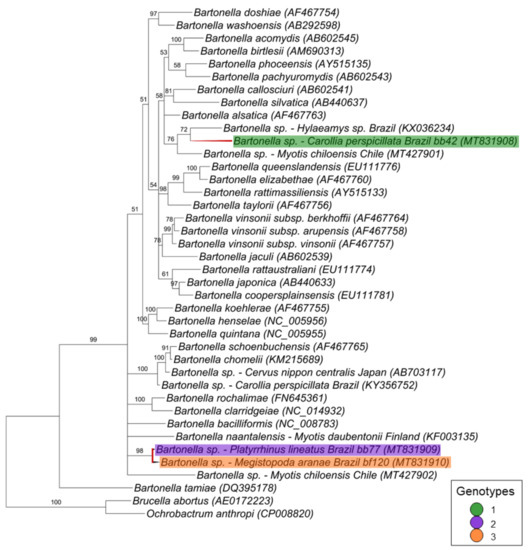
Figure 6.
Phylogenetic tree based on an alignment of 521 bp fragment of Bartonella spp. ftsZ sequences, using the Bayesian Inference method and the TIM1+I+G as evolutionary model. Numbers at nodes correspond to the posterior probability value support. Each sequence is highlighted according to its respective genotype. Each sample is designated with the type of sample (bb = bat blood and bf = bat fly) followed by the referred animal number.
4. Discussion
Genotypic diversity of Bartonella spp. in bats and associated ectoparasites have been widely observed and reported [6,34,35,36,37,38,39]. In Brazil, previous studies showed the genetic diversity of bartonellae among non-hematophagous bats [12], and Streblidae flies [41], by phylogenetic analyses based on the gltA gene as well among vampire bats by phylogenetic and genotypic analysis based on gltA and rpoB genes [40]. However, while the majority of studies in the literature has focused on the analysis of genetic diversity of bartonellae in bats and their associated ectoparasites by targeting different Bartonella housekeping genes, as previously proposed [42], the present work aimed to investigate such diversity within both vertebrate and invertebrate hosts at an intra-host perspective. For such qn investigation, the gltA gene was chosen as it is the most frequently used molecular marker and is considered a good tool to evaluate the genotype diversity [42,73]. Even more, the number of Bartonella gltA sequences in GenBank is high and most frequently updated. La Scola et al., (2003) [73] also indicated the gltA together with the rpoB gene as suitable molecular markers for Bartonella differentiation and characterization. So, in paralell, we also assessed the genetic diversity by targeting three additional molecular markers, namely rpoB, ftsZ and nuoG.
Interestingly, while 18 bat blood samples showed to be positive for Bartonella spp., all spleen samples were negative. Despite the fact that spleen samples are considered a pertinent option for Bartonella detection, the blood is the most suitable choice sample for diagnosis of Bartonella spp. due to the hemotropism of this group of bacteria. Nevertheless, it is important to keep in mind that even blood samples can show false-negative results because of the cyclic bacteremia of bartonellae [74].
Considering that Nycteribiidae and Streblidae flies have been suggested as putative vectors for Bartonella among bats [75,76,77], the assessment of the genetic diversity of bartonellae among vertebrate hosts and associated ectoparasites as well as the possible exchange of Bartonella-genotypes between bats and related ectoparasites [75,78] has been increasingly addressed. In addition, McKee et al., (2016) [69], when evaluating the host-parasite interaction of Bartonella in bats, pointed out the existence of frequent and bidirectional exchange of this group of bacteria between bats and associated ectoparasites, highlighting the influence of arthropod vectors on the pattern of genetic variants of Bartonella spp. in vertebrate hosts. Herein, we observed such matched relationship only when we considered the genotypic analysis based on the gltA gene. In this sense, we found two Bartonella genotypes (1 and 2) that comprised sequences obtained from both the vertebrate host and ectoparasites.
However, when analyzing the other targeted gene fragments, the detected genotypes showed to be exclusive only to the vertebrate host or only to the sampled flies. In the phylogenetic analysis based on the rpoB and nuoG genes, we also observed that sequences obtained from ectoparasites samples clustered together among themselves and in different branches from those obtained from blood samples collected from their respective bat hosts. Even though one sequence from a bat blood sample (P. lineatus #77) and one sequence from M. aranea (#120) clustered together in the ftsZ gene phylogeny, both represent one genotype each. In fact, all the three sequences obtained from this marker were separated in three different genotypes. These findings might be explained by the symbiotic character of some Bartonella spp. in arthropods that are not present in vertebrate hosts, as previously shown [38,79].
Interestingly, in the nuoG phylogenetic tree, four sequences obtained from bat flies were positioned as more ancient groups. Previously, McKee et al. (2020) [29] raised the hypothesis that Bartonella species evolved from symbionts found in blood-feeding ectoparasites once these arthropods depend on symbionts for additional nutrients [80].
Despite the high genetic diversity of Bartonella spp. previously reported in bat ectoparasites [35], the present work found a lower genetic diversity among the gltA sequences obtained from flies when compared to those found in blood samples from bats. Only one bat fly sample showed more than one Bartonella gltA genotype among the three clones obtained from the same sample, while eight bat blood samples showed two or three different genotypes. In addition, Bartonella sequences detected in bat blood samples showed a higher number of variable sites, but without significant statistical difference when compared to sequences obtained from bat flies. Sandór et al. (2018) [81] proposed that a lower genetic diversity of Bartonella in bat flies could be related to a high exchange of hosts, since they would not be able to maintain a diversity of genotypes, despite having more access to it. On the other hand, flies with a lower host exchange rate would present a greater capacity to maintain the acquired genotypes since they would maintain them for a longer time. Conversely, some authors pointed out the possibility that a greater diversity of bartonellae in bat flies would be the opposite and related to the frequent change of hosts [38], as well as their lack of specificity [79], since this kind of behavior would allow the infection with different genotypes. Finally, we cannot rule out the possibility of greater Bartonella diversity among bats than in flies due to the number of samples analyzed, i.e., gltA amplicons for cloning were obtained from only three flies whereas 14 amplicons were obtained from bat blood samples. In addition, Becker et al. (2018) [39] suggested that the different levels of bartonellae genetic diversity between bats and their associated ectoparasites may also be related to the dispersion of this group of bacteria by other arthropod vectors or other bat species involved in the dynamic transmission cycles. However, as pointed out by McKee et al. (2020) [29], while strict ectoparasites vectors may maintain enzootic Bartonella infections, generalist vectors can carry the bacteria to other phylogenetically distant hosts. Moreover, the same authors incriminated bats as the likely key for Bartonella diversification and spreading.
Mckee et al. (2019; 2020) [29,33] has shown that bat-associated Bartonella genotypes were scattered positioned in the phylogenetic trees versus those described in other mammal orders. Even though the Bartonella phylogeny is congruent with the host phylogeny, bat-related Bartonella clusters appeared to be polyphyletic, which is consistent to the topology observed in the phylogenetic analyses from the present work and from previous studies performed by our research group involving non-hematophagous bats [12], vampire bats [40], and Streblidae bat flies [41]. Herein, the sequences obtained from ectoparasites and bat samples appeared as branches distributed throughout the tree, showing that bartonellae detected in both bats and flies are very diverse. Even though Bartonella DNA detected in ectoparasites can be retrieved from the host as a product of blood meal, the cloned sequences obtained from the ectoparasite #79 clustered in a different branch from that where vertebrate host-associated sequences were positioned (bat blood sample #79) and therefore showed to be different from each other. Recently, Braga et al. (2020) [82] detected Bartonella DNA in 3 out 15 Trichobius spp. bat flies sampled in northeastern Brazil, despite the fact that all 29 blood samples from the associated bat hosts were negative. The gltA and rpoB Bartonella genotypes detected in that study were closely related to genotypes previously detected in bats from Brazil and Costa Rica [82]. It is important to highlight that the detection of Bartonella DNA in ectoparasites does not prove the vectorial competence and capacity of a certain vector for this group of bacteria. Also, we could observe that the found Bartonella sequences usually clustered together or were closely related to sequences previously obtained from bats and ectoparasites from Peru, Guatemala, French Guiana and Brazil. This clustering pattern was previously observed when Stuckey et al. (2017) [83] analyzed Bartonella sequences from bats from Mexico, which were positioned along with sequences obtained from previous works performed in Latin America. Similarly, Muller et al. (2020) [84] also showed a high diversity of Bartonella genotypes in bats from Chile, which also clustered with bartonellae detected in bats from Central and South America. Interestingly, some genotypes clustered with sequences obtained from bats from Asia, Africa and Europe. Herein, one sequence (from a bat P. lineatus blood sample #77) was positioned in the same branch with a rodent-related Bartonella sequence in the ftsZ phylogenic analysis. This phylogenetic relatedness between Bartonella sequences from bats and rodents has already been observed by our research group [12,41] and recently by Muller et al. 2020 [82].
Although Bartonella has already been detected in Macronyssidae mites from rodents [85], few works have reported the occurrence of bartonellae in bat-associated mites. Here, we found positivity to Bartonella spp. in five out of 21 pooled samples from Macronyssidae mites (Steatonyssus spp.) and none of the 46 pooled Spinturnicidae mites. Reeves et al. (2006) [86] reported positivity for a Bartonella spp. showing 96% identicalness to B. grahamii in a Steatonyssus spp. mite collected from a bat. In Hungary, Hornok et al. (2012) [87] detected Bartonella spp. in 25% (1/4) Macronyssidae (Steatonyssus ocidentalis) and 100% (3/3) Spinturnicidae (Spinturnix myoti) mite pooled samples collected from Myotis myotis specimens. Recently, 28% of the S. myoti (Spinturnicidae) mite pooled samples collected from bats from Poland were positive to Bartonella spp. [88]. Previously in Brazil, do Amaral et al. (2018) [41] did not find positivity among 200 Macronyssidae and Spinturnicidae collected from bats in the state of Rio de Janeiro. It is important to highlight that the pooled mite samples that were positive for Bartonella were collected in PCR negative animals and did not yield positive results in the additional conventional PCR assays.
None of the Ornithodoros tick larvae samples were positive to Bartonella spp. in the present study. Similarly, Tahir et al. (2016) [89] found all 107 larvae of Ornithodoros hasei from Noctilio albiventris collected in French Guiana to be negative for Bartonella spp. On the other hand, Davoust et al. (2016) [90] found four out of 105 O. hasei ticks positive for Bartonella spp. The ticks were collected from 4 N. albiventris also in French Guiana Noteworthy, zoonotic-related Bartonella has already been detected in ticks associated to bats previously. Lofti et al. (2005) [91] detected Bartonella henselae 16S-23S ITS in a single Carios kelleyi nymph sampled in residential and community buildings in Iowa, and Leulmi et al. (2016) [92] detected Bartonella tamiae in 12/19 Ixodes vespertilionis ticks collected from bats in Algeria. Even though Bartonella spp. has been detected in several tick species [93,94], there is no strong evidence that ticks can be competent vectors for Bartonella spp. [95,96]. So far, it has been shown that Ixodes ricinus tick may act as competent vectors for B. henselae in laboratory [97].
5. Conclusions
We showed the genetic diversity of Bartonella genotypes in bat blood and ectoparasite samples at both intra- and inter-hosts levels. Based on the gltA molecular marker, we found a high genotypic diversity in both bat and fly hosts, which may lead to a complex host-pathogen interaction and displays a new perspective on the role of these animals in the maintenance of Bartonella spp. in the environment. Herein, the results obtained for rpoB, nuoG and ftsZ markers highlight the consistency with previous works that showed the genetic diversity of Bartonella in bats and associated ectoparasites. Since bats represent a large and widespread group of mammals around the world and Bartonella appears to have high genotypic plasticity, these flying mammals have been pointed out as key hosts in the origin and spread of these important zoonotic bacteria.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1822/s1, Table S1. Identification of non-hematophagous bat species and associated ectoparasites sampled in Campo Grande city, state of Mato Grosso do Sul, Brazil. Table S2. Results of quantification and positivity of bat blood and ectoparasite samples tested for Bartonella spp. by qPCR (nuoG gene) and conventional (c) PCR assays for the gltA, rpoB, ftsZ and nuoG genes. The sequenced samples are highlighted in dark yellow. The sequenced samples are highlighted in dark yellow, Table S3. Results of nBLAST analysis with correspondent GenBank acession number found for each sequenced clone obtained from bat blood and fly samples with the correspondent genotype for each sample.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by P.I.; J.M.T.; L.P., H.M.H., C.E.d.O., R.Z.M., E.C.L. and M.R.A. The first draft of the manuscript was written by P.I. and M.R.A. and all authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FAPESP (The São Paulo Research Foundation—Process #2018/02753-0 and #2017/14124-4); FUNDECT (Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul, Case 59/300.187/2016); CNPq (National Council for Scientific and Technological Development) for the Productivity Grant to MRA (CNPq Process #302420/2017-7) and HMH (CNPq Process #308768/2017-5); FAPERJ/CAPES—E-26/202.158/2015—Rio de Janeiro Post-Doctoral Research Support Program for the stipends conceded to ECL. The funders had no role in the design of the study, in the sampling, analyses, interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
The authors are thankful to the Research Group InsanaHuna (Campo Grande-MS, Brazil) for assistance in the field work support; and to Imunodot Diagnósticos Ltda (Jaboticabal-SP, Brazil), specially to Márcia Mariza Jusi Merino, for the technical support in the cloning procedures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk Factors for Human Disease Emergence. Philos. Trans. R. Soc. B Biol. Sci. 2001, 356, 983–989. [Google Scholar] [CrossRef]
- Musso, T.; Badolato, R.; Ravarino, D.; Stornello, S.; Panzanelli, P.; Merlino, C.; Savoia, D.; Cavallo, R.; Ponzi, A.N.; Zucca, M. Interaction of Bartonella henselae with the murine macrophage cell line J774: Infection and proinflammatory response. Infect. Immun. 2001, 69, 5974–5980. [Google Scholar] [CrossRef][Green Version]
- Eicher, S.C.; Dehio, C. Bartonella entry mechanisms into mammalian host cells. Cell. Microbiol. 2012, 14, 1166–1173. [Google Scholar] [CrossRef]
- Schipper, J.; Chanson, J.S.; Chiozza, F.; Cox, N.A.; Hoffman, M.; Katariya, V.; Lamoreux, J.; Rodrigues, A.S.L.; Stuart, S.N.; Temple, H.J.; et al. The status of the world’s land and marine mammals: Diversity, threat, and knowledge. Science 2008, 322, 225–230. [Google Scholar] [CrossRef]
- Nogueira, M.R.; Lima, I.P.; Garbino, G.S.T.; Moratelli, R.; Tavares, V.C.; Gregorin, R.; Peracchi, A.L. Updated checklist of Brazilian bats: Version 2018.1. Comitê da Lista de Morcegos do Brasil - CLMB. Sociedade Brasileira para o Estudo de Quirópteros (Sbeq). 2018. Available online: http://www.sbeq.net/updatelist (accessed on 7 August 2020).
- Bai, Y.; Kosoy, M.; Recuenco, S.; Alvarez, D.; Moran, D.; Turmelle, A.; Ellison, J.; Garcia, D.L.; Estevez, A.; Lindblade, K.; et al. Bartonella spp. in bats, Guatemala. Emerg. Infect. Dis. 2011, 17, 1269–1272. [Google Scholar] [CrossRef]
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel Insights Into Immune Systems of Bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef]
- Mühldorfer, K. Bats and Bacterial Pathogens: A Review. Zoonoses Public Health 2013, 60, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of The United Nations. Investigating the Role of Bats in Emerging Zoonoses: Balancing Ecology, Conservation and Public Health Interests; Newman, S.H., Field, H.E., de Jong, C.E., Epstein, J.H., Eds.; FAO: Rome, Italy, 2011.
- D’Auria, S.R.; Camargo, M.C.; Pacheco, R.C.; Savani, E.S.M.M.; Dias, M.A.G.; da Rosa, A.R.; de Almeida, M.F.; Labruna, M.B. Serologic survey for rickettsiosis in bats from São Paulo city, Brazil. Vector Borne Zoonotic Dis. 2010, 10, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.Q.; Dos Reis, E.M.; Bezerra, A.V.A.; Cerva, C.; Rosa, J.; Cibulski, S.P.; Lima, F.E.S.; Pacheco, S.M.; Rodrigues, R.O. Pathogenic Leptospira spp. in bats: Molecular investigation in Southern Brazil. Comp. Immunol. Microbiol. Infect. Dis. 2017, 52, 14–18. [Google Scholar] [CrossRef]
- Ikeda, P.; Seki, M.C.; Carrasco, A.O.T.; Rudiak, L.V.; Miranda, J.M.D.; Gonçalves, S.M.M.; Hoppe, E.G.L.; Albuquerque, A.C.A.; Teixeira, M.M.G.; Passos, C.E.; et al. Evidence and molecular characterization of Bartonella spp. and hemoplasmas in neotropical bats in Brazil. Epidemiol. Infect. 2017, 145, 2038–2052. [Google Scholar] [CrossRef] [PubMed]
- Correia Dos Santos, L.; Vidotto, O.; Dos Santos, N.J.R.; Ribeiro, J.; Pellizzaro, M.; Dos Santos, A.P.; Haisi, A.; Wischral Jayme Vieira, T.S.; de Barros Filho, I.R.; Cubilla, M.P.; et al. Hemotropic mycoplasmas (hemoplasmas) in free-ranging bats from Southern Brazil. Comp. Immunol. Microbiol. Infect. Dis. 2020, 69, 101416. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.S.; Guterres, A.; Rozental, T.; Novaes, R.; Vilar, E.M.; Oliveira, R.C.; Fernandes, J.; Forneas, D.; Junior, A.A.; Brandão, M.L.; et al. Coxiella and Bartonella spp. in bats (Chiroptera) captured in the Brazilian Atlantic Forest biome. BMC Vet. Res. 2018, 14, 279. [Google Scholar] [CrossRef] [PubMed]
- Cabral, A.D.; D’Auria, S.R.; Camargo, M.C.; Rosa, A.R.; Sodré, M.M.; Galvão-Dias, M.A.; Jordão, L.R.; Dubey, J.P.; Gennari, S.M.; Pena, H.F. Seroepidemiology of Toxoplasma gondii infection in bats from São Paulo city, Brazil. Vet. Parasitol. 2014, 206, 293–296. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, R.F.; Rodrigues, G.M.; Silva, E.M.; Carneiro, A.J.; Franke, C.R.; de Magalhães Cunha, R.; Gondim, L.F. Toxoplasmatinae Parasites in Bats from Bahia State, Brazil. J. Wildl. Dis. 2017, 53, 144–147. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, F.M.; Costa, L.H.; de Barros, T.L.; Ito, P.K.; Colombo, F.A.; de Carvalho, C.; Pedro, W.A.; Queiroz, L.H.; Nunes, C.M. First detection of Leishmania spp. DNA in Brazilian bats captured strictly in urban areas. Acta Tropica 2015, 150, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, A.P.; Costa, F.B.; Soares, H.S.; Ramirez, D.G.; Mesquita, E.T.; Gennari, S.M.; Marcili, A. Trypanosoma cruzi and Leishmania infantum chagasi Infection in Wild Mammals from Maranhão State, Brazil. Vector Borne Zoonotic Dis. 2015, 15, 656–666. [Google Scholar] [CrossRef] [PubMed]
- De Rezende, M.B.; Herrera, H.M.; Carvalho, C.M.E.; Carvalho Anjos, E.A.; Ramos, C.A.N.; de Araújo, F.R.; Torres, J.M.; de Oliveira, C.E. Detection of Leishmania spp. in Bats from an Area of Brazil Endemic for Visceral Leishmaniasis. Transbound. Emerg. Dis. 2017, 64, e36–e42. [Google Scholar] [CrossRef]
- Gómez-Hernández, C.; Bento, E.C.; Rezende-Oliveira, K.; Nascentes, G.A.N.; Barbosa, C.G.; Batista, L.R.; Tiburcio, M.G.S.; Pedrosa, A.L.; Lages-Silva, E.; Ramírez, J.D.; et al. Leishmania infection in bats from a non-endemic region of Leishmaniasis in Brazil. Parasitology 2017, 144, 1980–1986. [Google Scholar] [CrossRef]
- Castro, L.S.; Dorval, M.E.C.; Matheus, L.M.D.; Bednaski, A.V.; Facco, G.G.; Silveira, M.; Santos, C.F.; Gontijo, C.M.F.; Oliveira, A.P.G.; Ferreira, E.C. Leishmania presence in bats in areas endemic for leishmaniasis in central-west Brazil. Int J. Parasitol. Parasites Wildl. 2020, 11, 261–267. [Google Scholar] [CrossRef]
- Dario, M.A.; Moratelli, R.; Schwabl, P.; Jansen, A.M.; Llewellyn, M.S. Small subunit ribosomal metabarcoding reveals extraordinary trypanosomatid diversity in Brazilian bats. PLoS Negl. Trop. Dis. 2017, 11, e0005790. [Google Scholar] [CrossRef]
- Szpeiter, B.B.; Ferreira, J.I.G.D.S.; Assis, F.F.V.; Stelmachtchuk, F.N.; Peixoto, K.D.C., Jr.; Ajzenberg, D.; Minervino, A.H.H.; Gennari, S.M.; Marcili, A. Bat trypanosomes from Tapajós-Arapiuns Extractive Reserve in Brazilian Amazon. Rev. Bras. Parasitol. Vet. 2017, 26, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Bento, E.C.; Gómez-Hernández, C.; Batista, L.R.; Anversa, L.; Pedrosa, A.L.; Lages-Silva, E.; Ramírez, J.D.; Ramirez, L.E. Identification of bat trypanosomes from Minas Gerais state, Brazil, based on 18S rDNA and Cathepsin-L-like targets. Parasitol Res. 2018, 117, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.H.S.; Lima, L.; Schubach, A.O.; Teixeira, M.M.G. Trypanosoma madeirae spp. n.: A species of the clade T. cruzi associated with the neotropical common vampire bat Desmodus rotundus. Int. J. Parasitol. Parasites Wildl. 2018, 8, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Rangel, D.A.; Lisboa, C.V.; Novaes, R.L.M.; Silva, B.A.; Souza, R.F.; Jansen, A.M.; Moratelli, R.; Roque, A.L.R. Isolation and characterization of trypanosomatids, including Crithidia mellificae, in bats from the Atlantic Forest of Rio de Janeiro, Brazil. PLoS Negl. Trop Dis. 2019, 13, e0007527. [Google Scholar] [CrossRef]
- Filgueiras, A.; Barros, J.H.D.S.; Xavier, S.C.C.; de Souza, S.F.; Medeiros, L.D.S.; Ribeiro, V.M.F.; Jansen, A.M.; Roque, A.L.R. Natural Trypanosoma (Trypanozoon) evansi (Steel, 1885) infection among mammals from Brazilian Amazon. Acta Tropica 2019, 190, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Chomel, B.B.; Stuckey, M.J.; Boulouis, H.J.; Aguilar-Setién, A. Bat-Related Zoonoses. In Zoonoses—Infections Affecting Humans and Animals: Focus on Public Health Aspects; Springer: Dordrecht, Germany, 2014; pp. 697–714. [Google Scholar] [CrossRef]
- McKee, C.D.; Bai, Y.; Webb, C.T.; Kosoy, M.Y. Bats are key hosts in the radiation of mammal-associated Bartonella bacteria. BioRxiv 2020. [Google Scholar] [CrossRef]
- Graciolli, G. Streblidae in Catálogo Taxonômico da Fauna do Brasil. PNUD. 2020. Available online: http://fauna.jbrj.gov.br/fauna/faunadobrasil/2624 (accessed on 7 August 2020.).
- Graciolli, G. Nycteribiidae in Catálogo Taxonômico da Fauna do Brasil. PNUD. 2020. Available online: http://fauna.jbrj.gov.br/fauna/faunadobrasil/2624 (accessed on 7 August 2020.).
- Moras, L.M.; Bernardi, L.F.O.; Graciolli, G.; Gregorin, R. Bat flies (Diptera: Streblidae, Nycteribiidae) and mites (Acari) associated with bats (Mammalia: Chiroptera) in a high-altitude region in southern Minas Gerais, Brazil. Acta Parasitologica 2013, 58, 556–563. [Google Scholar] [CrossRef]
- McKee, C.D.; Krawczyk, A.I.; Sándor, A.D.; Görföl6, T.; Földvári, M.; Földvári, G.; Dekeukeleire, D.; Haarsma, A.-J.; Kosoy, M.Y.; Webb, C.T.; et al. Host phylogeny, geographic overlap, and roost sharing shape parasite communities in European bats. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef]
- Bai, Y.; Recuenco, S.; Gilbert, A.T.; Osikowicz, L.M.; Go, J.; Rupprecht, C.; Kosoy, M.Y. Prevalence and diversity of Bartonella spp. in bats in Peru. Am. J. Trop. Med. Hyg. 2012, 87, 518–523. [Google Scholar] [CrossRef]
- Morse, S.F.; Olival, K.J.; Kosoy, M.; Billeter, S.; Patterson, B.D.; Dick, C.W.; Dittmar, K. Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect. Genet. Evol. 2012, 12, 1717–1723. [Google Scholar] [CrossRef]
- Olival, K.J.; Dittmar, K.; Bai, Y.; Rostal, M.K.; Lei, B.R.; Daszak, P.; Kosoy, M.Y. Bartonella spp. in a Puerto Rican Bat Community. J. Wildl. Dis. 2015, 51, 274–278, 2015. [Google Scholar] [CrossRef] [PubMed]
- McKee, C.D.; Kosoy, M.Y.; Bai, Y.; Osikowicz, L.M.; Franka, R.; Gilbert, A.T.; Boonmar, S.; Rupprecht, C.E.; Peruski, L.F. Diversity and phylogenetic relationships among Bartonella strains from Thai bats. PLoS ONE 2017, 20. [Google Scholar] [CrossRef] [PubMed]
- Moskaluk, A.E.; Stuckey, M.J.; Jaffe, D.A.; Kasten, R.W.; Aguilar-Setién, A.; Olave-Leyva, J.I.; Galvez-Romero, G.; Obregón-Morales, C.; Salas-Rojas, M.; García-Flores, M.M.; et al. Molecular Detection of Bartonella Species in Blood-Feeding Bat Flies from Mexico. Vector Borne Zoonotic Dis. 2018, 18, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.J.; Bergner, L.M.; Bentz, A.B.; Orton, R.J.; Altizer, S.; Streicker, D.G. Genetic diversity, infection prevalence, and possible transmission routes of Bartonella spp. in vampire bats. PLoS Negl. Trop. Dis. 2018, 9. [Google Scholar] [CrossRef]
- André, M.R.; Gutiérrez, R.; Ikeda, P.; Do Amaral, R.B.; De Sousa, K.C.M.; Nachum-Biala, Y.; Lima, L.; Teixeira, M.M.G.; Machado, R.Z.; Harrus, S. Genetic diversity of Bartonella spp. in vampire bats from Brazil. Transbound. Emerg. Dis. 2019, 66, 2329–2341. [Google Scholar] [CrossRef]
- do Amaral, R.B.; Lourenço, E.C.; Famadas, K.M.; Garcia, A.B.; Machado, R.Z.; André, M.R. Molecular detection of Bartonella spp. and Rickettsia spp. in bat ectoparasites in Brazil. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Kosoy, M.; McKee, C.; Albayrak, L.; Fofanov, Y. Genotyping of Bartonella bacteria and their animal hosts: Current status and perspectives. Parasitology 2018, 145, 543–562. [Google Scholar] [CrossRef]
- Reis, N.R.; Peracchi, A.L.; Pedro, W.A.; De Lima, I.P. Morcegos do Brasil; Universidade Estadual de Londrina: Londrina-PR, Brazil, 2007; p. 253. [Google Scholar]
- Reis, N.R.; Peracchi, A.L.; Batista, C.B.; Lima, I.P.; Pereira, A.D. História Natural de Morcegos Brasileiros: Chave de Identificação de Espécies; Technical Books: Rio de Janeiro-RJ, Brazil, 2017; p. 416. [Google Scholar]
- Herrin, C.S.; Tripton, V.J. Spinturnicid mites of Venezuela (Acarina: Spinturnicidae). Brigham Young Univ. Sci. Bull. Biol. Ser. 1975, 20, 1–72. Available online: https://scholarsarchive.byu.edu/byuscib/vol20/iss2/1 (accessed on 7 August 2020).
- Wenzel, R.L. The Streblidae batflies of Venezuela (Diptera: Streblidae). Brigh. Young Univ. Sci. Bull. Biol. Ser. 1976, 20, 1–183. Available online: https://scholarsarchive.byu.edu/byuscib/vol20/iss4/1 (accessed on 7 August 2020). [CrossRef]
- Guimarães, J.H.; Tucci, E.C.; Battesti, D.M.B. Ectoparasitos de Importância Veterinária, 1st ed.; FAPEPS: São Paulo-SP, Brazil, 2001. [Google Scholar]
- Birkenheuer, A.J.; Levy, M.G.; Breitschwerdt, E.B. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B. canis DNA in canine blood samples. J. Clin. Microbiol. 2003, 41, 4172–4177. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Mangold, A.J.; Bargues, A.M.D.; Mas-Coma, A.S. Mitochondrial 16S rDNA sequences Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef] [PubMed]
- André, M.R.; Dumler, J.S.; Herrera, H.M.; Goncalves, L.R.; De Sousa, K.C.; Scorpio, D.G.; De Santis, A.C.G.A.; Domingos, I.H.; De Macedo, G.C.; Machado, R.Z. Assessment of a quantitative 5′ nuclease real-time polymerase chain reaction using the nicotinamide adenine dinucleotide dehydrogenase gamma subunit (nuoG) for Bartonella species in domiciled and stray cats in Brazil. J. Feline Med. Surg. 2015, 8, 783–790. [Google Scholar] [CrossRef]
- Colborn, J.M.; Kosoy, M.Y.; Motin, V.L.; Telepnev, M.V.; Valbuena, G.; Myint, K.S.; Fofanov, Y.; Putonti, C.; Feng, C.; Peruski, L. Improved detection of Bartonella DNA in mammalian hosts and arthropod vectors by real-time PCR using the NADH dehydrogenase gamma subunit (nuoG). J. Clin. Microbiol. 2010, 48, 4630–4633. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.; Ayers, M.; Mcclure, S.C.; Richardson, S.E.; Tellier, R. Detection and identification of Bartonella species pathogenic for humans by PCR amplification targeting the riboflavin synthase gene (ribC). J. Clin. Microbiol. 2003, 41, 1069–1072. [Google Scholar] [CrossRef]
- Norman, A.F.; Regnery, R.; Jameson, P.; Greene, C.; Krause, D.C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 1995, 33, 1797–1803. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Paziewska, A.; Harris, P.D.; Zwolińska, L.; Bajer, A.; Siński, E. Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents. Microb. Ecol. 2011, 61, 134–145. [Google Scholar] [CrossRef]
- Maggi, R.G.; Breitschwerdt, E.B. Isolation of bacteriophages from Bartonella vinsonii sub spp. berkhoffii and the characterization of pap-31 gene sequences from bacterial and phage DNA. J. Mol. Microbiol. Biotechnol. 2005, 9, 44–51. [Google Scholar] [CrossRef]
- Zeaiter, Z.; Fournier, P.-E.; Ogata, H.; Raoult, D. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int. J. Syst. Evol. Microbiol. 2002, 52, 165–171. [Google Scholar] [CrossRef]
- Maggi, R.G.; Breitschwerdt, E.B. Potential limitations of the 16S-23S rRNA intergenic region for molecular detection of Bartonella species. J. Clin. Microbiol. 2005, 43, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Ewing, B.; Green, P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef]
- Ewing, B.; Hillier, L.; Wendl, M.; Green, P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walke, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium Fort Lauderdale, Fort Lauderdale, FL, USA, 15–19 April 2002; Werner, B., Ed.; IEEE Computer Society: Washington, DC, USA, 2002; p. 184. [Google Scholar]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies, Molecular Biology and Evolution. Bioinformatics 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. 1999, 41, 95–98. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches Over Likelihood Ratio Tests. Syst Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stover, B.C.; Muller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Zeaiter, Z.; Khamis, A.; Raoult, D. Gene-sequence-based criteria for species definition in bacteriology: The Bartonella paradigm. Trends Microbiol. 2003, 11, 318–321. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Vayssier-Taussat, M.; Buffet, J.P.; Harrus, S. Guidelines for the Isolation, Molecular Detection, and Characterization of Bartonella Species. Vector Borne Zoonotic Dis. 2017, 17, 42–50. [Google Scholar] [CrossRef]
- Judson, S.D.; Frank, H.K.; Hadly, E.A. Bartonellae are Prevalent and Diverse in Costa Rican Bats and Bat Flies. Zoonoses Public Health 2015, 62, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Brook, C.E.; Bai, Y.; Dobson, A.P.; Osikowicz, L.M.; Ranaivoson, H.C.; Zhu, Q.; Kosoy, M.Y.; Dittmar, K. Bartonella spp. in fruit bats and blood-feeding ectoparasites in Madagascar. PLoS Negl. Trop. Dis. 2015, 9, e0003532. [Google Scholar] [CrossRef] [PubMed]
- Szentiványi, T.; Christe, P.; Glaizot, O. Bat Flies and Their Microparasites: Current Knowledge and Distribution. Front. Vet. Sci. 2019, 6, 115. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Tjale, M.A.; Weyer, J.; Kearney, T.; Seamark, E.C.J.; Nel, L.H.; Monadjem, A.; Markotter, W. Diversity of Bartonella and Rickettsia spp. in Bats and Their Blood-Feeding Ectoparasites from South Africa and Swaziland. PLoS ONE 2016, 11, e0152077. [Google Scholar] [CrossRef] [PubMed]
- Mckee, C.D.; Hayman, D.T.S.; Kosoy, M.Y.; Webb, C.T. Phylogenetic and geographic patterns of bartonella host shifts among bat species. Infect. Genet. Evol. 2016, 44, 382–394. [Google Scholar] [CrossRef]
- Husnik, F. Host–symbiont–pathogen interactions in blood-feeding parasites: Nutrition, immune cross-talk and gene exchange. Parasitology 2018, 145, 1294–1303. [Google Scholar] [CrossRef]
- Sándor, A.D.; Földvári, M.; Krawczyk, A.I.; Sprong, H.; Corduneanu, A.; Barti, L.; Görföl, T.; Estók, P.; Kováts, D.; Szekeres, S.; et al. Eco-epidemiology of Novel Bartonella Genotypes from Parasitic Flies of Insectivorous Bats. Microb Ecol. 2018, 76, 1076–1088. [Google Scholar] [CrossRef]
- Braga, M.S.C.O.; Gonçalves, L.R.; Silva, T.M.V.; Costa, F.B.; Pereira, J.G.; Santos, L.S. Occurrence of Bartonella genotypes in bats and associated Streblidae flies from Maranhão state, northeastern Brazil. Braz. J. Vet. Parasitol. 2020, 29, e014420. [Google Scholar] [CrossRef]
- Stuckey, M.J.; Chomel, B.B.; Galvez-Romero, G.; Olave-Leyva, J.I.; Obregón-Morales, C.; Moreno-Sandoval, H.; Aréchiga-Ceballos, N.; Salas-Rojas, M.; Aguilar-Setién, A. Bartonella Infection in Hematophagous, Insectivorous, and Phytophagous Bat Populations of Central Mexico and the Yucatan Peninsula. Am. J. Trop. Med. Hyg. 2017, 97, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Sepúlveda, P.; Di Cataldo, S.; Cevidanes, A.; Lisón, F.; Millán, J. Molecular investigation of zoonotic intracellular bacteria in Chilean bats. Comp Immunol Microbiol Infect Dis. 2020, 73, 101541. [Google Scholar] [CrossRef] [PubMed]
- Reeves, W.K.; Loftis, A.D.; Szumlas, D.E.; Abbassy, M.M.; Helmy, I.M.; Hanafi, H.A.; Dasch, G.A. Rickettsial pathogens in the tropical rat mite Ornithonyssus bacoti (Acari: Macronyssidae) from Egyptian rats (Rattus spp.). Exp. Appl. Acarol. 2007, 41, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Reeves, W.K.; Dowling, A.P.G.; Dasch, G.A. Rickettsial agents from parasitic dermanyssoidea (Acari: Mesostigmata). Exp. Appl. Acarol. 2006, 38, 181–188. [Google Scholar] [CrossRef]
- Hornok, S.; Kovács, R.; Meli, M.L.; Gönczi, E.; Hofmann-Lehmann, R.; Kontschán, J.; Gyuranecz, M.; Dán, A.; Molnár, V. First detection of bartonellae in a broad range of bat ectoparasites. Vet. Microbiol. 2012, 159, 541–543. [Google Scholar] [CrossRef]
- Szubert-Kruszyńska, A.; Stańczak, J.; Cieniuch, S.; Podsiadły, E.; Postawa, T.; Michalik, J. Bartonella and Rickettsia Infections in Haematophagous Spinturnix myoti Mites (Acari: Mesostigmata) and their Bat Host, Myotis myotis (Yangochiroptera: Vespertilionidae), from Poland. Microb. Ecol. 2019, 77, 759–768. [Google Scholar] [CrossRef]
- Tahir, D.; Socolovschi, C.; Marié, J.-L.; Ganay, G.; Berenger, J.-M.; Bompar, J.-M.; Blanchet, D.; Cheuret, M.; Mediannikov, O.; Raoult, D.; et al. New Rickettsia species in soft ticks Ornithodoros hasei collected from bats in French Guiana. Ticks Tick-Borne Dis. 2016, 6, 1089–1096. [Google Scholar] [CrossRef]
- Davoust, B.; Marié, J.-L.; Dahmani, M.; Berenger, J.-M.; Bompar, J.-M.; Blanchet, D.; Cheuret, M.; Raoult, D.; Mediannikov, O. Evidence of Bartonella spp. in Blood and Ticks (Ornithodoros hasei) of Bats, in French Guiana. Vector Borne Zoonotic Dis. 2016, 16, 516–519. [Google Scholar] [CrossRef]
- Loftis, A.D.; Gill, J.S.; Schriefer, M.E.; Levin, M.L.; Eremeeva, M.E.; Gilchrist, M.J.; Dasch, G.A. Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Argasidae). J. Med. Entomol. 2005, 42, 473–480. [Google Scholar] [CrossRef]
- Leulmi, H.; Aouadi, A.; Bitam, I.; Bessas, A.; Benakhla, A.; Raoult, D.; Parola, P. Detection of Bartonella tamiae, Coxiella burnetii and rickettsiae in arthropods and tissues from wild and domestic animals in northeastern Algeria. Parasit. Vectors 2016, 9, 27. [Google Scholar] [CrossRef]
- Billeter, S.A.; Levy, M.G.; Chomel, B.B.; Breitschwerdt, E.B. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med. Vet. Entomol. 2008, 22, 1–15. [Google Scholar] [CrossRef]
- Okaro, U.; Addisu, A.; Casanas, B.; Anderson, B. Bartonella species, an emerging cause of blood-culture-negative endocarditis. Clin. Microbiol. Rev. 2017, 30, 709–746. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Billeter, S.A.; Breitschwerdt, E.B.; Chomel, B.B.; Raoult, D. Potential for Tick-borne Bartonelloses. Emerg. Infect. Dis. 2010, 16, 385–391. [Google Scholar] [CrossRef]
- Telford III, S.R.; Wormser, G.P. Bartonella spp. Transmission by Ticks Not Established. Emerg. Infect. Dis. 2010, 16, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Cotté, V.; Bonnet, S.; Le Rhun, D.; Le Naour, L.; Chauvin, A.; Boulouis, H.-J.; Lecuelle, B.; Lilin, T.; Vayssier-Taussat, M. Transmission of Bartonella henselae by Ixodes ricinus. Emerg. Infect. Dis. 2008, 14, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).