Performance of Population Pharmacokinetic Models in Predicting Polymyxin B Exposures
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Sites, and Patient Selection
2.2. Pharmacokinetic Assessment
2.3. Predictive Performance
2.4. Ethical Approval
3. Results
3.1. Demographics
3.2. Pharmacokinetics
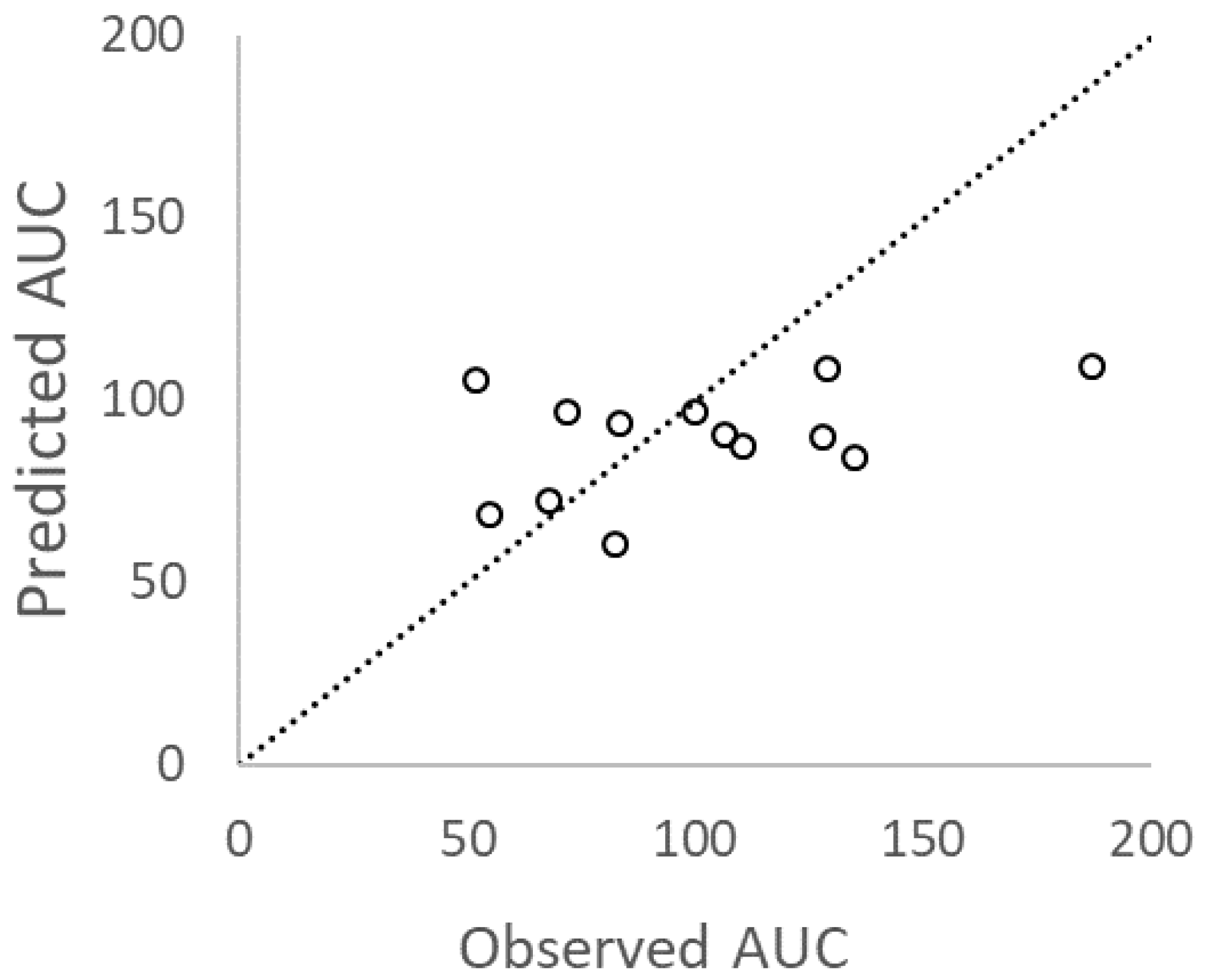
3.3. Predictive Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Model | Sandri | Manchandani | Avedissian |
|---|---|---|---|
| Equation | 0.0276 × 75 | N/A | (8.65 × (657.84))/((657.84) + (141.247.84)) + 1.43 |
| Predicted clearance (L/h) | 2.07 | 2.5 | 1.45 |
| Predicted AUC24 (mg.h/L) | 150/2.07 = 72 | 150/2.5 = 60 | 150/1.45 = 103 |
| Model | Sandri (2 comp) | Manchandani (1 comp) | Kubin (1 comp) | Avedissian (2 comp) | Miglis (2 comp) |
|---|---|---|---|---|---|
| 1- Compartment | 0.01 | 0.16 | 0.16 | 0.06 | 0.16 |
| 2- Compartment | 0.20 | 0.004 | 0.004 | 0.04 | 0.004 |
Appendix B
References
- Neuner, E.A.; Yeh, J.Y.; Hall, G.S.; Sekeres, J.; Endimiani, A.; Bonomo, R.A.; Shrestha, N.K.; Fraser, T.G.; van Duin, D. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagn. Microbiol. Infect. Dis 2011, 69, 357–362. [Google Scholar] [CrossRef]
- Tam, V.H.; Rogers, C.A.; Chang, K.T.; Weston, J.S.; Caeiro, J.P.; Garey, K.W. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob. Agents Chemother. 2010, 54, 3717–3722. [Google Scholar] [CrossRef]
- Tsuji, B.T.; Pogue, J.M.; Zavascki, A.P.; Paul, M.; Daikos, G.L.; Forrest, A.; Giacobbe, D.R.; Viscoli, C.; Giamarellou, H.; Karaiskos, I.; et al. International consensus guidelines for the optimal use of the polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 2019, 39, 10–39. [Google Scholar] [PubMed]
- Nelson, B.C.; Eiras, D.P.; Gomez-Simmonds, A.; Loo, A.S.; Satlin, M.J.; Jenkins, S.G.; Whittier, S.; Calfee, D.P.; Furuya, E.Y.; Kubin, C.J. Clinical outcomes associated with polymyxin B dose in patients with bloodstream infections due to carbapenem-resistant Gram-negative rods. Antimicrob. Agents Chemother. 2015, 59, 7000–7006. [Google Scholar] [CrossRef] [PubMed]
- Dubrovskaya, Y.; Prasad, N.; Lee, Y.; Esaian, D.; Figueroa, D.A.; Tam, V.H. Risk factors for nephrotoxicity onset associated with polymyxin B therapy. J. Antimicrob. Chemother. 2015, 70, 1903–1907. [Google Scholar] [CrossRef] [PubMed]
- Ouderkirk, J.P.; Nord, J.A.; Turett, G.S.; Kislak, J.W. Polymyxin B nephrotoxicity and efficacy against nosocomial infections caused by multiresistant gram-negative bacteria. Antimicrob. Agents Chemother. 2003, 47, 2659–2662. [Google Scholar] [CrossRef] [PubMed]
- Hee, K.H.; Leaw, Y.K.J.; Ong, J.L.; Lee, L.S. Development and validation of liquid chromatography tandem mass spectrometry method quantitative determination of polymyxin B1, polymyxin B2, polymyxin B3 and isoleucine-polymyxin B1 in human plasma and its application in clinical studies. J. Pharm. Biomed. Anal. 2017, 140, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Manchandani, P.; Dubrovskaya, Y.; Gao, S.; Tam, V.H. Comparative pharmacokinetic profiling of different polymyxin B components. Antimicrob. Agents Chemother. 2016, 60, 6980–6982. [Google Scholar] [CrossRef] [PubMed]
- Manchandani, P.; Thamlikitkul, V.; Dubrovskaya, Y.; Babic, J.T.; Lye, D.C.; Lee, L.S.; Tam, V.H. Population pharmacokinetics of polymyxin B. Clin. Pharmacol. Ther. 2018, 104, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Kubin, C.J.; Nelson, B.C.; Miglis, C.; Scheetz, M.H.; Rhodes, N.J.; Avedissian, S.N.; Cremers, S.; Yin, M.T. Population Pharmacokinetics of intravenous polymyxin B from clinical samples. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Miglis, C.; Rhodes, N.J.; Avedissian, S.N.; Kubin, C.J.; Yin, M.T.; Nelson, B.C.; Pai, M.P.; Scheetz, M.H. Population pharmacokinetics of polymyxin B in acutely ill adult patients. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Sandri, A.M.; Landersdorfer, C.B.; Jacob, J.; Boniatti, M.M.; Dalarosa, M.G.; Falci, D.R.; Behle, T.F.; Bordinhao, R.C.; Wang, J.; Forrest, A.; et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: Implications for selection of dosage regimens. Clin. Infect. Dis. 2013, 57, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Avedissian, S.; Miglis, C.; Kubin, C.J.; Rhodes, N.J.; Yin, M.T.; Cremers, S.; Prickett, M.; Scheetz, M.H. Polymyxin B pharmacokinetics in adult cystic fibrosis patients. Pharmacotherapy 2018, 38, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.H.; Schilling, A.N.; Vo, G.; Kabbara, S.; Kwa, A.L.; Wiederhold, N.P.; Lewis, R.E. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 3624–3630. [Google Scholar] [CrossRef] [PubMed]
- Landersdorfer, C.B.; Wang, J.; Wirth, V.; Chen, K.; Kaye, K.S.; Tsuji, B.T.; Li, J.; Nation, R.L. Pharmacokinetics/pharmacodynamics of systemically administered polymyxin B against Klebsiella pneumoniae in mouse thigh and lung infection models. J. Antimicrob. Chemother. 2018, 73, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Satlin, M.J.; Kubin, C.J.; Blumenthal, J.S.; Cohen, A.B.; Furuya, E.Y.; Wilson, S.J.; Jenkins, S.G.; Calfee, D.P. Comparative effectiveness of aminoglycosides, polymyxin B, and tigecycline for clearance of carbapenem-resistant Klebsiella pneumoniae from urine. Antimicrob. Agents Chemother. 2011, 55, 5893–5899. [Google Scholar] [CrossRef] [PubMed]
- Thamlikitkul, V.; Dubrovskaya, Y.; Manchandani, P.; Ngamprasertchai, T.; Boonyasiri, A.; Babic, J.T.; Tam, V.H. Dosing and pharmacokinetics of polymyxin B in patients with renal insufficiency. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.L.; Altshuler, J.; Guervil, D.J.; Hirsch, E.B.; Ostrosky-Zeichner, L.L.; Ericsson, C.D.; Tam, V.H. Cefepime free minimum concentration to minimum inhibitory concentration (fCmin/MIC) ratio predicts clinical failure in patients with Gram-negative bacterial pneumonia. Int. J. Antimicrob. Agents 2015, 45, 541–544. [Google Scholar] [CrossRef] [PubMed]

| Study Reference | Sandri [12] | Manchandani [9] | Kubin [10] | Avedissian [13] | Miglis [11] |
|---|---|---|---|---|---|
| Year published | 2013 | 2017 | 2018 | 2018 | 2018 |
| Sample size | 24 | 35 | 43 | 9 | 52 |
| Compartment model | 2 | 1 | 1 | 2 | 2 |
| % Male | 54.2 | 65.7 | 70.0 | N/A | 64.0 |
| Mean/Median age (years) | 61.5 | 58.7 | 58.0 | 55.5 | 47.0 |
| Patient origins | Brazil | U.S., Thailand, Singapore | U.S. | U.S. | U.S. |
| Patient type | Intensive care | N/A | N/A | Cystic fibrosis | Acutely ill |
| Clearance covariate | Body weight | None | None | Creatinine clearance | None |
| Mean % Bias | −0.7 | −16.6 | −12.0 | −5.2 | −30.0 |
| Mean % Precision | 28.5 | 22.3 | 20.5 | 43.6 | 31.2 |
| % subject AUC captured * | 76.9 | 84.6 | 84.6 | 84.6 ** | 84.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tam, V.H.; Lee, L.S.; Ng, T.-M.; Lim, T.-P.; Cherng, B.P.Z.; Adewusi, H.; Hee, K.H.; Ding, Y.; Chung, S.J.; Ling, L.-M.; et al. Performance of Population Pharmacokinetic Models in Predicting Polymyxin B Exposures. Microorganisms 2020, 8, 1814. https://doi.org/10.3390/microorganisms8111814
Tam VH, Lee LS, Ng T-M, Lim T-P, Cherng BPZ, Adewusi H, Hee KH, Ding Y, Chung SJ, Ling L-M, et al. Performance of Population Pharmacokinetic Models in Predicting Polymyxin B Exposures. Microorganisms. 2020; 8(11):1814. https://doi.org/10.3390/microorganisms8111814
Chicago/Turabian StyleTam, Vincent H., Lawrence S. Lee, Tat-Ming Ng, Tze-Peng Lim, Benjamin P. Z. Cherng, Hafeez Adewusi, Kim H. Hee, Ying Ding, Shimin Jasmine Chung, Li-Min Ling, and et al. 2020. "Performance of Population Pharmacokinetic Models in Predicting Polymyxin B Exposures" Microorganisms 8, no. 11: 1814. https://doi.org/10.3390/microorganisms8111814
APA StyleTam, V. H., Lee, L. S., Ng, T.-M., Lim, T.-P., Cherng, B. P. Z., Adewusi, H., Hee, K. H., Ding, Y., Chung, S. J., Ling, L.-M., Chlebicki, P., Kwa, A. L. H., & Lye, D. C. (2020). Performance of Population Pharmacokinetic Models in Predicting Polymyxin B Exposures. Microorganisms, 8(11), 1814. https://doi.org/10.3390/microorganisms8111814





