Comparison of the Performance and Microbial Community Structure of Two Outdoor Pilot-Scale Photobioreactors Treating Digestate
Abstract
1. Introduction
2. Materials and Methods
2.1. Pilot Plant System and Operation
2.2. Analytical Methods
2.3. Operational Data Processing
2.4. DNA Extraction and Sequencing
2.5. Bacterial Quantification by Real-Time Polymerase Chain Reaction
2.6. Bioinformatics
2.7. Statistical Analysis
3. Results
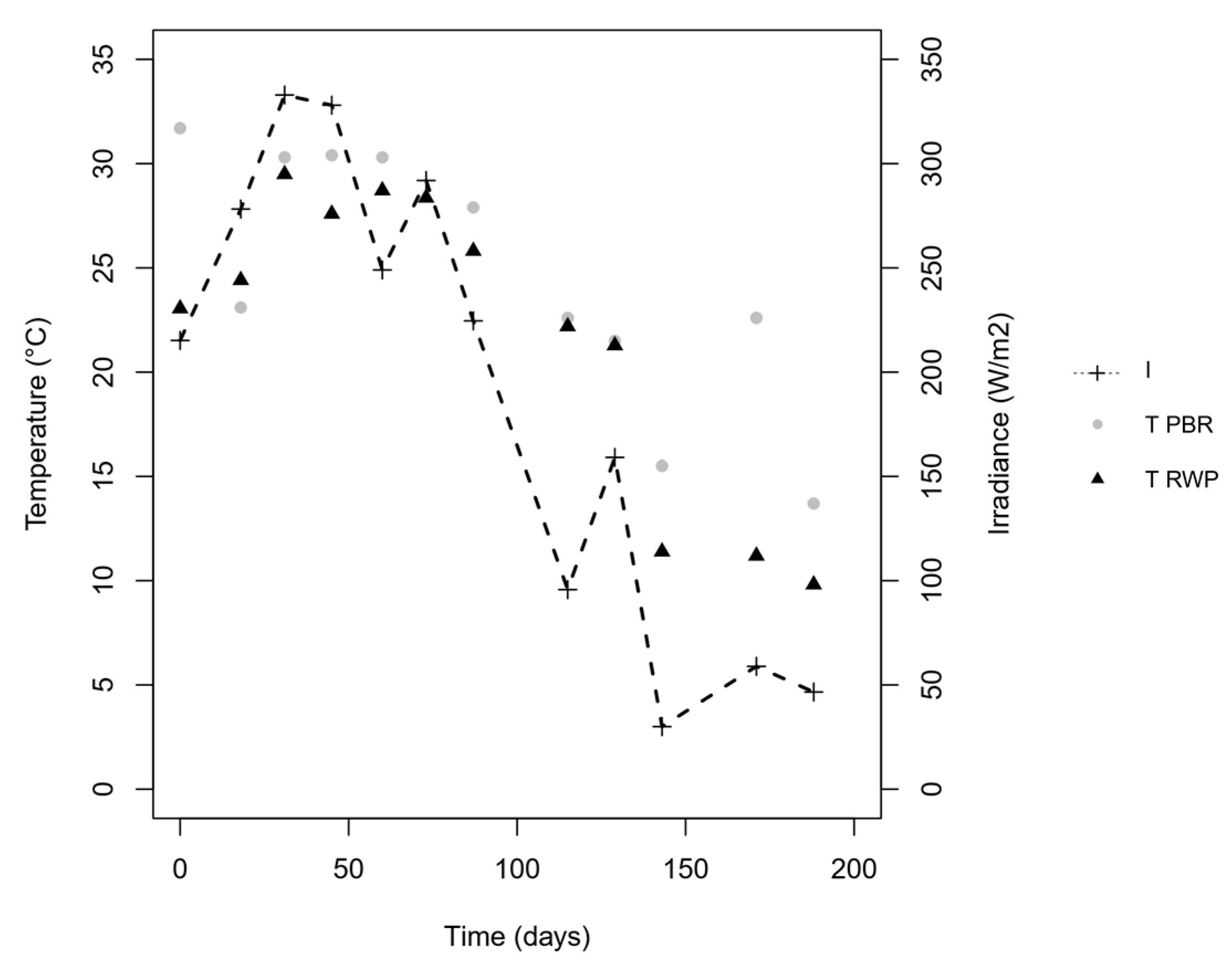
3.1. Performance of the Bioreactors
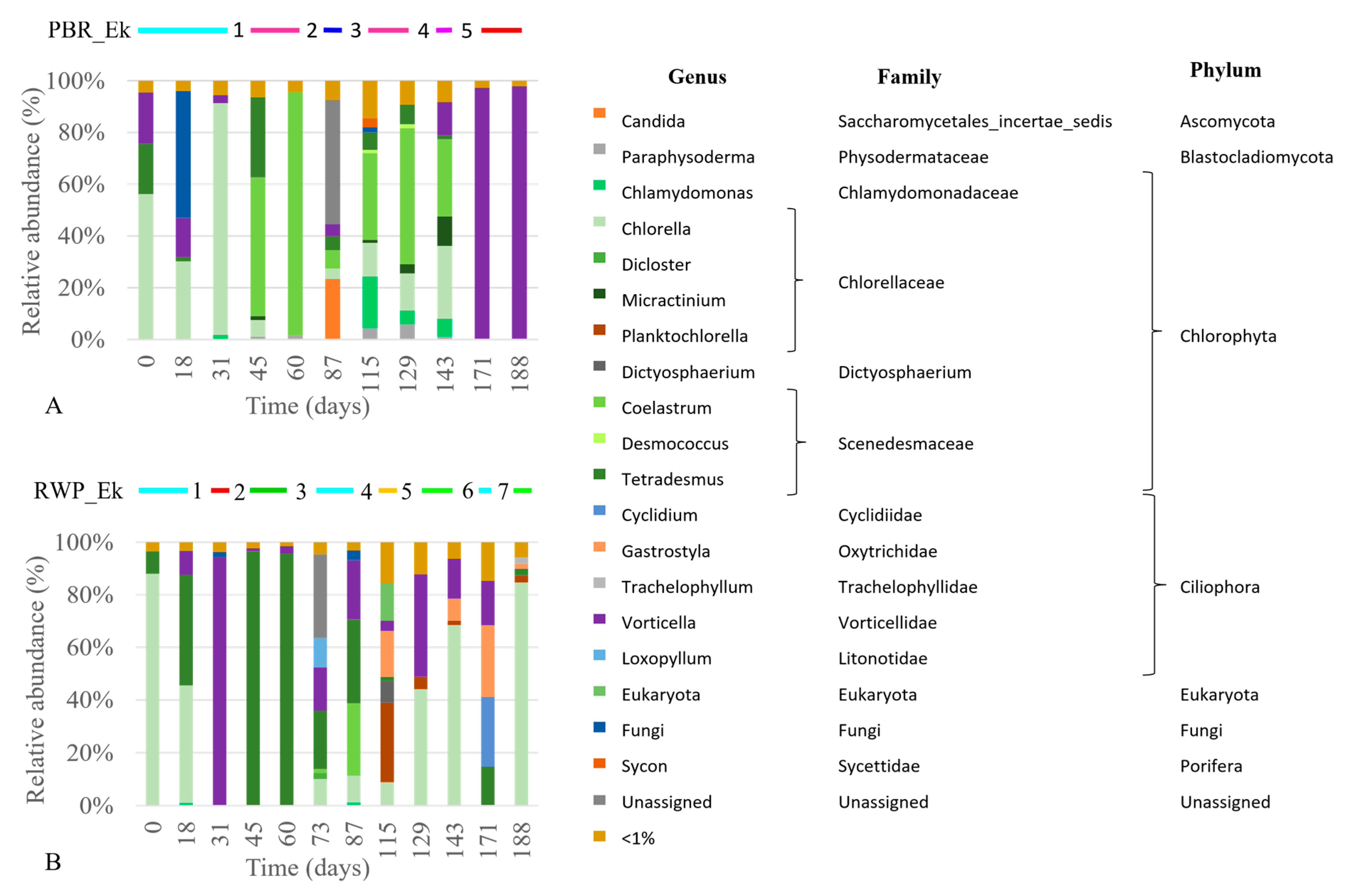
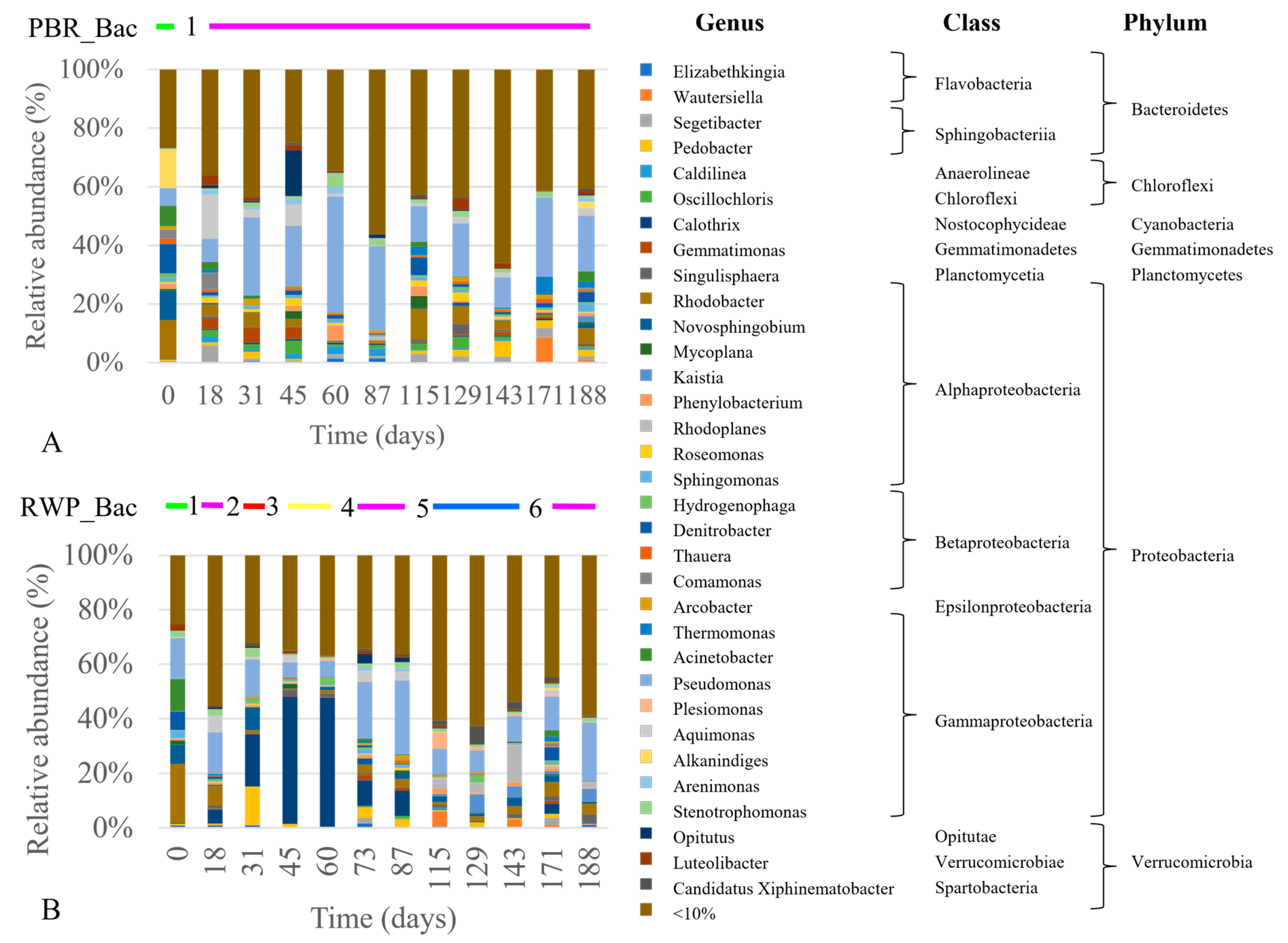
3.2. Biomass Productivity and Abundance of the Microbial Populations
3.3. Composition of the Eukaryotic and Bacterial Communities
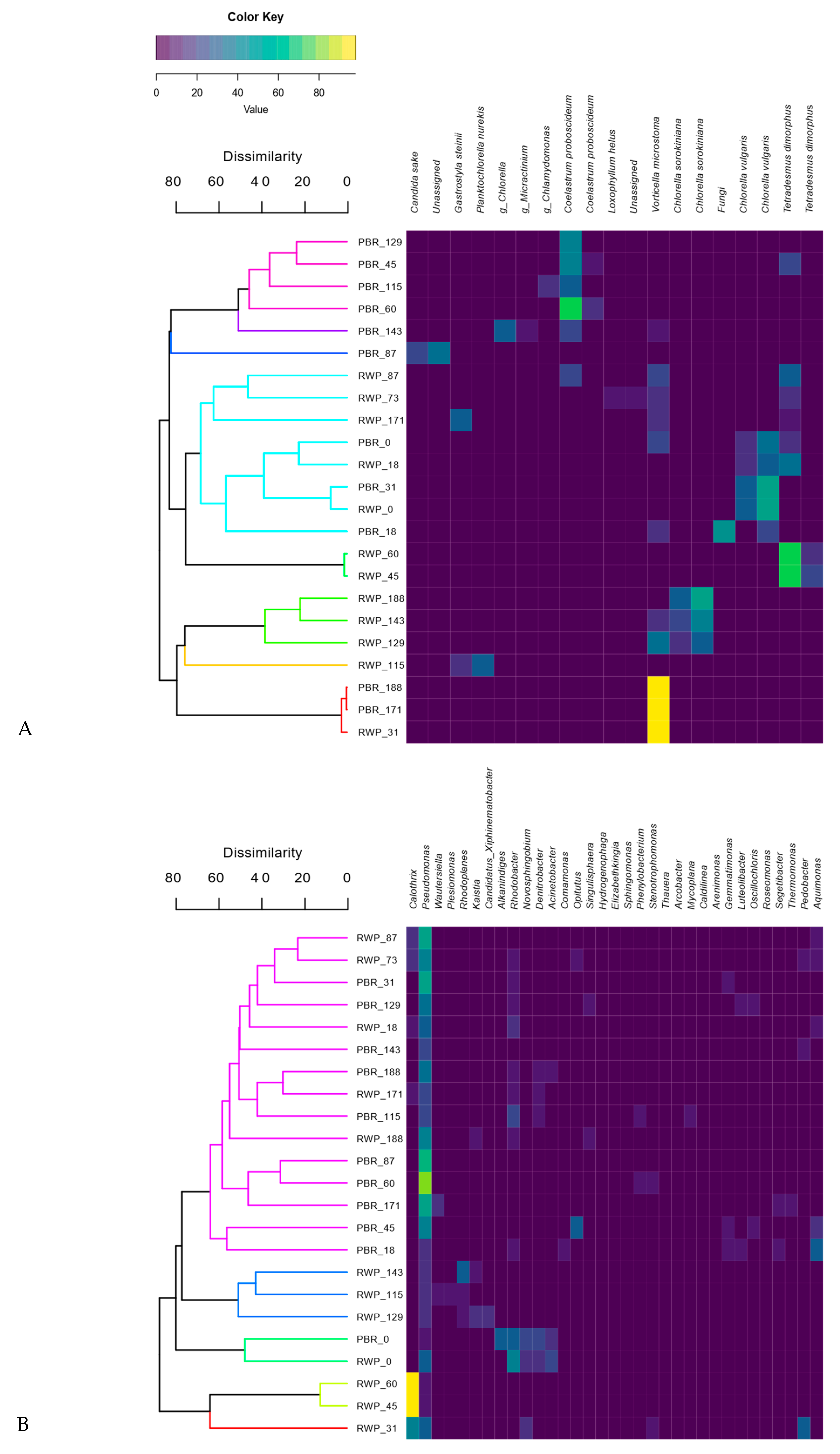
3.4. Diversity of the Eukaryotic and Bacterial Communities
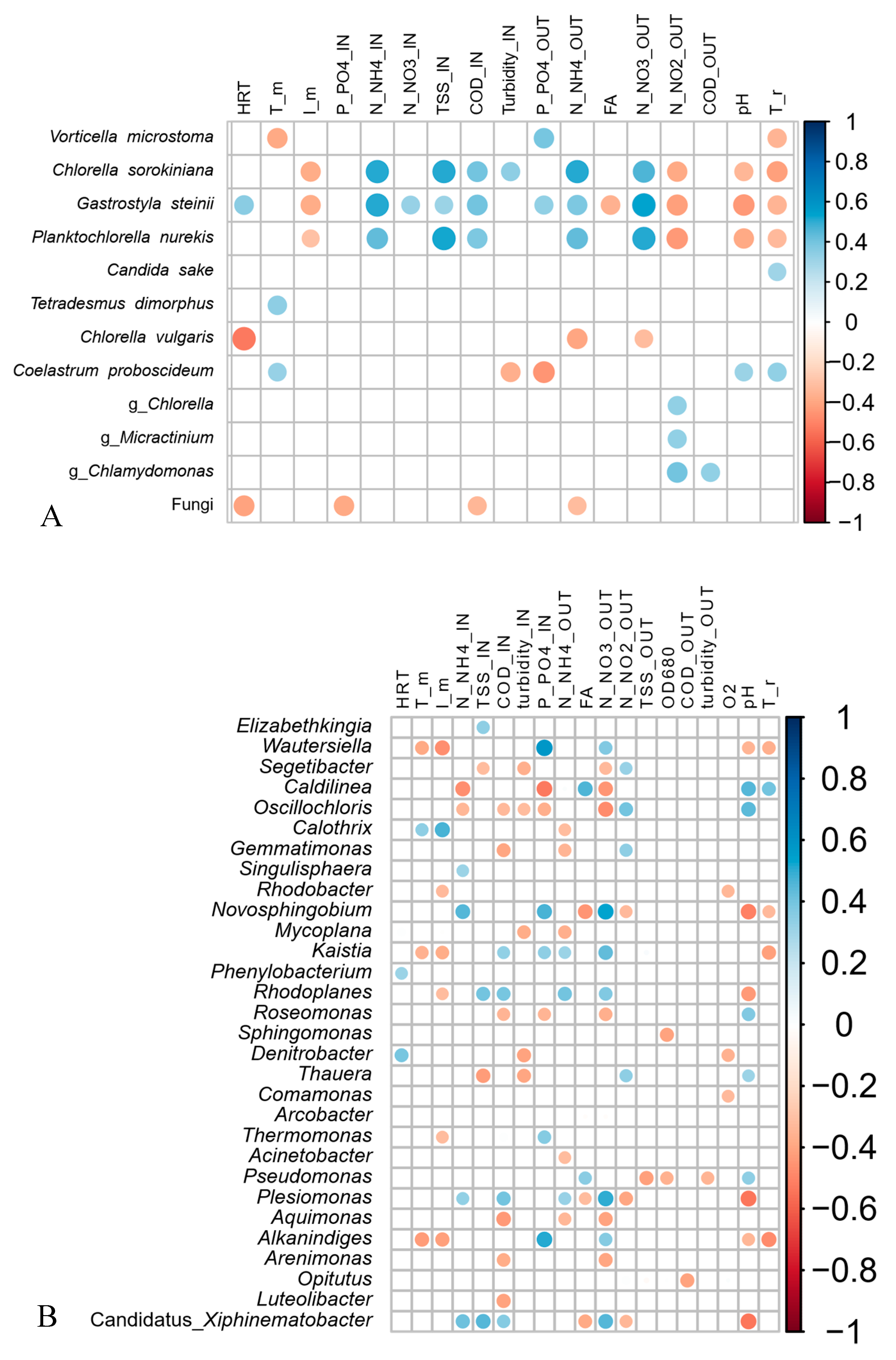
3.5. Associations between the Microbial Community Structure and the Abiotic and Biotic Parameters
3.6. Network Analyses
4. Discussion
4.1. Environmental Parameters, Biomass Productivity and Nutrients Removal in the Photobioreactors
4.2. Microbial Community Structure
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Puyol, D.; Batstone, D.J.; Hülsen, T.; Astals, S.; Peces, M.; Krömer, J.O. Resource Recovery from Wastewater by Biological Technologies: Opportunities, Challenges, and Prospects. Front. Microbiol. 2017, 7, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Subashchandrabose, S.R.; Ramakrishnan, B.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. Consortia of cyanobacteria/microalgae and bacteria: Biotechnological potential. Biotechnol. Adv. 2011, 29, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, M.; Marazzi, F.; Naddeo, L.S.; Piergiacomo, F.; Beneduce, L.; Ficara, E.; Mezzanotte, V. Disinfection and nutrient removal in laboratory-scale photobioreactors for wastewater tertiary treatment. J. Chem. Technol. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Marazzi, F.; Bellucci, M.; Fantasia, T.; Ficara, E.; Mezzanotte, V. Interactions between Microalgae and Bacteria in the Treatment of Wastewater from Milk Whey Processing. Water 2020, 12, 297. [Google Scholar] [CrossRef]
- Huy, M.; Kumar, G.; Kim, H.W.; Kim, S.H. Photoautotrophic cultivation of mixed microalgae consortia using various organic waste streams towards remediation and resource recovery. Bioresour. Technol. 2018, 247, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.L.; Chu, W.L.; Phang, S.M. Use of Chlorella vulgaris for bioremediation of textile wastewater. Bioresour. Technol. 2010, 101, 7314–7322. [Google Scholar] [CrossRef]
- Trentin, G.; Bertucco, A.; Sforza, E. Mixotrophy in Synechocystis sp. for the treatment of wastewater with high nutrient content: Effect of CO2 and light. Bioprocess Biosyst. Eng. 2019, 42, 1661–1669. [Google Scholar] [CrossRef]
- Xia, A.; Murphy, J.D. Microalgal cultivation in treating liquid digestate from biogas systems. Trends Biotechnol. 2016, 34, 264–275. [Google Scholar] [CrossRef]
- Lavrinovičs, A.; Juhna, T. Review on Challenges and Limitations for Algae-Based Wastewater Treatment. Constr. Sci. 2018, 20, 17–25. [Google Scholar] [CrossRef]
- Fulbright, S.P.; Dean, M.K.; Wardle, G.; Lammers, P.J.; Chisholm, S. Molecular diagnostics for monitoring contaminants in algal cultivation. Algal Res. 2014, 4, 41–51. [Google Scholar] [CrossRef]
- Ofiţeru, I.D.; Lunn, M.; Curtis, T.P.; Wells, G.F.; Criddle, C.S.; Francis, C.A.; Sloan, W.T. Combined niche and neutral effects in a microbial wastewater treatment community. Proc. Natl. Acad. Sci. USA 2010, 107, 15345–15350. [Google Scholar] [CrossRef] [PubMed]
- Van Der Gast, C.J.; Ager, D.; Lilley, A.K. Temporal scaling of bacterial taxa is influenced by both stochastic and deterministic ecological factors. Environ. Microbiol. 2008, 10, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Day, J.G.; Gong, Y.; Hu, Q. Microzooplanktonic grazers—A potentially devastating threat to the commercial success of microalgal mass culture. Algal Res. 2017, 27, 356–365. [Google Scholar] [CrossRef]
- Mark Ibekwe, A.; Murinda, S.E.; Murry, M.A.; Schwartz, G.; Lundquist, T. Microbial community structures in high rate algae ponds for bioconversion of agricultural wastes from livestock industry for feed production. Sci. Total Environ. 2017, 580, 1185–1196. [Google Scholar] [CrossRef]
- Sutherland, D.L.; Turnbull, M.H.; Craggs, R.J. Environmental drivers that influence microalgal species in fullscale wastewater treatment high rate algal ponds. Water Res. 2017, 124, 504–512. [Google Scholar] [CrossRef]
- Cho, D.-H.; Choi, J.-W.; Kang, Z.; Kim, B.-H.; Oh, H.-M.; Kim, H.; Ramanan, R. Microalgal diversity fosters stable biomass productivity in open ponds treating wastewater. Sci. Rep. 2017, 7, 1979. [Google Scholar] [CrossRef]
- Rodolfi, L.; Chini Zittelli, G.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yuan, D.; Li, Y.; Ma, M.; Hu, Q.; Gong, Y. Contaminating microzooplankton in outdoor microalgal mass culture systems: An ecological viewpoint. Algal Res. 2016, 20, 258–266. [Google Scholar] [CrossRef]
- González-Camejo, J.; Aparicio, S.; Ruano, M.V.; Borrás, L.; Barat, R.; Ferrer, J. Effect of ambient temperature variations on an indigenous microalgae-nitrifying bacteria culture dominated by Chlorella. Bioresour. Technol. 2019, 290, 121788. [Google Scholar] [CrossRef]
- González-Camejo, J.; Montero, P.; Aparicio, S.; Ruano, M.V.; Borrás, L.; Seco, A.; Barat, R. Nitrite inhibition of microalgae induced by the competition between microalgae and nitrifying bacteria. Water Res. 2020, 172, 115499. [Google Scholar] [CrossRef]
- Xin, L.; Hong-ying, H.; Ke, G.; Jia, Y. Growth and nutrient removal properties of a freshwater microalga Scenedesmus sp. LX1 under different kinds of nitrogen sources. Ecol. Eng. 2010, 36, 379–381. [Google Scholar] [CrossRef]
- Lian, J.; Wijffels, R.H.; Smidt, H.; Sipkema, D. The effect of the algal microbiome on industrial production of microalgae. Microb. Biotechnol. 2018, 11, 806–818. [Google Scholar] [CrossRef] [PubMed]
- Krustok, I.; Odlare, M.; Shabiimam, M.A.; Truu, J.; Truu, M.; Ligi, T.; Nehrenheim, E. Characterization of algal and microbial community growth in a wastewater treating batch photo-bioreactor inoculated with lake water. Algal Res. 2015, 11, 421–427. [Google Scholar] [CrossRef]
- Lakaniemi, A.-M.; Hulatt, C.J.; Wakeman, K.D.; Thomas, D.N.; Puhakka, J. A Eukaryotic and prokaryotic microbial communities during microalgal biomass production. Bioresour. Technol. 2012, 124, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Beyter, D.; Tang, P.; Becker, S.; Hoang, T.; Bilgin, D.; Lim, W.; Peterson, T.C.; Mayfield, S.; Haerizadeh, F.; Shurin, J.B.; et al. Diversity, Productivity, and Stability of an Industrial Microbial Ecosystem. Appl. Environ. Microbiol. 2016, 82, 2494–2505. [Google Scholar] [CrossRef]
- Eland, L.E.; Davenport, R.J.; dos Santos, A.B.; Mota Filho, C.R. Molecular evaluation of microalgal communities in full-scale waste stabilisation ponds. Environ. Technol. 2019, 40, 1969–1976. [Google Scholar] [CrossRef]
- Ferro, L.; Hu, Y.O.O.; Gentili, F.G.; Andersson, A.F.; Funk, C. DNA metabarcoding reveals microbial community dynamics in a microalgae-based municipal wastewater treatment open photobioreactor. Algal Res. 2020, 51, 102043. [Google Scholar] [CrossRef]
- Pizzera, A.; Scaglione, D.; Bellucci, M.; Marazzi, F.; Mezzanotte, V.; Parati, K.; Ficara, E. Digestate treatment with algae-bacteria consortia: A field pilot-scale experimentation in a sub-optimal climate area. Bioresour. Technol. 2019, 274, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Anthonisen, A.; Loehr, R.; Prakasam, T.; Srinath, E. Inhibition of Nitrification by Ammonia and Nitrous Acid. J. Water Pollut. Control Fed. 1976, 48, 835–852. [Google Scholar]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Ferris, M.J.; Muyzer, G.; Ward, D.M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 1996, 62, 340–346. [Google Scholar] [CrossRef]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef] [PubMed]
- Marazzi, F.; Bellucci, M.; Fornaroli, R.; Bani, A.; Ficara, E.; Mezzanotte, V. Lab-scale testing of operation parameters for algae based treatment of piggery wastewater. J. Chem. Technol. Biotechnol. 2020, 95, 967–974. [Google Scholar] [CrossRef]
- Bani, A.; Borruso, L.; Matthews Nicholass, K.J.; Bardelli, T.; Polo, A.; Pioli, S.; Gómez-Brandón, M.; Insam, H.; Dumbrell, A.J.; Brusetti, L. Site-Specific Microbial Decomposer Communities Do Not Imply Faster Decomposition: Results from a Litter Transplantation Experiment. Microorganisms 2019, 7, 349. [Google Scholar] [CrossRef]
- Bellucci, M.; Ofiţeru, I.D.; Beneduce, L.; Graham, D.W.; Head, I.M.; Curtis, T.P. A preliminary and qualitative study of resource ratio theory to nitrifying lab-scale bioreactors. Microb. Biotechnol. 2015, 8, 590–603. [Google Scholar] [CrossRef]
- Klappenbach, J.A. rrndb: The Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef]
- McTavish, H.; Fuchs, J.A.; Hooper, A.B. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 1993, 175, 2436–2444. [Google Scholar] [CrossRef]
- Bellucci, M.; Ofiteru, I.D.; Graham, D.W.; Head, I.M.; Curtis, T.P. Low-dissolved-oxygen nitrifying systems exploit ammonia-oxidizing bacteria with unusually high yields. Appl. Environ. Microbiol. 2011, 77, 7787–7796. [Google Scholar] [CrossRef]
- Coskuner, G.; Ballinger, S.J.; Davenport, R.J.; Pickering, R.L.; Solera, R.; Head, I.M.; Curtis, T.P. Agreement between theory and measurement in quantification of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2005, 71, 6325–6334. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- R Core Team; R Development Core Team. R: A Language and Environment for Statistical Computing [Internet]; 2019. Available online: https://www.r-project.org/ (accessed on 20 October 2020).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Packag Version 2.4-1. 2016. Available online: https://cran.r-project.org/web/packa (accessed on 20 October 2020).
- Borruso, L.; Sannino, C.; Selbmann, L.; Battistel, D.; Zucconi, L.; Azzaro, M.; Turchetti, B.; Buzzini, P.; Guglielmin, M. A thin ice layer segregates two distinct fungal communities in Antarctic brines from Tarn Flat (Northern Victoria Land). Sci. Rep. 2018, 8, 6582. [Google Scholar] [CrossRef]
- Whitaker, D.; Christman, M. Package ‘Clustsig’. R Top Doc. 2015; pp. 1–7. Available online: http://www.douglaswhitaker.com (accessed on 20 October 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix. 2017. Available online: https://github.com/taiyun/corrplot (accessed on 20 October 2020).
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Research 2016, 5, 1519. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Inter J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Daims, H.; Ramsing, N.B.; Schleifer, K.H.; Wagner, M. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 2001, 67, 5810–5818. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, M.; Marazzi, F.; Fornaroli, R.; Bellucci, M.; Ficara, E.; Mezzanotte, V. Outdoor pilot-scale raceway as a microalgae-bacteria sidestream treatment in a WWTP. Sci. Total Environ. 2020, 710, 135583. [Google Scholar] [CrossRef] [PubMed]
- Marazzi, F.; Bellucci, M.; Rossi, S.; Fornaroli, R.; Ficara, E.; Mezzanotte, V. Outdoor pilot trial integrating a sidestream microalgae process for the treatment of centrate under non optimal climate conditions. Algal Res. 2019, 39, 101430. [Google Scholar] [CrossRef]
- Krohn-molt, I.; Alawi, M.; Förstner, K.U.; Wiegandt, A.; Burkhardt, L.; Indenbirken, D.; Thieß, M.; Grundhoff, A.; Kehr, J.; Tholey, A.; et al. Insights into Microalga and Bacteria Interactions of Selected Phycosphere Biofilms Using and Proteomic Approaches. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Tilman, D. Resource Competition between Plankton Algae: An Experimental and Theoretical Approach. Ecology 1977, 58, 338–348. [Google Scholar] [CrossRef]
- Hodaifa, G.; Martínez, M.E.; Sánchez, S. Influence of temperature on growth of Scenedesmus obliquus in diluted olive mill wastewater as culture medium. Eng. Life Sci. 2010, 10, 257–264. [Google Scholar] [CrossRef]
- Masojídek, J.; Torzillo, G.; Koblížek, M.M.; Kopecký, J.; Bernardini, P.; Sacchi, A.; Komenda, J. Photoadaptation of two members of the Chlorophyta (Scenedesmus and Chlorella) in laboratory and outdoor cultures: Changes in chlorophyll fluorescence quenching and the xanthophyll cycle. Planta 1999, 209, 126–135. [Google Scholar] [CrossRef]
- Rosenberg, J.N.; Kobayashi, N.; Barnes, A.; Noel, E.A.; Betenbaugh, M.J.; Oyler, G.A. Comparative analyses of three Chlorella species in response to light and sugar reveal distinctive lipid accumulation patterns in the Microalga C. sorokiniana. PLoS ONE 2014, 9, e92460. [Google Scholar] [CrossRef]
- Mayeli, S.M.; Nandini, S.; Sarma, S.S.S. The efficacy of Scenedesmus morphology as a defense mechanism against grazing by selected species of rotifers and cladocerans. Aquat. Ecol. 2005, 38, 515–524. [Google Scholar] [CrossRef]
- Litchman, E.; Pinto, P.D.T.; Klausmeier, C.A.; Thomas, M.K.; Yoshiyama, K. Linking traits to species diversity and community structure in phytoplankton. Hydrobiologia 2010, 15–28. [Google Scholar] [CrossRef]
- Issa, A.; Mohamed, H.A.; Ohyama, T. Nitrogen fixing cyanobacteria: Future prospect. In Advances in Biology and Ecology of Nitrogen Fixation; Ohyama, T., Ed.; InTech: Leeds, UK, 2014; pp. 23–48. [Google Scholar]
- Yang, G.; Peng, M.; Tian, X.; Dong, S. Molecular ecological network analysis reveals the effects of probiotics and florfenicol on intestinal microbiota homeostasis: An example of sea cucumber. Sci. Rep. 2017, 7, 4778. [Google Scholar] [CrossRef]
- Marcilhac, C.; Sialve, B.; Pourcher, A.M.; Ziebal, C.; Bernet, N.; Béline, F. Control of nitrogen behaviour by phosphate concentration during microalgal-bacterial cultivation using digestate. Bioresour. Technol. 2015, 175, 224–230. [Google Scholar] [CrossRef]
- Bellucci, M.; Marazzi, F.; Ficara, E.; Mezzanotte, V. Effect of N:P ratio on microalgae/nitrifying bacteria community in agro-digestate treatment. Environ. Clim. Technol. 2020, 24, 136–148. [Google Scholar] [CrossRef]
- Montes, C.; Altimira, F.; Canchignia, H.; Castro, Á.; Sánchez, E.; Miccono, M.; Tapia, E.; Sequeida, Á.; Valdés, J.; Tapia, P.; et al. A draft genome sequence of Pseudomonas veronii R4: A grapevine (Vitis vinifera L.) root-associated strain with high biocontrol potential. Stand. Genom. Sci. 2016, 11, 76. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Y.; Zhang, Q.; Liu, X.; Xu, J.; Li, Y.; Di, H. Heterotrophic nitrification and denitrification are the main sources of nitrous oxide in two paddy soils. Plant Soil 2019, 445, 39–53. [Google Scholar] [CrossRef]
- Trung Tran, T.; Bott, N.J.; Dai Lam, N.; Trung Nguyen, N.; Hoang Thi Dang, O.; Hoang Le, D.; Tung Le, L.; Hoang Chu, H. The Role of Pseudomonas in Heterotrophic Nitrification: A Case Study on Shrimp Ponds (Litopenaeus vannamei) in Soc Trang Province. Microorganisms 2019, 7, 155. [Google Scholar] [CrossRef]
- Ramanan, R.; Kim, B.-H.H.; Cho, D.-H.H.; Oh, H.-M.M.; Kim, H.-S.S. Algae-bacteria interactions: Evolution, ecology and emerging applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Amavizca, E.; Bashan, Y.; Ryu, C.M.; Farag, M.A.; Bebout, B.M.; De-Bashan, L.E. Enhanced performance of the microalga Chlorella sorokiniana remotely induced by the plant growth-promoting bacteria Azospirillum brasilense and Bacillus pumilus. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Saranraj, P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)—Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]
- Seymour, J.R.; Amin, S.A.; Raina, J.-B.; Stocker, R. Zooming in on the phycosphere: The ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef]
- Cardinale, B.J. Biodiversity improves water quality through niche partitioning. Nature 2011, 472, 86–89. [Google Scholar] [CrossRef]
- Godwin, C.M.; Lashaway, A.R.; Hietala, D.C.; Savage, P.E.; Cardinale, B.J. Biodiversity improves the ecological design of sustainable biofuel systems. GCB Bioenergy 2018, 10, 752–765. [Google Scholar] [CrossRef]
- Stockenreiter, M.; Graber, A.K.; Haupt, F.; Stibor, H. The effect of species diversity on lipid production by micro-algal communities. J. Appl. Phycol. 2012, 24, 45–54. [Google Scholar] [CrossRef]



| PBR | RWP | p-value | |
|---|---|---|---|
| COD (mg L−1) | 298 ± 48.7 | 376 ± 111 | * |
| NH4+-N (mg L−1) | 260.1 ± 61.9 | 288.2 ± 86.7 | |
| NO3--N (mg L−1) | 7.9 ± 3.1 | 11.3 ± 5.1 | |
| PO43--P (mg L−1) | 14.6 ± 4 | 15 ± 9.8 |
| PBR | RWP | p-value | |
|---|---|---|---|
| pH | 8.5 ± 0.5 | 7.2 ± 0.6 | * |
| Tr § | 24.5± 6.2 | 21.9 ± 7.2 | * |
| rrCOD (mg L−1 d −1) | −4 ± 10.7 | 5.7 ± 6.1 | |
| rrTAN (mg L−1 d −1) | 21.9 ± 7.3 | 19.5 ± 6.6 | |
| rpNOx-N (mg L−1 d −1) | 14.4 ± 12.1 | 18.1 ± 6.9 | |
| rpNO2--N (mg L−1 d −1) | 13.5 ± 11.2 | 1.4 ± 2.2 | * |
| rpNO3--N (mg L−1 d −1) | 1 ± 1.3 | 16.7 ± 7.9 | * |
| Free Ammonia (mg L−1) | 5.7 ± 8.5 | 1.8 ± 3.3 | |
| ηTAN (%) | 84.1 ± 13.1 | 79 ± 14.8 | |
| rrPO43−-P (mg L−1 d −1) | 0.6 ± 0.9 | 0.3 ± 0.9 | |
| rpVSS (mg L−1 d −1) | 27 ± 20.3 | 23 ± 20.3 |
| Total | Shared | Only PBR | Only RWP | |
|---|---|---|---|---|
| Eukaryotes | 63 | 10 | 18 | 35 |
| Non-microalgal | 40 | 2 | 11 | 27 |
| Microalgae | 23 | 8 | 7 | 8 |
| Eukariotes | |||
|---|---|---|---|
| Code | PBR | Code | RWP |
| Inoculum |
| Inoculum |
|
| PBR_Ek_1 (31–45) |
| RWP_Ek_1 (18–31) |
|
| PBR_Ek_2 (60–87) |
| RWP_Ek_2 (31–45) |
|
| PBR_Ek_3 (87–115) |
| RWP_Ek_3 (60–73) |
|
| PBR_Ek_4 (129–143) |
| RWP_Ek_4 (87–115) |
|
| PBR_Ek_5 (143–171) |
| RWP_Ek_5 (115–129) |
|
| RWP_Ek_6 (143–171) |
| ||
| RWP_Ek_7 (171–188) |
| ||
| Bacteria | |||
| Code | PBR | Code | RWP |
| Inoculum |
| Inoculum |
|
| PBR_Bac_1 (0_18) |
| RWP_Bac_1 (0_18) |
|
| RWP_Bac_2 (18_31) |
| ||
| RWP_Bac_3 (31_45) |
| ||
| RWP_Bac_4 (60_73) |
| ||
| RWP_Bac_5 (87_115) |
| ||
| RWP_Bac_6 (143_188) |
| ||
| Network Properties | PBR | RWP |
|---|---|---|
| Modularity | 0.415 | 0.508 |
| Connected component | 101 | 117 |
| Average degree | 9.373 | 8.048 |
| Average path length | 1.982 | 2.314 |
| Network centralization | 0.2078 | 0.167 |
| Network heterogeneity | 0.7014 | 0.590 |
| Number nodes | 110 | 125 |
| Network diameter | 6 | 7 |
| Network density | 0.086 | 0.065 |
| Interaction | 2480 | 2588 |
| Co-occurrence | 1492 | 1488 |
| Mutual exclusion | 988 | 1100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bani, A.; Parati, K.; Pozzi, A.; Previtali, C.; Bongioni, G.; Pizzera, A.; Ficara, E.; Bellucci, M. Comparison of the Performance and Microbial Community Structure of Two Outdoor Pilot-Scale Photobioreactors Treating Digestate. Microorganisms 2020, 8, 1754. https://doi.org/10.3390/microorganisms8111754
Bani A, Parati K, Pozzi A, Previtali C, Bongioni G, Pizzera A, Ficara E, Bellucci M. Comparison of the Performance and Microbial Community Structure of Two Outdoor Pilot-Scale Photobioreactors Treating Digestate. Microorganisms. 2020; 8(11):1754. https://doi.org/10.3390/microorganisms8111754
Chicago/Turabian StyleBani, Alessia, Katia Parati, Anna Pozzi, Cristina Previtali, Graziella Bongioni, Andrea Pizzera, Elena Ficara, and Micol Bellucci. 2020. "Comparison of the Performance and Microbial Community Structure of Two Outdoor Pilot-Scale Photobioreactors Treating Digestate" Microorganisms 8, no. 11: 1754. https://doi.org/10.3390/microorganisms8111754
APA StyleBani, A., Parati, K., Pozzi, A., Previtali, C., Bongioni, G., Pizzera, A., Ficara, E., & Bellucci, M. (2020). Comparison of the Performance and Microbial Community Structure of Two Outdoor Pilot-Scale Photobioreactors Treating Digestate. Microorganisms, 8(11), 1754. https://doi.org/10.3390/microorganisms8111754






