Abstract
Under a one health perspective and the worldwide antimicrobial resistance concern, we investigated extraintestinal pathogenic Escherichia coli (ExPEC), uropathogenic E. coli (UPEC), and multidrug resistant (MDR) E. coli from 197 isolates recovered from healthy dogs in Spain between 2013 and 2017. A total of 91 (46.2%) isolates were molecularly classified as ExPEC and/or UPEC, including 50 clones, among which (i) four clones were dominant (B2-CH14-180-ST127, B2-CH52-14-ST141, B2-CH103-9-ST372 and F-CH4-58-ST648) and (ii) 15 had been identified among isolates causing extraintestinal infections in Spanish and French humans in 2015 and 2016. A total of 28 (14.2%) isolates were classified as MDR, associated with B1, D, and E phylogroups, and included 24 clones, of which eight had also been identified among the human clinical isolates. We selected 23 ST372 strains, 21 from healthy dogs, and two from human clinical isolates for whole genome sequencing and built an SNP-tree with these 23 genomes and 174 genomes (128 from canine strains and 46 from human strains) obtained from public databases. These 197 genomes were segregated into six clusters. Cluster 1 comprised 74.6% of the strain genomes, mostly composed of canine strain genomes (p < 0.00001). Clusters 4 and 6 also included canine strain genomes, while clusters 2, 3, and 5 were significantly associated with human strain genomes. Finding several common clones and clone-related serotypes in dogs and humans suggests a potentially bidirectional clone transfer that argues for the one health perspective.
Keywords:
Escherichia coli; dogs; virulence genes; antibiotic resistance; WGS; ST372; clonal structure 1. Introduction
Escherichia coli is a common commensal of the gastrointestinal tract. However, E. coli is also the main bacterial pathogen responsible for extraintestinal infections in humans and dogs, including urinary tract infections (UTIs) [1,2,3,4,5]. Most UTIs are thought to result from ascending infections. The two theories for the origin of uropathogenic isolates are the “prevalence” and the “special pathogenicity”. The prevalence hypothesis postulates that most UTIs are opportunistic infections caused by bacteria that predominate in the faecal microbiota, whereas the special pathogenicity hypothesis suggests that most UTI are caused by pathogenic strains that possess appropriate virulence factor (VF)-encoding genes [5,6]. More than 50 E. coli genes associated with extraintestinal infections have been identified, encoding adhesins, toxins, siderophores, capsular antigens, and invasines [7,8]. Isolates are designed presumptively as extraintestinal pathogenic E. coli (ExPEC) if they contained ≥2 of 5 of the following VF-encoding genes: papAH and/or papC, sfa/focDE, afa/draBC, kpsM II, and iutA [7], and as uropathogenic E. coli (UPEC) if they are positive for ≥3 of the four following VF-encoding genes: chuA, fyuA, vat, and yfcV [8].
The majority of ExPEC and UPEC isolates belong to B2 phylogenetic group. Although there is a notable diversity of phylogenetic groups among E. coli isolates causing human and animal extraintestinal infections, some epidemiological studies indicate that certain O:H serotypes, sequence types (STs) and clonotypes are more predominant and especially successful [3,9,10,11,12,13,14,15]. Three recent studies showed the dominance of some STs in dogs in Australia, the United States, and France, such as ST372, assessed to be specifically associated with dogs, and ST12, ST73, ST127, and ST141, assessed to be specifically associated with humans [10,11,14]. On the other hand, within-household sharing of ExPEC ST73 and ST95 strains, those with same serotypes and VF-encoding genes have been documented in the United States among humans and dogs [16]. Furthermore, in Australia and the United States, human and canine E. coli ST127, ST131, and ST1193 that exhibited identical virulence genotypes and highly similar PFGE profiles have been identified [17,18,19]. These findings suggest that some E. coli infections may sometimes be a zoonosis in either direction (human to pet or pet to human).
The antimicrobial resistance of human ExPEC and UPEC isolates has increased dramatically due to the emergence of the pandemic clone ST131 and more especially to subclade C2 (also known as subclone H30Rx) [20,21,22,23,24,25,26]. This subclone has also been occasionally isolated from dogs in several countries [27,28,29,30,31]. The emergence of multidrug resistance (MDR) among E. coli causing infections in dogs is of great concern and increases the risk of treatment failure [32,33,34,35,36,37,38,39,40,41,42]. Additionally, exposure to dogs and/or dog faeces has been identified as a risk factor for the development of drug-resistant E. coli UTI in women [43].
As relatively little is known on the clonal structure of canine ExPEC, UPEC and MDR isolates, the present study was carried out (i) to establish which clones (defined by the association of phylogroup, clonotype and ST) dominate in dogs and (ii) to compare these clones with those causing extraintestinal infections in humans. To our knowledge, this is the first study that uses whole genome sequencing (WGS) to define the genetic relatedness between the ST372 E. coli lineage, which we found dominant among the Spanish canine faecal E. coli populations, and human E. coli ST372 that cause extra-intestinal infections.
2. Materials and Methods
2.1. E. coli Isolates
A total of 197 non-duplicate E. coli isolated from faecal samples of 104 healthy dogs collected in Spain between 2013 and 2017 were characterized.
2.2. Phylogenetic Grouping
Assignment to the main phylogroups (A, B1, B2, C, D, E, and F) was based on the protocol of Clermont et al. [44].
2.3. Serotyping
The determination of O and H antigens was carried out using the method previously described by Guinée et al. [45] with all available O (O1 to O181) and H (H1 to H56) antisera. Isolates that did not react with any antisera were classified as O non-typeable (ONT) or H non typeable (HNT) and those non motile were denoted as HNM.
2.4. Multilocus Sequence Typing (MLST)
The sequence types (STs) were established following the MLST scheme of Achtman by gene amplification and sequencing of the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) according to the protocol and primers specified at the E. coli MLST web site (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) [46].
2.5. CH Typing
Clonotype identification was determined by fumC and fimH (CH) sequencing [47,48].
2.6. Virulence Genotyping
VF-encoding genes of E. coli causing extraintestinal infections were screened by PCR [4,49]. The virulence gene score was the number of extraintestinal virulence-associated genes detected. The isolates were designed presumptively as extraintestinal pathogenic E. coli (ExPEC) if positive for ≥2 of 5 markers, including papAH and/or papC, sfa/focDE, afa/draBC, kpsM II, and iutA [7], and as uropathogenic E. coli (UPEC) if positive for ≥3 of 4 markers, including chuA, fyuA, vat, and yfcV [8].
2.7. Antimicrobial Susceptibility and ESBL and pAmpC Typing
Antimicrobial susceptibility was determined by the minimal inhibitory concentrations (MICs) and/or the disc diffusion method. Resistance was interpreted based on the recommended breakpoints of the CLSI [50]. Fifteen classes of antimicrobial agents were analyzed: penicillins (ampicillin), penicillins and β-lactamase inhibitors (amoxicillin-clavulanic acid), 1st and 2nd generation of non-extended spectrum cephalosporins (cefazolin and cefuroxime), extended-spectrum cephalosporins (cefotaxime, ceftazidime and cefepime), cephamycins (cefoxitin), monobactams (aztreonam), carbapenems (imipenem), aminoglycosides (gentamicin, tobramycin, amikacin), tetracyclines (doxycycline), phenicols (chloramphenicol), nitrofurans (nitrofurantoin), quinolones (nalidixic acid and ciprofloxacin), folate pathway inhibitors (trimethoprim-sulphamethoxazole), phosphonic acids (fosfomycin), and polymyxins (colistin). E. coli MDR was defined as resistance to one or more agents in three or more classes of tested drugs [51]. Genetic identification of ESBL and pAmpC types was carried out by PCR followed by amplicon sequencing [52,53,54].
2.8. Whole Genome Sequencing (WGS)
The WGS of 23 ST372 isolates from our LREC collection was performed under the protocol of the Genomics and Bioinformatics Core Facility (Centre for Biomedical Research of La Rioja) as it was described previously [55]. The assembly information of draft genomes, database sources and input parameters can be found in Table S1 (NCBI Bioproject accession PRJNA627579).
PLACNET webserver [56,57] was used for the genome reconstruction after which Prokka [58] was used to annotate the assembled genetic elements. Primary in silico analyses were carried out using the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/) services with home-made databases, the CGE databases, and other complementary databases to explore the resistance and virulence factors. Plasmid typing was complemented by subtyping relaxases with the method defined by Alvarado et al. [59] and integrative conjugative elements (ICEs) typing was complemented by in silico analyzing the ICE-harbouring contigs with ICEberg (ICEfinder and VRprofile) (information provided in Table S1). Additionally, the ICE-harbouring contigs were analyzed with Easyfig, a comparative genomic tool that allows for visualizing homologies and similarities between contigs using BLAST [60].
Besides, we performed a single nucleotide position (SNP) tree analysis of the 23 ST372 genomes sequenced in this study plus 174 ST372 full-genome references retrieved from NCBI bioproject and EnteroBase. The SNP-tree was done using the CSI Phylogeny 1.4 server from the CGE with J22 strain as reference (ID: GCA_009497315). After analyzing the SNP matrix, we took all the ST372 genomes from human strains plus some representative genomes from canine strains to make a tree visualization using EnteroBase [61]. The accession number of all the genomes included in this study can be found in Table S2.
2.9. Statistical Analysis
All the p values were calculated using Fisher’s exact test, except for the comparison of the means that was performed using the one-way ANOVA test. p values < 0.05 were considered statistically significant.
3. Results
3.1. Phylogenetic Groups of the 197 Canine Isolates
The most common phylogenetic group displayed by the 197 canine faecal E. coli isolates was B2 (42.6%), followed by A (16.2%), B1 (13.2%), F (9.1%), E (7.1%), C (5.1%), and D (3.0%) (Table S3).
3.2. Virulence Factor (VF)-Encoding Genes in the 197 Canine Isolates
Of the 28 VF-encoding genes analyzed, eight (fimH, yfcV, vat, iroN, fyuA, chuA, malX, and usp) were detected in more than 40% of the 197 canine isolates and nine (papAH, papC, sfa/focDE, cnf1, hlyA, kpsM II, kpsM II-K5, traT, ibeA) in at least 20%. In contrast, six VF-encoding genes (afa/draBC, sat, cdtB, neuC-K1, kpsM II-K2, kpsM III) were found in less than 10% of these isolates (Table 1).

Table 1.
Virulence factor (VF)-encoding genes detected in the 197 canine E. coli isolates. Relationship with phylogenetic groups.
A higher mean of VF-encoding gene score was observed in the 84 canine isolates belonging to the dominant B2-phylogenetic group (mean of 12.79) (p < 0.05) compared with the isolates belonging to phylogroups A (2.31), B1 (2.96), C (6.60), D (5.67), E (4.14), and F (7.94) (Table 1).
Of the 197 canine isolates, 74 (37.6%) were presumptively classified as ExPEC and 82 (41.6%) as UPEC (Table 1) resulting in 91 ExPEC and/or UPEC isolates. The majority (85.7%; 78 of 91) of ExPEC and/or UPEC isolates belonged to phylogenetic group B2. In contrast, only 5.7% (6 of 106) of non-ExPEC and non-UPEC isolates were assigned to this phylogenetic group (p < 0.00001). The A, B1, C, and E phylogenetic groups were significantly associated with non-ExPEC and non-UPEC isolates (Table S4).
3.3. Antimicrobial Resistance in the 197 Canine Isolates
In total, 28 (14.2%) of the 197 analyzed canine faecal E. coli isolates were classified as MDR. Multidrug resistance was significantly associated with isolates belonging to B1, D, and E phylogenetic groups (Table S5). Furthermore, only eight (28.6%) of MDR isolates showed the ExPEC and/or the UPEC status (Table S6).
In total, 10 of the 28 MDR isolates produced an ESBL enzyme: CTX-M-1 (four isolates), CTX-M-14 (four isolates), CTX-M-55 (one isolate) and SHV12 (one isolate). Besides, 10 other isolates produced a plasmid-mediated AmpC β-lactamase of CMY-2 type.
3.4. Sequence Types, Clones and Serotypes Displayed by the 91 Canine ExPEC and/or UPEC Isolates and 28 MDR Isolates
Sequences types (STs), clones (defined by the association of phylogroup, clonotype and ST) and O:H serotypes were established only for the 91 canine isolates classified as ExPEC and/or UPEC and the 28 canine MDR isolates.
A total of 34 STs were identified in the canine ExPEC and/or UPEC isolates and 22 in the MDR isolates. Among these STs, 18 were previously undescribed (Table S7). Each of these 18 new STs were displayed by one isolate. Seven dominant STs (ST12, ST38, ST73, ST127, ST141, ST372, and ST648) were observed among the 91 canine ExPEC and/or UPEC and the 28 canine MDR isolates. There was a strong correlation between VF-encoding gene profiles and the dominant STs (Table 2).

Table 2.
Virulence factor (VF)-encoding genes detected in the 65 canine E. coli isolates included in the 7 most frequent sequence types identified in ExPEC, UPEC and MDR isolates.
A total of 50 clones were identified among the 91 canine isolates classified as ExPEC and/or UPEC, with 11 of them including at least two isolates and only four, at least four isolates i.e., B2-CH14-180-ST127 (four isolates), B2-CH52-14-ST141 (four isolates), B2-CH103-9-ST372 (25 isolates), and F-CH4-58-ST648 (five isolates) (Table 3). In recent studies conducted by our research group [3,21,62], we had identified, as indicated in Table 3, 15 of the 50 canine ExPEC/UPEC clones comprising 49 isolates among the 261 human ExPEC and/or UPEC isolates included in a collection of 394 E. coli isolates causing extraintestinal infections. However, only 31 of the 49 human ExPEC and/or UPEC isolates presented the same clone-related O:H serotypes as the canine isolates (Table 3). Among these 31 human isolates, 28 belonged to B2 phylogroup clones and three to F phylogroup clones identified among canine isolates. These B2 clones were distributed into five ST lineages, including four lineages currently dominant in humans (ST73, ST127, ST141, and ST1193,) and the lineage currently established as the dominant lineage in dogs, namely lineage ST372. In dogs, we found three clones in the lineage ST73 with the same serotype (O6:H1). The eight human isolates sharing this lineage with dogs were distributed into the same three clones and displayed serotype O6:H1. In dogs, we found two clones in the lineage ST127 displaying two serotypes with one (O6:HNM) of them present in the two clones. The four human isolates sharing this lineage with dogs were distributed into the same two clones but displayed the common serotype (O6:HNM). In dogs, we found two clones in the lineage ST141 with the same serotype (O2:H6). The 11 human isolates that shared this lineage with dogs were distributed into the same two clones and displayed the same serotype as human isolates. In dogs, we found one clone in lineage ST1193 with one serotype (O75:HNM). Three human isolates shared this clone and serotype with dogs. Concerning the lineage ST372, we found five clones in dogs with one of them including isolates displaying six serotypes. The two human isolates sharing the lineage ST372 with dogs belonged to this multiple-serotype clone and both displayed one of the six serotypes (O83:H31). Concerning the three F group human isolates, they belonged to one of the three F group clones (F-CH32-41-ST59) identified in dogs and showed the same serotype (O1:H7).

Table 3.
Clones and clonal-related serotypes of 91 canine ExPEC and/or UPEC isolates. Prevalence of the canine clones and clonal-related serotypes among ExPEC and/or UPEC isolates causing extraintestinal infections in humans [3,21,62].
Among the 28 canine MDR isolates, we observed 24 different clones, of which nine had also been identified among the above cited 394 isolates causing infections in humans (Table 4) [3,21,62].

Table 4.
Clones and clonal-related serotypes of 28 canine multidrug resistant (MDR) E. coli isolates. Prevalence of the canine clones and clone-related serotypes among E. coli isolates causing extraintestinal infections in humans [3,21,62].
3.5. Whole Genome Sequencing (WGS) and Molecular Characterisation of ST372 Isolates
For WGS, we selected 23 of the above studied ST372 isolates. They comprised 21 of the 29 Spanish canine faecal ST372 strains that were isolated in 2013 (n = 9) and 2017 (n = 12) and two previously published human ST372 strains isolated in 2016 [3,61]: strains LREC_341 isolated in Spain from an abscess and LREC_342 isolated in France from a bone infection. Both human strains showed serotype O18:H31 and clonotype CH103-9, whereas the 21 canine strains showed six different serotypes (O4:H31 (seven isolates), O83:H31 (four isolates), O25:H31 (four isolates), O15:H31 (three isolates), O21:H31 (two isolates) and O117:H28 (one isolate)) and four clonotypes (CH103-9 (18 isolates), CH103-10, CH103-17, and CH103-240).
The main objectives were to get more insights into the E. coli ST372 lineage that appears as one of the most prevalent E coli lineages among the canine faeces E. coli populations and to elucidate if there is any relation between canine and human ST372 strains.
To infer the phylogeny, we performed an SNP-tree with 197 genomes of ST372 strains (23 from this study (labelled LREC strains) and 174 obtained from public databases) corresponding to 151 genomes from canine strains and 46 genomes from human strains. A total of 70% of these genomes corresponded to strains collected between 2017 and 2019 while the remaining 30% corresponded to strains isolated between 1995 and 2016. Regarding geographical distribution, 46 genomes (23.4%) were from strains collected in Europe and 143 (72.6%) from strains collected in North America.
The SNP analysis of the E. coli ST372 lineage revealed a wide and heterogeneous population, allowing us to describe six clusters. Figure 1 only includes 97 representative genomes (including the 23 LREC genomes sequenced in this study and the 46 genomes from human strains) of the 197 analyzed so that it is possible to visualize all the information.
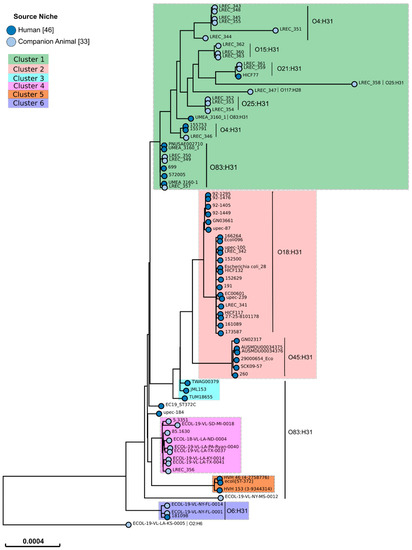
Figure 1.
SNP-tree of 79 representative ST372 E. coli genomes from 46 human strains and 33 canine strains. Tree visualization by EnteroBase [61]. The 33 genomes from canine strains are representatives of the different clusters identified in the SNP matrix of a previous SNP-tree performed with the 197 genomes analyzed in this study. The identified serotypes are listed beside the vertical line.
The criterion established to define a cluster was that it should include genomes with less than 200 SNPs distance between them. An exception to this rule was the inclusion of the genome ECOL-19-VL-SD-MI-0018, with a maximum of 391 SNP distance, in cluster 4. Five genomes did not reach this criterion, having more than 400 SNP distance between them and could form five other clusters. However, we have included those genomes in only one category (undefined) to simplify the following analysis.
According to the phylogenetic tree built from the genome of the 197 strains, cluster 1 comprised 147 (74.6%) of the 197 analyzed genomes. This cluster was mostly composed of genomes of canine strains (138 genomes; 93.9%). Genomes of canine strains were also included in clusters 4 (nine genomes) and 6 (two genomes) while only human strain genomes were included in clusters 2 (28 genomes), 3 (three genomes) and 5 (three genomes) (Table 5). Thus, cluster 1 comprised significantly more canine strain genomes (p < 0.00001) while clusters 2 (p < 0.00001), 3 (p = 0.01209), and 5 (p = 0.01209) comprised significantly more human strain genomes. A total of 20 of the 21 genomes of the Spanish canine strains belonged to cluster 1, whereas, the genome of the remaining Spanish canine strain (LREC_356) belonged to cluster 4. The genomes of the Spanish and French human strains (LREC_341 and LREC_342) belonged to cluster 2.

Table 5.
Distribution into the phylogenetic clusters of the 197 canine and human ST372 strains.
Both clusters 1 and 2 were the most frequent clusters observed among the studied E. coli ST372 strains (canine and human) isolated in Europe and North America. However, cluster 1 was significantly associated with North America strains (p = 0.02476), while cluster 2 was especially associated with Europe strains (p = 0.01233) (Table 6).

Table 6.
Cluster distribution of the 197 studied ST372 strains according to countries.
To compare the virulence profile of the 197 canine and human ST372 strains, we in silico investigated the presence of 32 VF-encoding genes in the 197 strains and defined their ExPEC and UPEC status. We also investigated the distribution of those VF-encoding genes according to the classification of the strains into the six defined clusters. Table 7 summarizes the results obtained from the mentioned analysis. Microbiological, geographical, and genomic data of each of the 197 studied strains are available in Table S2.

Table 7.
Distribution of the VF-encoding genes detected among the 197 ST372 E. coli genomes according to strain origins (canine/human) and cluster types.
The canine ST372 strains showed a higher VF-encoding gene score (mean 16.79) compared with the human ST372 strains (mean 13.76). However, three human stains belonging to cluster 5 were those with the highest number of VF-encoding genes (mean 21.67). Eight VF-encoding genes (papAH, papC, papEF, focCD, focG, cnf1, hlyA, and iroN) were significantly associated with canine ST372 isolates, whereas five (hlyF, iutA, kpsM II, kpsM II-K5, and iss1) were significantly associated with human ST372 isolates. Interestingly, the ExPEC status was found more frequently among canine ST372 strains (74.8%) than human strains (21.7%) (p < 0.00001) (Table 7).
The more prevalent serotype was O83:H31, which represents 36.0% of the 197 ST372 strains, followed by O4:H31 (17.8%), O15:H31 (15.2%), O18:H31 (10.2%), O45:H31 (3.0%), O117:H28 (3.0%), O21:H14 (2.5%), O21:H31 (2.5%), O75:H31 (2.5%), O-unknown:H31 (2.5%), O25:H31 (2.0%), O2:H6 (0.5%), and O-unknown:H28 (0.5%). The serotypes O4:H31 (p = 0.02631) and O15:H31 (p = 0.00062) were significantly associated with canine ST372 strains, whereas the serotypes O18:H31 (p < 0.00001) and O45:H31 (p = 0.00012) were significantly more frequent among human ST372 strains. The 65 canine strains of serotypes O4:H31 and O15:H31 belonged to cluster 1 and the 26 human strains of serotypes O18:H31 and O45:H31 belonged to cluster 2 (Table 8). In contrast, the dominant serotype O83:H31 was frequently identified among canine (38.4%) and human (28.3%) strains, and, although the majority of the strains with this serotype belonged to cluster 1, O83:H31 strains were also found in clusters 3, 4, and 5.

Table 8.
Distribution of serotypes among the 197 ST372 strains according to origins (canine and human) and cluster types.
The three most prevalent serotypes in Europe were O4:H31 (23.9%), O83:H31 (21.7%) and O18:H31 (19.6%), while in North America, they were O83:H31 (39.9%), O15:H31 (18.2%) and O4:H31 (16.8%). The serotypes O18:H31 (p = 0.02203) and O25:H31 (p = 0.00317) were more frequently observed in Europe, whereas the serotype O83:H31 (p = 0.03291) was more prevalent in North America (Table 9).

Table 9.
Serotypes of the 197 ST372 strains according to countries.
The 23 LRCE genomes sequenced in this study were investigated in greater depth. These genomes were reconstructed to analyze the chromosome and plasmidome separately. The size of the chromosomes had an average of 5,043,308 pb and were encompassed in 55 to 178 contigs. We found an integrative conjugative element (ICE) with relaxase type MOBQ in all the genomes except for LREC_347 genome. These ICEs belong to the ICEKp1 family, a yersiniabactin synthesis-associated ICE type (similar to ICEEcoUMN026-1). The contigs that harboured the ICE region were revised, allowing us to detect the presence of a pathogenic island (PAI) and some VF-encoding genes. The ICE contig retrieved from LREC_356 genome was the longest (2,540,863 pb) and showed a high percentage of homology with the contigs of the other genomes harbouring the ICE region (Figure S1). Interestingly, the contig from LREC_356 harboured the ompT, iss, vat, fyvA, and yfcV virulence- encoding genes, the last three mentioned genes being those used (in addition to chuA) to define UPEC status. We also identified the secretion system effector homolog type T6SS and the PAI_AET37190. In second place, in terms of ICE contig length, was LREC_357 (1,677,509 pb) genome that harboured the iroN, iss, fyvA, and yfcV virulence-encoding genes. The ICE contigs had not the same length [varying from 2,540,863 pb to 307,789 pb (short read sequencing limitations)] and all the genes mentioned as found in the ICE contig from LREC_356 were not retrieved in the other 20 genomes harbouring ICE contigs: three harboured the fyvA and yfcV genes (LREC_359, LREC_361, and LREC_344) and the remaining 17 genomes harboured the fyvA gene. We concluded that the presence of this type of ICE was a common feature in the ST372 genomes from the 22 of 23 studied Spanish strains and may be involved in the acquisition of their UPEC status.
We also described 11 plasmids (four conjugative plasmids, six mobilizable plasmids and one plasmid with no relaxase suggesting that it is not mobilizable) which belonged to the following relaxase families (MOB) and incompatibility groups (Inc.): MOBP3/IncX1 (n = 3); MOBP1/nd (n = 2); MOBF12/IncFII-pCD1 (n = 2); MOBF12/IncFII-IncFIB (n = 1); MOBH11/IncHI2 (n = 1); MOBQu/ColRNAI (n = 1); nd/p0111 (n = 1). To predict plasmid transferability, we investigated the presence of mating pair formation (Mpf) system proteins. These proteins were present in all the previously described MOBF12 and MOBH11 conjugative plasmids. Furthermore, in silico analysis showed that these plasmids did not carry resistance or virulence encoding genes except for the cba and cma genes that were found in plasmid pLREC354_1 and a blaTEM gene found in pLREC346_1. Table 10 summarizes the MGE content of the 23 ST372 genomes.

Table 10.
Description of mobile genetic elements (MGE)s found in the 23 ST372 strain genomes sequenced in this study.
We in silico investigated the presence of 189 VF-encoding genes, 87 antibiotic-resistance encoding genes (ARGs), and 18 types of point mutations (Table S8). Through this analysis, the 23 ST372 strains were shown with an UPEC status and harbouring a wide variety of VF-encoding genes, reaching an average number of 80. In contrast, these 23 ST372 strains were shown as carrying very few ARGs. However, genes encoding drug efflux were detected but only in the two human strain genomes (LREC_341 and LREC_342) that also harboured antibiotic-resistance encoding genes: blaTEM-1A, sul1, aadA1, dfrA1, and mdf(A). These results were in agreement with those previously obtained by conventional methods.
4. Discussion
To get more insights into the population structure of canine E coli, we investigated those harboured in the intestinal tract of 104 healthy Spanish dogs by using different approaches, knowing that the gut is the reservoir of the great majority of E. coli causing extraintestinal infections. The phylogenetic group, VF-encoding gene and antibiotic susceptibility analyses, showed that among the 197 canine faecal isolates obtained from the 104 dogs, 84 (42.6%) belonged to B2 phylogroup, 91 (46.2%), mostly B2 group isolates, were classified as ExPEC and/or UPEC, and 28 (14.2%), mostly non-B2 group isolates, as MDR. This strongly suggests that the intestinal tract of healthy dogs might be an important reservoir of ExPEC and/or UPEC isolates, and in a lesser extent, of MDR E. coli isolates. However, some studies that focused on antibiotic-resistant canine isolates suggested that dogs might also be an important reservoir for antibiotic-resistant strains [63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81], notably for those producing ESBLs or CMY-2 [64,66,67,70,72]. Although there was a low prevalence of MDR isolates among the 197 studied isolates, we found, as previously described that they produced ESBLs or CMY-2.
MLST assigned the 91 Spanish canine faecal isolates with an ExPEC and/or UPEC status to 34 STs. among which six were displayed by 67% of the 91 isolates: ST372 (31.9%), ST12 (9.9%), ST127 (8.8%), ST648 (6.6%), ST141 (5.5%), and ST73 (4.4%). Few studies have been carried out so far to characterize the ST structure of canine ExPEC and/or UPEC isolates. In the USA, LeCuyer et al. [11] analyzed 295 E. coli isolates from canine UTI. They found that ST372, which is uncommon among the human E. coli pathogens [3,12,62,82,83], was the predominant ST in canine UTI isolates (21.7%), and this was well ahead of the five other most frequent STs: ST12 (6.4%), ST73 (6.4%), ST127 (4.1%), ST131 (4.1%), and ST297 (3.7%). A total of 170 (57.4%) of these isolates met the criterion to be classified as ExPEC, and, except for ST297, the most prevalent STs were associated with ExPEC status. In France, Valat et al. [14] analyzed 618 canine E. coli isolates collected from diagnostic laboratories, including 403 (65.2%) from UTIs. B2 phylogroup was over-represented (79.6%) and positively associated with the presence of numerous VFs, including those defining the ExPEC status. MLST of a randomly chosen subset of 89 isolates belonging to B2 phylogroup revealed five dominant STs: ST372 (17.9%), ST73 (17.9%), ST12 (10.1%), ST141 (7.9%), and ST961 (5.6%). In Australia, Kidsley et al. [10] focused their study on the canine fluroquinolone-susceptible E. coli clinical isolates (n = 449) that were identified during a nation-wide survey of antibiotic resistance in Australian animals between January 2013 and January 2014. They found that these isolates mostly (n = 317; 71%) belonged to B2 phylogroup. By using the RAPD typing system, they found a distribution of the 317 B2 group isolates into 35 main clusters. To pursue their molecular investigation, they sequenced and analyzed the whole genome of 77 representatives of the B2 group fluoroquinolone-susceptible isolates. Thus, they found that the 77 sequenced isolates were assigned to 24 STs, among which four were dominant: ST372 (31%), ST73 (17%), ST12 (7%), and ST80 (7%). In sum, the present study and those previously published show that three STs (ST372, ST12, and ST73) are the dominant ST in healthy and infected dogs irrespective of the countries (the USA, France, Australia and Spain) and strain sources (clinical samples and faeces). Such a finding might argue for the “prevalence” theory with regard to UTI pathogenesis (most UTIs are opportunistic infections caused by bacteria that predominate in the faecal microbiota) in dogs. Nevertheless, the fact that the canine isolates belonging to the most dominant ST, either present in all studied countries (ST372, ST12, and ST73) or present in some studied countries (ST127 in the USA and Spain, and ST141 in France and Spain) were shown to harbour numerous VF-encoding genes might also argue for the “special pathogenicity” theory in dogs.
Concerning the ST structure of canine MDR isolates, there seems to exist a more important difference between the countries than for the non-MDR isolates. MLST assigned the 28 Spanish canine MDR isolates to a great diversity of STs comprising 15 established STs (ST10, ST12, ST38, ST57, ST58, ST88, ST93, ST155, ST457, ST648, ST695, ST1011, ST1140, ST3774, and ST8953) and seven new STs. The first 10 STs here listed have been identified in canine MDR isolates from different countries [1,11,14,15,28,29,30,33,34,37,38,39,40,41,42,66,68,71,72,73,75,76,77,80,81,84,85,86,87,88]. In contrast to some studies, we did not detect either canine MDR isolates displaying the five important emerging MDR STs in humans: ST69, ST127, ST131, ST410, and ST1193 [1,2,11,14,15,17,27,28,29,30,31,32,33,34,35,36,38,39,40,41,70,71,74,78], canine isolates harbouring the mcr-1 gene encoding resistance to colistin as described in China [89], or canine isolates producing carbapeneamases, notably OXA-48, as described in Germany [90], France [91], and United States [38]. Finally, we found that none of the 29 here studied ST372 was MDR while previous studies have found ST372 isolates producing different types of ESBLs and CMY-2 [1,11,14,30,33,38,40,41,42,79].
To get more insight into the potential link between the canine ExPEC and/or UPEC and the E. coli isolates causing extra-intestinal infections in humans, we determined which clone (defined by the association of phylogroup, clonotype and ST) and which serotypes characterized the 91 canine ExPEC and/or UPEC in order to compare them with human E. coli clinical isolates collected in 2015 and 2016 in Spain and France and characterized for these two traits [3,21,62]. This approach allowed us to found that among the 50 clones identified in the 91 Spanish canine ExPEC and/or UPEC isolates, 15 were present in the human collection accounting for 49 (18,8%) of the human isolates. However, only 31 of the 49 human ExPEC and/or UPEC isolates presented the same O:H serotype as the canine ones. By coupling clonal type and serotype for each E. coli ST lineage shared by dogs and humans, we observed various features about the distribution of the human isolates when the lineages included several clones and several clone-related serotype in dogs. This feature shows that it is difficult to make hypotheses about the relationship between canine and human isolates sharing a given clone-serotype couple in a given lineage without knowing the structure of the clone-serotype couples in the given lineage in humans. For example, we had found [3] that the human ST73 isolates were distributed into four clones, of which two here were identified in dogs. In human, three of the four clones comprised, each, isolates with different serotypes but one serotype (O6:H1) was exhibited by isolates distributed into the four clones. In dogs, the ST73 isolates exhibited only serotype O6:H1. This suggests that serotype might be an ecological niche marker, meaning, in this case, that isolates of the lineage ST73 exhibiting serotype O6:H1 are adapted to both dogs and humans. However, the Kidsley et al.’s study [10] in which a phylogenetic tree was built with the genome of ST73 strains from dogs, cats, and humans, seems to contradict this hypothesis. Indeed, the 13 studied Australian canine isolates of the lineage ST73 exhibited four serotypes among which serotype O6:H1 was exhibited by only one isolate that formed an animal-specific cluster (containing cat O6:H1 ST73 isolates) distinct from the four main clusters of human O6:H1 ST73 isolates. Nevertheless, the hypothesis that we made for serotype O6:H1 with regard to Spanish canine and human ST73 isolates could be made for serotype O2:H1 with regard to Australian canine and human ST73 isolates as this serotype was shared by clustered canine and human isolates. By extending the comparison of the structure of clone-serotype couples to the other human-specific-human ST lineages (ST127, ST141 and ST1193) shared by the Spanish studied dogs and humans, we observed that the clone-serotype couples shared by dogs and humans comprised mostly the serotype the most frequent in the human clones. This feature seems to indicate that serotype frequency might be a variable involved in the E. coli exchanges between dogs and humans. Concerning the dog-specific lineage ST372, we had found only one clone comprising to isolates in Spanish humans, while we found here five clones in the 29 Spanish dogs. Among the six serotypes exhibited by these 29 canine isolates, the serotype exhibited by the two human ST372 isolates corresponded to one (O18:H31) of the two dominant serotypes in dogs that was, on the other hand, exhibited by canine isolates belonging to three different clones. Thus, the suggestions that we made about the fact that serotype could be an ecological niche marker and that serotype frequency could shape the E coli exchange between dogs and humans seems to be able to be applied to the lineage ST 372.
Interestingly, concerning the 24 clones identified in the 28 canine MDR isolates, which were mostly non-B2-group isolates, we observed that if there were some clones (n = 9) shared by the Spanish human (35 of 394) and canine (10 of 197) isolates there was only one isolate that shared the same clone and the same serotype as one canine isolate.
To better understand the potential relationship between canine and human E. coli isolates with regard to the lineage ST372, we turned to the whole genome sequencing and analysis of 197 ST372 strains (151 from dogs and 46 from humans). The SNP analysis of the core genome of these 197 strains revealed an extensive phylogenetic diversity of the ST372 isolates that was segregated into six clusters. Cluster 1 comprised 91.4% of canine strains while cluster 2 comprised 60.9% of human strains. Cluster 2 was specific of human strains associated with serotypes O18:H31 and O45:H31, the latter serotype being exclusively found in human ST372 strains. Three other serotypes were the most prevalent serotype among strains belonging to cluster 1, including O4:H31 and O15:H31 associated with canine strains, and O83:H31 identified in similar proportion among canine and human strains. Overall, the WGS analysis suggests that canine strains of clone B2-CH103-9-ST372, belonging to cluster 1 and having serotype O83:H31 might cause extraintestinal infections in humans and dogs, as already suggested by the clone-serotype couple analysis, whereas strains of this clone belonging to cluster 2 and having serotypes O18:H31 and O45:H31 might cause only human extraintestinal infections. Molecular epidemiological studies on E. coli ST372 in human extra-intestinal infections are required to confirm these suggestions.
Furthermore, we localized ICEs in the chromosome of 22 of the ST372 sequenced genomes and confirmed that all ICEs belong to a yersiniabactin synthesis-associated ICE type (ICEKp1 family) with relaxase type MOBQ. In contrast, we found very few plasmids. Moreover, we found that the number of plasmids retrieved from the human ST372 strains was higher than that of plasmids found in the canine strains (four plasmids in the two human strains versus seven plasmids in the 21 canine strains). Interestingly, the genome of canine LREC_356 strain from cluster 4 carried two plasmids and was the canine strain genome the most similar to human strain genomes. Those plasmids were not rich with genes encoding of antibiotic resistance and virulence-factors. Nonetheless, a high number of virulence factor encoding-genes were found in the chromosome of the ST372 genomes and we hypothesized that the origin of the UPEC status of ST372 strains is due to the acquisition of ICEs harbouring the genes associated with this status. Although there is still limited knowledge about the origin of genomic islands, like ICEs or pathogenicity islands (PAIs), it has been speculated that they derive from the integration of plasmids or phages into the chromosome. Further, genomic research has shown that genomic islands have played a major role in the transformation of avirulent into virulent bacteria. Besides, most VFs of ExPEC are encoded by ICEs and PAIs [92,93,94,95].
5. Conclusions
The intestinal tract of healthy dogs appears as an important reservoir of ExPEC and/or UPEC, and, in a lesser extent, of MDR E. coli isolates. However, the canine MDR isolates could be a good reservoir of ESBLs and CMY-2 because most of them produce these enzymes. Among the canine isolates displaying an ExPEC and/or UPEC status, clone B2-CH103-9-ST372 was dominant. This canine clone and 14 others, also displaying an ExPEC and/or UPEC status, had been identified in isolates previously published as causing extraintestinal infections in human suggesting a zoonotic potential of these clones. WGS analysis suggests that canine strains of clone B2-CH103-9-ST372, belonging to cluster 1 and having serotype O83:H31 might cause extraintestinal infections in both humans and dogs, whereas those strains of this clone belonging to cluster 2 and serotypes O18:H31 and O45:H31 might cause only human infections. Taking into consideration that Kidsley et al. have recently characterized the phylogenetic relationship between canine, cat, and human isolates of the lineage ST73 [10], such studies are still required for the other ST lineages and clones that we showed in this study to be shared by canine and human isolates in order to clarify their potential role in infection occurrence in both dogs and humans.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1712/s1, Figure S1: Comparison of contigs harbouring integrative conjugative elements (ICEs) from 22 ST372 E. coli genomes, Table S1: Bioproject accession (PRJNA627579) and assembly genome information, Table S2: SNP matrix and VF-encoding genes of 197 ST372 genomes, Table S3: Prevalence of the phylogenetic groups in the 197 canine E. coli isolates, Table S4: Comparison of the distribution of the phylogenetic groups among the 197 canine isolates according to the strain ExPEC and UPEC status, Table S5: Comparison of the distribution of the phylogenetic groups among the 197 canine isolates according to the strain multidrug resistant (MDR) status, Table S6: Comparison of the strain ExPEC and UPEC status among canine multidrug resistant (MDR) and non-MDR isolates, Table S7: New sequence types observed in 18 canine E. coli isolates, Table S8: In silico determination of VF-encoding genes, antibiotic-resistance encoding genes (ARGs) and point mutations in 23 ST372 E. coli genomes.
Author Contributions
S.-C.F.-S., M.d.T., V.G., J.E.B., M.B., M.P.A., A.G., J.D.-G., M.-H.N.-C., and J.B. undertook the laboratory work and the genome analysis. S.-C.F.-S. and J.B. conceived the concept for the paper and designed the experiments. All authors provided critical input and contributed to the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by projects: PI16/01477 from Plan Estatal de I+D+I 2013-2016, Instituto de Salud Carlos III (ISCIII), Subdirección General de Evaluación y Fomento de la Investigación, Ministerio de Economía y Competitividad (Gobierno de España) and Fondo Europeo de Desarrollo Regional (FEDER); ED431C2017/57 from the Consellería de Cultura, Educación e Ordenación Universitaria, (Xunta de Galicia) and FEDER.
Acknowledgments
S.-C.F.-S. acknowledges the FPU programme for her grant (FPU15/02644) from the Secretaría General de Universidades, Spanish Ministerio de Educación, Cultura y Deporte. V.G. acknowledges the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia for her postdoctoral grant (ED481B2018/018).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boehmer, T.; Vogler, A.J.; Thomas, A.; Sauer, S.; Hergenroether, M.; Straubinger, R.K.; Birdsell, D.; Keim, P.; Sahl, J.W.; Williamson, C.H.D.; et al. Phenotypic characterization and whole genome analysis of extended-spectrum beta-lactamase-producing bacteria isolated from dogs in Germany. PLoS ONE 2018, 13, e0206252. [Google Scholar] [CrossRef]
- Bourne, J.A.; Chong, W.L.; Gordon, D.M. Genetic structure, antimicrobial resistance and frequency of human associated Escherichia coli sequence types among faecal isolates from healthy dogs and cats living in Canberra, Australia. PLoS ONE 2019, 14, e0212867. [Google Scholar] [CrossRef]
- Flament-Simon, S.C.; Nicolas-Chanoine, M.H.; García, V.; Duprilot, M.; Mayer, N.; Alonso, M.P.; García-Meniño, I.; Blanco, J.E.; Blanco, M.; Blanco, J. Clonal structure, virulence factor-encoding genes and antibiotic resistance of Escherichia coli, causing urinary tract infections and other extraintestinal infections in humans in Spain and France during 2016. Antibiotics 2020, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Mamani, R.; Flament-Simon, S.C.; García, V.; Mora, A.; Alonso, M.P.; López, C.; García-Meniño, I.; Díaz-Jiménez, D.; Blanco, J.E.; Blanco, M.; et al. Sequence types, clonotypes, serotypes, and virotypes of extended-spectrum β-lactamase-producing Escherichia coli causing bacteraemia in a Spanish hospital over a 12-year period (2000 to 2011). Front. Microbiol. 2019, 10, 1530. [Google Scholar] [CrossRef]
- Thompson, M.F.; Litster, A.L.; Platell, J.L.; Trott, D.J. Canine bacterial urinary tract infections: New developments in old pathogens. Vet. J. 2011, 190, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Whittam, T.S.; Wolfe, M.L.; Wilson, R.A. Genetic relationships among Escherichia coli isolates causing urinary tract infections in humans and animals. Epidemiol. Infect. 1989, 102, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Porter, S.; Johnston, B.; Kuskowski, M.A.; Spurbeck, R.R.; Mobley, H.L.T.; Williamson, D.A. Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect. Dis. 2015, 2, ofv083. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Dinh, P.C.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L.T. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef] [PubMed]
- Kallonen, T.; Brodrick, H.J.; Harris, S.R.; Corander, J.; Brown, N.M.; Martin, V.; Peacock, S.J.; Parkhill, J. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017, 27, 1437–1449. [Google Scholar] [CrossRef]
- Kidsley, A.K.; O’Dea, M.; Saputra, S.; Jordan, D.; Johnson, J.R.; Gordon, D.M.; Turni, C.; Djordjevic, S.P.; Abraham, S.; Trott, D.J. Genomic analysis of phylogenetic group B2 extraintestinal pathogenic E. coli causing infections in dogs in Australia. Vet. Microbiol. 2020, 248, 108783. [Google Scholar] [CrossRef] [PubMed]
- LeCuyer, T.E.; Byrne, B.A.; Daniels, J.B.; Diaz-Campos, D.V.; Hammac, G.K.; Miller, C.B.; Besser, T.E.; Davis, M.A. Population structure and antimicrobial resistance of canine uropathogenic Escherichia coli. J. Clin. Microbiol. 2018, 56, e00788-18. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global extraintestinal pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Valat, C.; Drapeau, A.; Beurlet, S.; Bachy, V.; Boulouis, H.-J.; Pin, R.; Cazeau, G.; Madec, J.-Y.; Haenni, M. Pathogenic Escherichia coli in dogs reveals the predominance of ST372 and the human-associated ST73 extra-intestinal lineages. Front. Microbiol. 2020, 11, 580. [Google Scholar] [CrossRef] [PubMed]
- Zogg, A.L.; Zurfluh, K.; Schmitt, S.; Nüesch-Inderbinen, M.; Stephan, R. Antimicrobial resistance, multilocus sequence types and virulence profiles of ESBL producing and non-ESBL producing uropathogenic Escherichia coli isolated from cats and dogs in Switzerland. Vet. Microbiol. 2018, 216, 79–84. [Google Scholar] [CrossRef]
- Johnson, J.R.; Clabots, C.; Kuskowski, M.A. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. J. Clin. Microbiol. 2008, 46, 4078–4082. [Google Scholar] [CrossRef]
- Johnson, J.R.; Johnston, B.; Clabots, C.R.; Kuskowski, M.A.; Roberts, E.; DebRoy, C. Virulence genotypes and phylogenetic background of Escherichia coli serogroup O6 isolates from humans, dogs, and cats. J. Clin. Microbiol. 2008, 46, 417–422. [Google Scholar] [CrossRef]
- Platell, J.L.; Cobbold, R.N.; Johnson, J.R.; Heisig, A.; Heisig, P.; Clabots, C.; Kuskowski, M.A.; Trott, D.J. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob. Agents Chemother. 2011, 55, 3782–3787. [Google Scholar] [CrossRef]
- Platell, J.L.; Trott, D.J.; Johnson, J.R.; Heisig, P.; Heisig, A.; Clabots, C.R.; Johnston, B.; Cobbold, R.N. Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob. Agents Chemother. 2012, 56, 3898–3904. [Google Scholar] [CrossRef]
- De Toro, M.; Fernández, J.; García, V.; Mora, A.; Blanco, J.; de la Cruz, F.; Rodicio, M.R. Whole genome sequencing, molecular typing and in vivo virulence of OXA-48-producing Escherichia coli isolates including ST131 H30-Rx, H22 and H41 subclones. Sci. Rep. 2017, 7, 12103. [Google Scholar] [CrossRef]
- Flament-Simon, S.C.; García, V.; Duprilot, M.; Mayer, N.; Alonso, M.P.; García-Meniño, I.; Blanco, J.E.; Blanco, M.; Nicolas-Chanoine, M.H.; Blanco, J. High Prevalence of ST131 subclades C2-H30Rx and C1-M27 among extended-spectrum β-lactamase-producing Escherichia coli causing human extraintestinal infections in patients from two hospitals of Spain and France during 2015. Front. Cell. Infect. Microbiol. 2020, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Jamborova, I.; Johnston, B.D.; Papousek, I.; Kachlikova, K.; Micenkova, L.; Clabots, C.; Skalova, A.; Chudejova, K.; Dolejska, M.; Literak, I.; et al. Extensive genetic commonality among wildlife, wastewater, community, and nosocomial isolates of Escherichia coli sequence type 131 (H30R1 and H30Rx subclones) that carry blaCTX-M-27 or blaCTX-M-15. Antimicrob. Agents Chemother. 2018, 62, e00519-18. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Pitout, J.D.D.; Peirano, G.; DeVinney, R.; Noguchi, T.; Yamamoto, M.; Gomi, R.; Matsuda, T.; Nakano, S.; Nagao, M.; et al. Rapid identification of different Escherichia coli sequence type 131 clades. Antimicrob. Agents Chemother. 2017, 61, e00179-17. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Canica, M.M.; Park, Y.-J.; Lavigne, J.-P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal Group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 2013, 4, e00377-13. [Google Scholar] [CrossRef] [PubMed]
- Belas, A.; Marques, C.; Aboim, C.; Pomba, C. Emergence of Escherichia coli ST131 H30/H30-Rx subclones in companion animals. J. Antimicrob. Chemother. 2018, 74, 266–269. [Google Scholar] [CrossRef]
- Bortolami, A.; Zendri, F.; Maciuca, E.I.; Wattret, A.; Ellis, C.; Schmidt, V.; Pinchbeck, G.; Timofte, D. Diversity, virulence, and clinical significance of extended-spectrum β-lactamase- and pAmpC-producing Escherichia coli from companion animals. Front. Microbiol. 2019, 10, 1260. [Google Scholar] [CrossRef]
- Kawamura, K.; Sugawara, T.; Matsuo, N.; Hayashi, K.; Norizuki, C.; Tamai, K.; Kondo, T.; Arakawa, Y. Spread of CTX-type extended-spectrum β-lactamase-producing Escherichia coli isolates of epidemic clone B2-O25-ST131 among dogs and cats in Japan. Microb. Drug Resist. 2017, 23, 1059–1066. [Google Scholar] [CrossRef]
- Maeyama, Y.; Taniguchi, Y.; Hayashi, W.; Ohsaki, Y.; Osaka, S.; Koide, S.; Tamai, K.; Nagano, Y.; Arakawa, Y.; Nagano, N. Prevalence of ESBL/AmpC genes and specific clones among the third-generation cephalosporin-resistant Enterobacteriaceae from canine and feline clinical specimens in Japan. Vet. Microbiol. 2018, 216, 183–189. [Google Scholar] [CrossRef]
- Melo, L.C.; Haenni, M.; Saras, E.; Duprilot, M.; Nicolas-Chanoine, M.H.; Madec, J.Y. Emergence of the C1-M27 cluster in ST131 Escherichia coli from companion animals in France. J. Antimicrob. Chemother. 2019, 74, 3111–3113. [Google Scholar] [CrossRef]
- Bogaerts, P.; Huang, T.-D.; Bouchahrouf, W.; Bauraing, C.; Berhin, C.; El Garch, F.; Glupczynski, Y. Characterization of ESBL- and AmpC-producing Enterobacteriaceae from diseased companion animals in Europe. Microb. Drug Resist. 2015, 21, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.; Zhang, Y.; Zhang, Z.; Lei, L.; Xia, Z. Increasing prevalence of ESBL-producing multidrug resistance Escherichia coli from diseased pets in Beijing, China from 2012 to 2017. Front. Microbiol. 2019, 10, 2852. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Imirzalioglu, C.; Ghosh, H.; Gwozdzinski, K.; Schmiedel, J.; Gentil, K.; Bauerfeind, R.; Kämpfer, P.; Seifert, H.; Michael, G.B.; et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents 2016, 47, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Nakai, Y.; Kataoka, Y. Mechanisms of resistance to cephalosporin and emergence of O25b-ST131 clone harboring CTX-M-27 β-lactamase in extraintestinal pathogenic Escherichia coli from dogs and cats in Japan. Microbiol. Immunol. 2012, 56, 480–485. [Google Scholar] [CrossRef]
- Johnson, J.R.; Kuskowski, M.A.; Owens, K.; Clabots, C.; Singer, R.S. Virulence genotypes and phylogenetic background of fluoroquinolone-resistant and susceptible Escherichia coli urine isolates from dogs with urinary tract infection. Vet. Microbiol. 2009, 136, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Li, Y.; Hao, C. High prevalence of β-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in Shaanxi, China. Front. Microbiol. 2016, 7, 1843. [Google Scholar] [CrossRef]
- Liu, X.; Thungrat, K.; Boothe, D.M. Occurrence of OXA-48 carbapenemase and other β-lactamase genes in ESBL-producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009–2013. Front. Microbiol. 2016, 7, 1057. [Google Scholar] [CrossRef]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef]
- Pepin-Puget, L.; El Garch, F.; Bertrand, X.; Valot, B.; Hocquet, D. Genome analysis of Enterobacteriaceae with non-wild type susceptibility to third-generation cephalosporins recovered from diseased dogs and cats in Europe. Vet. Microbiol. 2020, 242, 108601. [Google Scholar] [CrossRef]
- Wagner, S.; Gally, D.L.; Argyle, S.A. Multidrug-resistant Escherichia coli from canine urinary tract infections tend to have commensal phylotypes, lower prevalence of virulence determinants and ampC-replicons. Vet. Microbiol. 2014, 169, 171–178. [Google Scholar] [CrossRef][Green Version]
- Zhang, P.L.C.; Shen, X.; Chalmers, G.; Reid-Smith, R.J.; Slavic, D.; Dick, H.; Boerlin, P. Prevalence and mechanisms of extended-spectrum cephalosporin resistance in clinical and fecal Enterobacteriaceae isolates from dogs in Ontario, Canada. Vet. Microbiol. 2018, 213, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Ukah, U.V.; Glass, M.; Avery, B.; Daignault, D.; Mulvey, M.R.; Reid-smith, R.J.; Parmley, E.J.; Portt, A.; Boerlin, P.; Manges, A.R. Risk factors for acquisition of multidrug-resistant Escherichia coli and development of community-acquired urinary tract infections. Epidemiol. Infect. 2018, 146, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Guinée, P.A.M.; Jansen, W.H.; Wadström, T.; Sellwood, R. Escherichia coli associated with neonatal diarrhoea in piglets and calves. In Laboratory Diagnosis in Neonatal Calf and Pig Diarrhoea; Springer: Dordrecht, The Netherlands, 1981; pp. 126–162. [Google Scholar]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Tchesnokova, V.; Billig, M.; Chattopadhyay, S.; Linardopoulou, E.; Aprikian, P.; Roberts, P.L.; Skrivankova, V.; Johnston, B.; Gileva, A.; Igusheva, I.; et al. Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J. Clin. Microbiol. 2013, 51, 2991–2999. [Google Scholar] [CrossRef]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesøe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a web tool for subtyping of extraintestinal pathogenic Escherichia coli based on the fumC and fimH alleles. J. Clin. Microbiol. 2018, 56, e00063-18. [Google Scholar] [CrossRef]
- Dahbi, G.; Mora, A.; Mamani, R.; López, C.; Alonso, M.P.; Marzoa, J.; Blanco, M.; Herrera, A.; Viso, S.; García-Garrote, F.; et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: Comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. 2014, 304, 1247–1257. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Blanco, M.; Alonso, M.P.; Nicolas-Chanoine, M.-H.; Dahbi, G.; Mora, A.; Blanco, J.E.; López, C.; Cortés, P.; Llagostera, M.; Leflon-Guibout, V.; et al. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): Dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2009, 63, 1135–1141. [Google Scholar] [CrossRef]
- Leflon-Guibout, V.; Jurand, C.; Bonacorsi, S.; Espinasse, F.; Guelfi, M.C.; Duportail, F.; Heym, B.; Bingen, E.; Nicolas-Chanoine, M.H. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 2004, 48, 3736–3742. [Google Scholar] [CrossRef]
- Perez-Perez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Flament-Simon, S.C.; de Toro, M.; Mora, A.; García, V.; García-Meniño, I.; Díaz-Jiménez, D.; Herrera, A.; Blanco, J. Whole genome sequencing and characteristics of mcr-1–harboring plasmids of porcine Escherichia coli isolates belonging to the high-risk clone O25b:H4-ST131 clade B. Front. Microbiol. 2020, 11, 387. [Google Scholar] [CrossRef] [PubMed]
- Lanza, V.F.; de Toro, M.; Garcillán-Barcia, M.P.; Mora, A.; Blanco, J.; Coque, T.M.; de la Cruz, F. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 2014, 10, e1004766. [Google Scholar] [CrossRef] [PubMed]
- Vielva, L.; de Toro, M.; Lanza, V.F.; de la Cruz, F. PLACNETw: A web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 2017, 33, 3796–3798. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Alvarado, A.; Garcillán-Barcia, M.P.; de la Cruz, F. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS ONE 2012, 7, e40438. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Flament-Simon, S.C.; Duprilot, M.; Mayer, N.; García, V.; Alonso, M.P.; Blanco, J.; Nicolas-Chanoine, M.H. Association between kinetics of early biofilm formation and clonal lineage in Escherichia coli. Front. Microbiol. 2019, 10, 1183. [Google Scholar] [CrossRef]
- Abreu-Salinas, F.; Díaz-Jiménez, D.; García-Meniño, I.; Lumbreras, P.; López-Beceiro, A.M.; Fidalgo, L.E.; Rodicio, M.R.; Mora, A.; Fernández, J. High prevalence and diversity of cephalosporin-resistant Enterobacteriaceae including extraintestinal pathogenic E. coli CC648 lineage in rural and urban dogs in Northwest Spain. Antibiotics 2020, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Belas, A.; Salazar, A.S.; Gama, L.T.d.; Couto, N.; Pomba, C. Risk factors for faecal colonisation with Escherichia coli producing extended-spectrum and plasmid-mediated AmpC-lactamases in dogs. Vet. Rec. 2014, 175, 202. [Google Scholar] [CrossRef] [PubMed]
- Damborg, P.; Morsing, M.K.; Petersen, T.; Bortolaia, V.; Guardabassi, L. CTX-M-1 and CTX-M-15-producing Escherichia coli in dog faeces from public gardens. Acta Vet. Scand. 2015, 57, 83. [Google Scholar] [CrossRef] [PubMed]
- Dupouy, V.; Abdelli, M.; Moyano, G.; Arpaillange, N.; Bibbal, D.; Cadiergues, M.-C.; Lopez-Pulin, D.; Sayah-Jeanne, S.; de Gunzburg, J.; Saint-Lu, N.; et al. Prevalence of beta-lactam and quinolone/fluoroquinolone resistance in Enterobacteriaceae from dogs in France and Spain—Characterization of ESBL/pAmpC isolates, genes, and conjugative plasmids. Front. Vet. Sci. 2019, 6, 279. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Métayer, V.; Médaille, C.; Madec, J.-Y. High prevalence of blaCTX-M-1 /IncI1/ST3 and blaCMY-2 /IncI1/ST2 plasmids in healthy urban dogs in France. Antimicrob. Agents Chemother. 2014, 58, 5358–5362. [Google Scholar] [CrossRef]
- Hansen, K.H.; Bortolaia, V.; Nielsen, C.A.; Nielsen, J.B.; Schønning, K.; Agersø, Y.; Guardabassi, L. Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 β-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl. Environ. Microbiol. 2016, 82, 4705–4714. [Google Scholar] [CrossRef]
- Hong, J.S.; Song, W.; Park, H.-M.; Oh, J.-Y.; Chae, J.-C.; Shin, S.; Jeong, S.H. Clonal spread of extended-spectrum cephalosporin-resistant Enterobacteriaceae between companion animals and humans in South Korea. Front. Microbiol. 2019, 10, 1371. [Google Scholar] [CrossRef]
- Hordijk, J.; Schoormans, A.; Kwakernaak, M.; Duim, B.; Broens, E.; Dierikx, C.; Mevius, D.; Wagenaar, J.A. High prevalence of fecal carriage of extended-spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 2013, 4, 242. [Google Scholar] [CrossRef]
- Karkaba, A.; Hill, K.; Benschop, J.; Pleydell, E.; Grinberg, A. Carriage and population genetics of extended spectrum β-lactamase-producing Escherichia coli in cats and dogs in New Zealand. Vet. Microbiol. 2019, 233, 61–67. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Betts, J.; La Ragione, R.; van Essen-Zandbergen, A.; Ceccarelli, D.; Petinaki, E.; Koutinas, C.K.; Mevius, D.J. Occurrence and characterization of extended-spectrum cephalosporin-resistant Enterobacteriaceae in healthy household dogs in Greece. J. Med. Microbiol. 2018, 67, 931–935. [Google Scholar] [CrossRef]
- Melo, L.C.; Oresco, C.; Leigue, L.; Netto, H.M.; Melville, P.A.; Benites, N.R.; Saras, E.; Haenni, M.; Lincopan, N.; Madec, J.Y. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 2018, 221, 59–66. [Google Scholar] [CrossRef]
- Pires, J.; Bernasconi, O.J.; Kasraian, S.; Hilty, M.; Perreten, V.; Endimiani, A. Intestinal colonisation with extended-spectrum cephalosporin-resistant Escherichia coli in Swiss pets: Molecular features, risk factors and transmission with owners. Int. J. Antimicrob. Agents 2016, 48, 759–760. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Gracia, R.C.; Cortés-Cortés, G.; Lozano-Zarain, P.; Bello, F.; Martínez-Laguna, Y.; Torres, C. Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 2015, 203, 315–319. [Google Scholar] [CrossRef]
- Schaufler, K.; Bethe, A.; Lübke-Becker, A.; Ewers, C.; Kohn, B.; Wieler, L.H.; Guenther, S. Putative connection between zoonotic multiresistant extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in dog feces from a veterinary campus and clinical isolates from dogs. Infect. Ecol. Epidemiol. 2015, 5, 25334. [Google Scholar] [CrossRef]
- Seni, J.; Falgenhauer, L.; Simeo, N.; Mirambo, M.M.; Imirzalioglu, C.; Matee, M.; Rweyemamu, M.; Chakraborty, T.; Mshana, S.E. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front. Microbiol. 2016, 7, 142. [Google Scholar] [CrossRef]
- Sevilla, E.; Mainar-Jaime, R.C.; Moreno, B.; Martín-Burriel, I.; Morales, M.; Andrés-Lasheras, S.; Chirino-Trejo, M.; Badiola, J.J.; Bolea, R. Antimicrobial resistance among canine enteric Escherichia coli isolates and prevalence of attaching–effacing and extraintestinal pathogenic virulence factors in Spain. Acta Vet. Hung. 2020, 68, 1–7. [Google Scholar] [CrossRef]
- Suay-García, B.; Galán, F.; Rodríguez-Iglesias, M.A.; Pérez-Gracia, M.T. Detection and characterization of extended-spectrum beta-lactamases-producing Escherichia coli in animals. Vector Borne Zoonotic Dis. 2019, 19, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Umeda, K.; Hase, A.; Matsuo, M.; Horimoto, T.; Ogasawara, J. Prevalence and genetic characterization of cephalosporin-resistant Enterobacteriaceae among dogs and cats in an animal shelter. J. Med. Microbiol. 2019, 68, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Wedley, A.L.; Dawson, S.; Maddox, T.W.; Coyne, K.P.; Pinchbeck, G.L.; Clegg, P.; Nuttall, T.; Kirchner, M.; Williams, N.J. Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 2017, 199, 23–30. [Google Scholar] [CrossRef]
- Adler, A.; Gniadkowski, M.; Baraniak, A.; Izdebski, R.; Fiett, J.; Hryniewicz, W.; Malhotra-Kumar, S.; Goossens, H.; Lammens, C.; Lerman, Y.; et al. Transmission dynamics of ESBL-producing Escherichia coli clones in rehabilitation wards at a tertiary care centre. Clin. Microbiol. Infect. 2012, 18, E497–E505. [Google Scholar] [CrossRef]
- Izdebski, R.; Baraniak, A.; Fiett, J.; Adler, A.; Kazma, M.; Salomon, J.; Lawrence, C.; Rossini, A.; Salvia, A.; Vidal Samso, J.; et al. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 2013, 57, 309–316. [Google Scholar] [CrossRef]
- Dierikx, C.M.; van Duijkeren, E.; Schoormans, A.H.W.; van Essen-Zandbergen, A.; Veldman, K.; Kant, A.; Huijsdens, X.W.; van der Zwaluw, K.; Wagenaar, J.A.; Mevius, D.J. Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 2012, 67, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Stamm, I.; Grobbel, M.; Kopp, P.A.; Guerra, B.; Stubbe, M.; Doi, Y.; Zong, Z.; Kola, A.; et al. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: Another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 2014, 69, 1224–1230. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Li, Y.; Hao, C. Association between virulence profile and fluoroquinolone resistance in Escherichia coli isolated from dogs and cats in China. J. Infect. Dev. Ctries. 2017, 11, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Loncaric, I.; Misic, D.; Szostak, M.P.; Künzel, F.; Schäfer-Somi, S.; Spergser, J. Broad-spectrum cephalosporin-resistant and/or fluoroquinolone-resistant Enterobacterales associated with canine and feline urogenital infections. Antibiotics 2020, 9, 387. [Google Scholar] [CrossRef] [PubMed]
- Kidsley, A.K.; White, R.T.; Beatson, S.A.; Saputra, S.; Schembri, M.A.; Gordon, D.; Johnson, J.R.; O’Dea, M.; Mollinger, J.L.; Abraham, S.; et al. Companion animals are spillover hosts of the multidrug-resistant human extraintestinal Escherichia coli pandemic clones ST131 and ST1193. Front. Microbiol. 2020, 11, 1968. [Google Scholar] [CrossRef]
- Wang, J.; Huang, X.-Y.; Xia, Y.-B.; Guo, Z.-W.; Ma, Z.-B.; Yi, M.-Y.; Lv, L.-C.; Lu, P.-L.; Yan, J.-C.; Huang, J.-W.; et al. Clonal spread of Escherichia coli ST93 carrying mcr-1-harboring IncN1-IncHI2/ST3 plasmid among companion animals, China. Front. Microbiol. 2018, 9, 2989. [Google Scholar] [CrossRef]
- Pulss, S.; Stolle, I.; Stamm, I.; Leidner, U.; Heydel, C.; Semmler, T.; Prenger-Berninghoff, E.; Ewers, C. Multispecies and clonal dissemination of OXA-48 carbapenemase in Enterobacteriaceae from companion animals in Germany, 2009–2016. Front. Microbiol. 2018, 9, 1265. [Google Scholar] [CrossRef]
- Melo, L.C.; Boisson, M.N.G.; Saras, E.; Médaille, C.; Boulouis, H.-J.; Madec, J.-Y.; Haenni, M. OXA-48-producing ST372 Escherichia coli in a French dog. J. Antimicrob. Chemother. 2016, 72, 1256–1258. [Google Scholar] [CrossRef][Green Version]
- Schubert, S.; Rakin, A.; Karch, H.; Carniel, E.; Heesemann, J. Prevalence of the “High-Pathogenicity Island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 1998, 66, 480–485. [Google Scholar] [CrossRef]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nature Rev. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M.; Dalmasso, G.; Beyrouthy, R.; Barnich, N.; Delmas, J.; Bonnet, R. Pathogenicity factors of genomic islands in intestinal and extraintestinal Escherichia coli. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).