Understanding High-Salt and Cold Adaptation of a Polyextremophilic Enzyme
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Strains, Plasmids, Media, and Culture Conditions
2.3. Expression of the β-Galactosidase Gene in Haloferax Volcanii
2.4. β-Galactosidase Purification
2.5. Tryptic Digest and LC-MS/MS Analysis
2.6. MALDI-TOF
2.7. β-Galactosidase Activity Assay
2.8. Enzyme Characterization
2.9. X-ray Crystallography
2.10. Data Collection, Structure Solution and Refinement
2.11. Molecular Modeling
2.12. Structural Analysis
2.13. Molecular Dynamics Simulations
2.14. Accession Numbers
3. Results
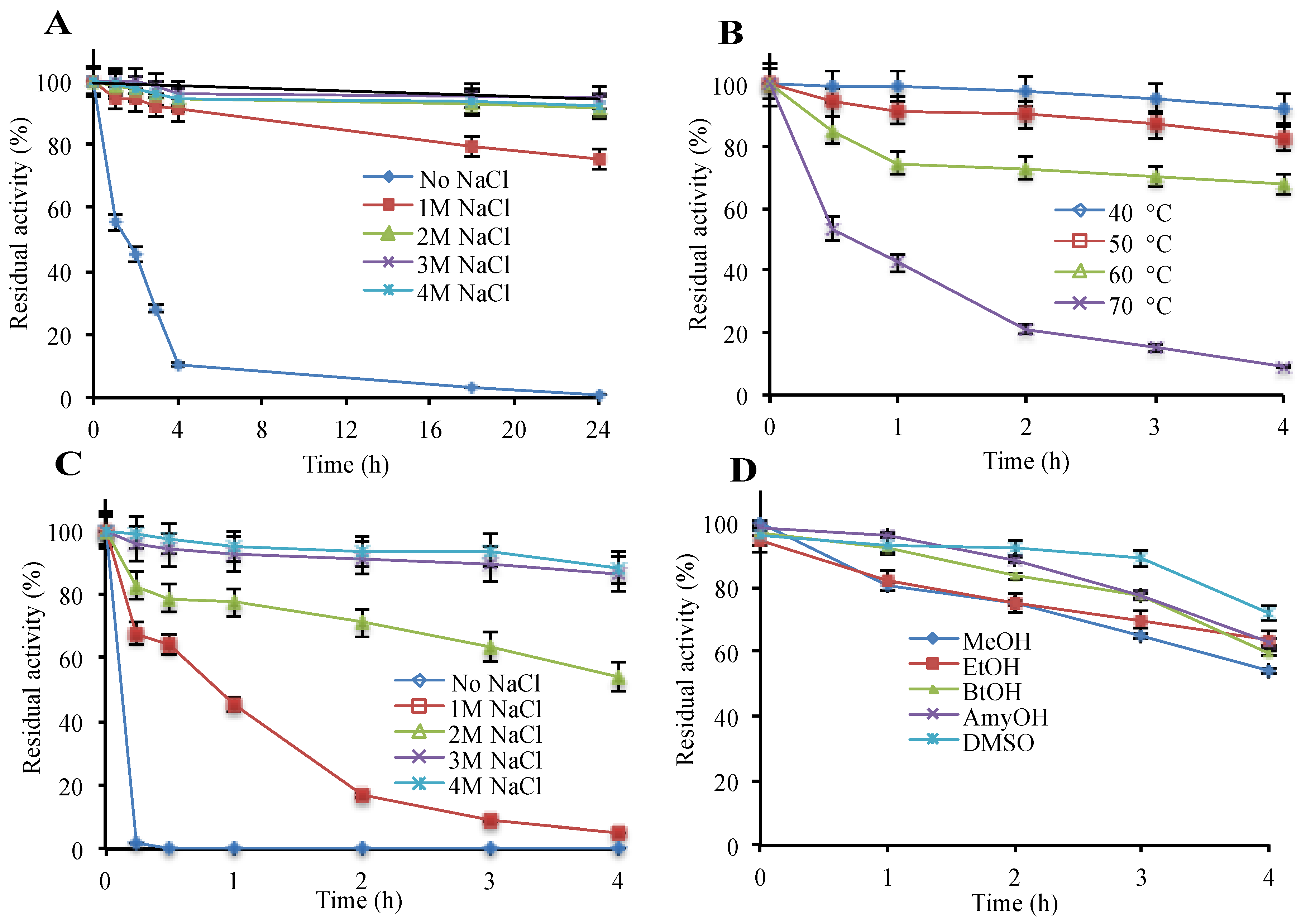
3.1. Recombinant Production and Biochemical Assessment of hla_bga
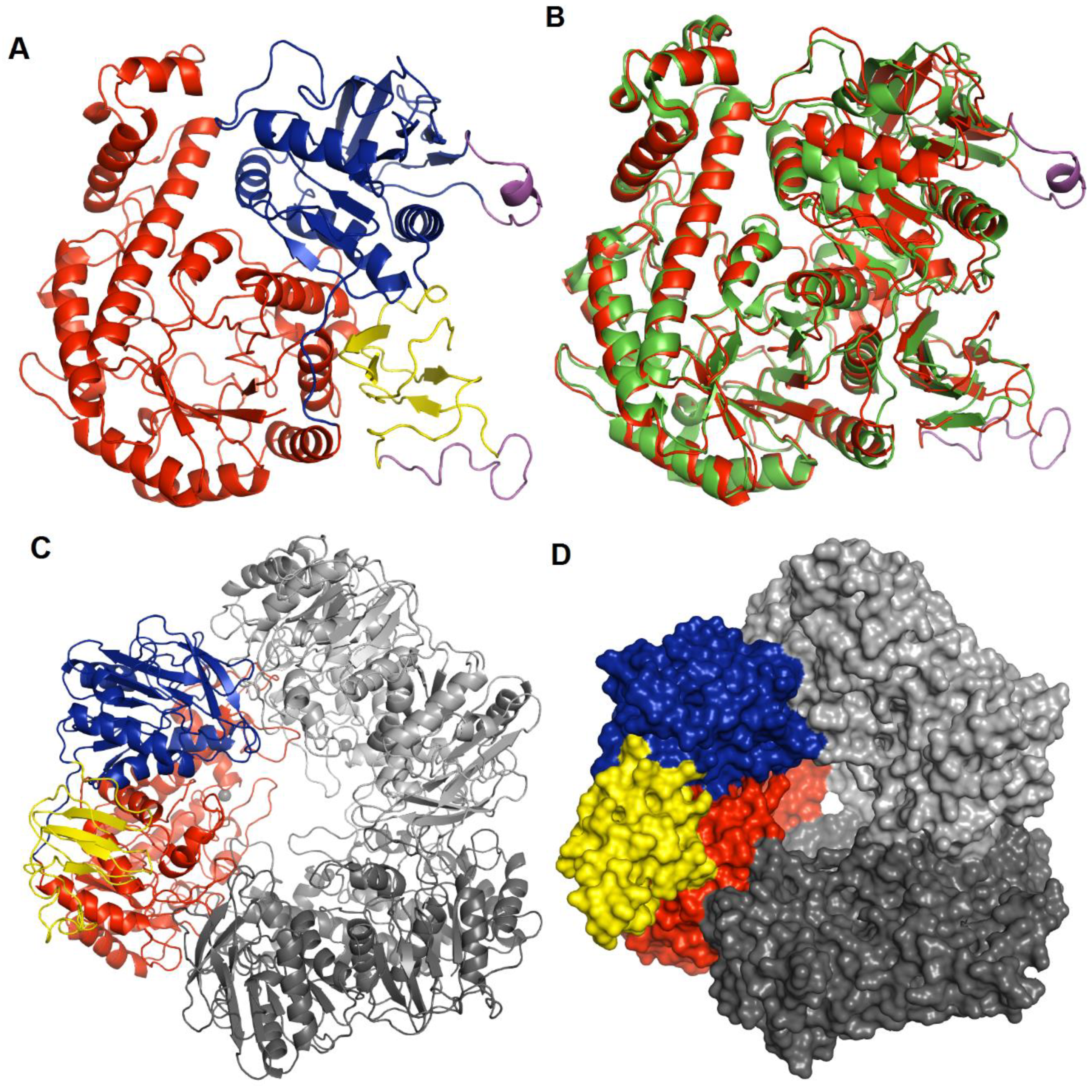
3.2. Overall Structure of hla_bga and Subunit Interactions
3.3. Catalytic Site Architecture
3.4. Quaternary Structure
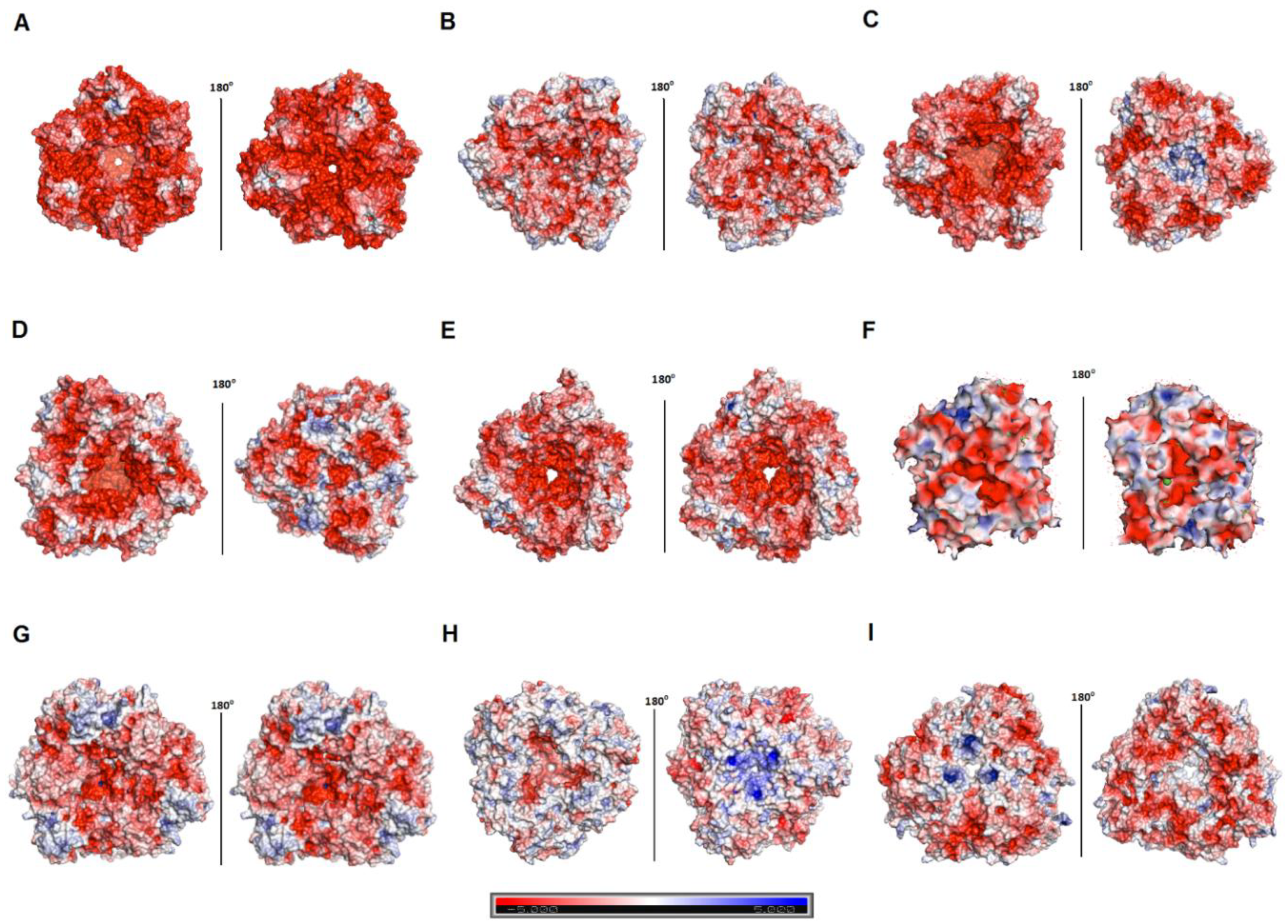
3.5. Hla_bga Combines Adaptive Features for Halo- and Psychrophily
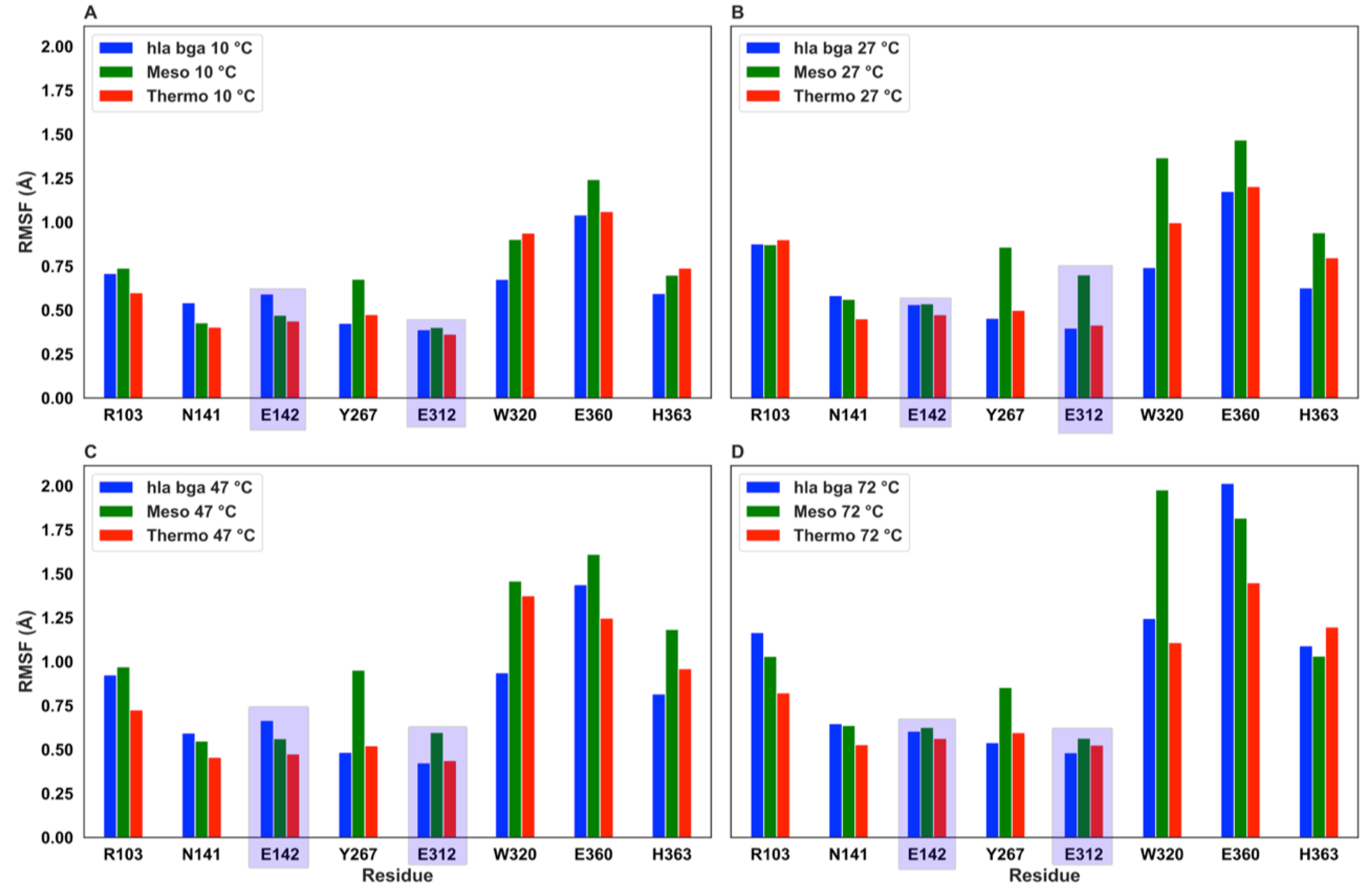
3.6. Molecular Dynamics Simulations Show Residue-Level Flexibility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Sharma, S. Enzymes in green chemistry: The need for environment and sustainability. Int. J. Appl. Res. 2016, 2, 337–341. [Google Scholar]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Deng, C.; Huang, T.; Jiang, Z.; Lv, X.; Liu, L.; Chen, J.; Du, G. Enzyme Engineering and Industrial Bioprocess. In Current Developments in Biotechnology and Bioengineering; Singh, S.P., Pandey, A., Du, G., Kumar, S., Eds.; Elsevier: New York, NY, USA, 2019; pp. 165–188. [Google Scholar]
- Elleuche, S.; Schroder, C.; Sahm, K.; Antranikian, G. Extremozymes--biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 2014, 29, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. What are the limitations of enzymes in synthetic organic chemistry? Chem. Rec. 2016, 16, 2449–2459. [Google Scholar] [CrossRef] [PubMed]
- Kaul, P.; Asano, Y. Strategies for discovery and improvement of enzyme function: State of the art and opportunities. Microb. Biotechnol. 2012, 5, 18–33. [Google Scholar] [CrossRef]
- Liszka, M.J.; Clark, M.E.; Schneider, E.; Clark, D.S. Nature versus nurture: Developing enzymes that function under extreme conditions. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 77–102. [Google Scholar] [CrossRef]
- Jimenez-Rosales, A.; Flores-Merino, M.V. Tailoring proteins to re-evolve Nature: A short review. Mol. Biotechnol. 2018, 60, 946–974. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and hot extremozymes: Industrial relevance and current trends. Front. Bioeng. 2015, 3, 148. [Google Scholar] [CrossRef]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F1000Research 2016, 5. F1000 Faculty Rev.396. [Google Scholar] [CrossRef]
- Gomes, J.; Steiner, W. The biocatalytic potential of extremophiles and extremozymes. Food Technol. 2004, 42, 223–225. [Google Scholar]
- Karan, R.; Kumar, S.; Sinha, R.; Khare, S. Halophilic microorganisms as sources of novel enzymes. In Microorganisms in Sustainable Agriculture and Biotechnology; Satyanarayana, T., Johri, B., Prakash, A., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 555–579. [Google Scholar]
- Karan, R.; Capes, M.D.; DasSarma, S. Function and biotechnology of extremophilic enzymes in low water activity. Aquat. Biosyst. 2012, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Grotzinger, S.W.; Karan, R.; Strillinger, E.; Bader, S.; Frank, A.; Al Rowaihi, I.S.; Akal, A.; Wackerow, W.; Archer, J.A.; Rueping, M.; et al. Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes. ACS Chem. Biol. 2018, 13, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Karan, R.; Khare, S. Stability of haloalkaliphilic Geomicrobium sp. protease modulated by salt. Biochemistry (Moscow) 2011, 76, 686. [Google Scholar] [CrossRef]
- Karan, R.; Singh, S.; Kapoor, S.; Khare, S. A novel organic solvent tolerant protease from a newly isolated Geomicrobium sp. EMB2 (MTCC 10310): Production optimization by response surface methodology. New Biotechnol. 2011, 28, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Vogler, M.; Karan, R.; Renn, D.; Vancea, A.; Vielberg, M.-T.; Grötzinger, S.W.; DasSarma, P.; DasSarma, S.; Eppinger, J.; Groll, M. Crystal structure and active site engineering of a halophilic γ-carbonic anhydrase. Front. Microbiol. 2020, 11, 742. [Google Scholar] [CrossRef]
- Cabrera, M.Á.; Blamey, J.M. Biotechnological applications of archaeal enzymes from extreme environments. J. Biol. Res. 2018, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Karan, R.; Khare, S. Purification and characterization of a solvent-stable protease from Geomicrobium sp. EMB2. Environ. Technol. 2010, 31, 1061–1072. [Google Scholar] [CrossRef]
- Joseph, B.; Kumar, V.; Ramteke, P.W. Psychrophilic Enzymes: Potential Biocatalysts for Food Processing. In Enzymes in Food Biotechnology; Kuddus, M., Ed.; Elsevier: New York, NY, USA, 2019; pp. 817–825. [Google Scholar]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and Applications of Extremozymes from Deep-Sea Extremophilic Microorganisms: A Mini Review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef]
- Mangiagalli, M.; Brocca, S.; Orlando, M.; Lotti, M. The “cold revolution”. Present and future applications of cold-active enzymes and ice-binding proteins. New Biotechnol. 2020, 55, 5–11. [Google Scholar] [CrossRef]
- Al-Maqtari, Q.A.; Waleed, A.A.; Mahdi, A.A. Cold-active enzymes and their applications in industrial fields-A review. Int. J. Res. Stud. Agric. Sci. 2019, 6, 107–123. [Google Scholar]
- Britton, K.L.; Baker, P.J.; Fisher, M.; Ruzheinikov, S.; Gilmour, D.J.; Bonete, M.J.; Ferrer, J.; Pire, C.; Esclapez, J.; Rice, D.W. Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei. Proc. Natl. Acad. Sci. USA 2006, 103, 4846–4851. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.M.; Lee, J.; Blaber, M. Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein. Proc. Natl. Acad. Sci. USA 2013, 110, 2135–2139. [Google Scholar] [CrossRef]
- Karan, R.; Capes, M.D.; DasSarma, P.; DasSarma, S. Cloning, overexpression, purification, and characterization of a polyextremophilic beta-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnol. 2013, 13, 3. [Google Scholar] [CrossRef]
- DasSarma, S.; Capes, M.D.; Karan, R.; DasSarma, P. Amino acid substitutions in cold-adapted proteins from Halorubrum lacusprofundi, an extremely halophilic microbe from Antarctica. PLoS ONE 2013, 8, e58587. [Google Scholar] [CrossRef]
- Laye, V.J.; Karan, R.; Kim, J.-M.; Pecher, W.T.; DasSarma, P.; DasSarma, S. Key amino acid residues conferring enhanced enzyme activity at cold temperatures in an Antarctic polyextremophilic β-galactosidase. Proc. Natl. Acad. Sci. USA 2017, 114, 12530–12535. [Google Scholar] [CrossRef]
- Laye, V.J.; DasSarma, S. An Antarctic extreme halophile and its polyextremophilic enzyme: Effects of perchlorate salts. Astrobiology 2018, 18, 412–418. [Google Scholar] [CrossRef]
- Cavicchioli, R. Cold-adapted archaea. Nat. Rev. Microbiol. 2006, 4, 331. [Google Scholar] [CrossRef]
- Orosei, R.; Ding, C.; Fa, W.; Giannopoulos, A.; Hérique, A.; Kofman, W.; Lauro, S.E.; Li, C.; Pettinelli, E.; Su, Y. The Global Search for Liquid Water on Mars from Orbit: Current and Future Perspectives. Life 2020, 10, 120. [Google Scholar] [CrossRef]
- Franzmann, P.; Stackebrandt, E.; Sanderson, K.; Volkman, J.; Cameron, D.; Stevenson, P.; McMeekin, T.; Burton, H. Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst. Appl. Microbiol. 1988, 11, 20–27. [Google Scholar] [CrossRef]
- Strillinger, E.; Grötzinger, S.W.; Allers, T.; Eppinger, J.; Weuster-Botz, D. Production of halophilic proteins using Haloferax volcanii H1895 in a stirred-tank bioreactor. Appl. Microbiol. Biotechnol. 2016, 100, 1183–1195. [Google Scholar] [CrossRef] [PubMed]
- Dyall-Smith, M. The Halohandbook: Protocols for Haloarchaeal Genetics. Available online: http://http://www.haloarchaea.com/resources/halohandbook (accessed on 1 October 2020).
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. J. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359. [Google Scholar] [CrossRef] [PubMed]
- Hungler, A.; Momin, A.; Diederichs, K.; Arold, S.T. ContaMiner and ContaBase: A webserver and database for early identification of unwantedly crystallized protein contaminants. J. Appl. Crystallogr. 2016, 49, 2252–2258. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. Section D: Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Long, F.; Vagin, A.A.; Young, P.; Murshudov, G.N. BALBES: A molecular-replacement pipeline. Acta Crystallog. Section D-Biol. Crystallogr. 2008, 64, 125–132. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Section D-Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Hung, L.-W.; Terwilliger, T.C.; Zwart, P.H. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Cryst. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Systemes, D. BIOVIA, Discovery Studio Modeling Environment. Release 4.5; Dassault Systemes: San Diego, CA, USA, 2015. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S. The protein data bank. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Maksimainen, M.; Paavilainen, S.; Hakulinen, N.; Rouvinen, J. Structural analysis, enzymatic characterization, and catalytic mechanisms of β-galactosidase from Bacillus circulans sp. alkalophilus. FEBS J. 2012, 279, 1788–1798. [Google Scholar] [CrossRef]
- Hidaka, M.; Fushinobu, S.; Ohtsu, N.; Motoshima, H.; Matsuzawa, H.; Shoun, H.; Wakagi, T. Trimeric crystal structure of the glycoside hydrolase family 42 β-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J. Mol. Biol. 2002, 322, 79–91. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Van Der Spoel, D. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Lemkul, J. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1. 0]. Living J. Comput. Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction models for water in relation to protein hydration. In Intermolecular Forces, Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry Held in Jerusalem, Israel; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. [Google Scholar]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Herrou, J.; Crosson, S. Molecular structure of the Brucella abortus metalloprotein RicA, a Rab2-binding virulence effector. Biochemistry 2013, 52, 9020–9028. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2008, 37, D233–D238. [Google Scholar] [CrossRef]
- Holm, L. Benchmarking fold detection by DaliLite v. 5. Bioinformatics 2019, 35, 5326–5327. [Google Scholar] [CrossRef]
- Mangiagalli, M.; Lapi, M.; Maione, S.; Orlando, M.; Brocca, S.; Pesce, A.; Barbiroli, A.; Camilloni, C.; Pucciarelli, S.; Lotti, M.; et al. The co-existence of cold activity and thermal stability in an Antarctic GH42 β-galactosidase relies on its hexameric quaternary arrangement. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.T.; Dutta, C. Molecular signature of hypersaline adaptation: Insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef]
- Li, Y.; Reilly, P.J.; Ford, C. Effect of introducing proline residues on the stability of Aspergillus awamori. Protein Eng. 1997, 10, 1199–1204. [Google Scholar] [CrossRef]
- Agah, S.; Larson, J.D.; Henzl, M.T. Impact of proline residues on parvalbumin stability. Biochemistry 2003, 42, 10886–10895. [Google Scholar] [CrossRef]
- Dong, Y.-W.; Liao, M.-L.; Meng, X.-L.; Somero, G.N. Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc. Natl. Acad. Sci. USA 2018, 115, 1274–1279. [Google Scholar] [CrossRef]
- Tan, T.-C.; Mijts, B.N.; Swaminathan, K.; Patel, B.K.; Divne, C. Crystal structure of the polyextremophilic α-amylase AmyB from Halothermothrix orenii: Details of a productive enzyme–substrate complex and an N domain with a role in binding raw starch. J. Mol. Biol. 2008, 378, 852–870. [Google Scholar] [CrossRef] [PubMed]
- Bezsudnova, E.Y.; Boyko, K.M.; Polyakov, K.M.; Dorovatovskiy, P.V.; Stekhanova, T.N.; Gumerov, V.M.; Ravin, N.V.; Skryabin, K.G.; Kovalchuk, M.V.; Popov, V.O. Structural insight into the molecular basis of polyextremophilicity of short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Thermococcus sibiricus. Biochimie 2012, 94, 2628–2638. [Google Scholar] [CrossRef]
- DasSarma, S.; DasSarma, P. Halophiles and their enzymes: Negativity put to good use. Curr. Opin. Microbiol. 2015, 25, 120–126. [Google Scholar] [CrossRef]
- Garg, R.; Srivastava, R.; Brahma, V.; Verma, L.; Karthikeyan, S.; Sahni, G. Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome. Sci. Rep. 2016, 6, 39634. [Google Scholar] [CrossRef]
- Hutcheon, G.W.; Vasisht, N.; Bolhuis, A. Characterisation of a highly stable α-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 2005, 9, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure 1998, 6, 351–361. [Google Scholar] [CrossRef]
- Yang, J.; Niu, T.; Zhang, A.; Mishra, A.K.; Zhao, Z.J.; Zhou, G.W. Relation between the flexibility of the WPD loop and the activity of the catalytic domain of protein tyrosine phosphatase SHP-1. J. Cell. Biochem. 2002, 84, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H.; Wintrode, P.L.; Miyazaki, K.; Gershenson, A. How enzymes adapt: Lessons from directed evolution. Trends Biochem. Sci. 2001, 26, 100–106. [Google Scholar] [CrossRef]
- Nilmeier, J.; Hua, L.; Coutsias, E.A.; Jacobson, M.P. Assessing protein loop flexibility by hierarchical Monte Carlo sampling. J. Chem. Theory Comput. 2011, 7, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.; Schwaneberg, U.; Roccatano, D. Temperature effects on structure and dynamics of the psychrophilic protease subtilisin S41 and its thermostable mutants in solution. Protein Eng. Des. Sel. 2011, 24, 533–544. [Google Scholar] [CrossRef][Green Version]
- Nestl, B.M.; Hauer, B. Engineering of flexible loops in enzymes. ACS Catal. 2014, 4, 3201–3211. [Google Scholar] [CrossRef]
- Dym, O.; Mevarech, M.; Sussman, J.L. Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium. Science 1995, 267, 1344–1346. [Google Scholar] [CrossRef]
- Mevarech, M.; Frolow, F.; Gloss, L.M. Halophilic enzymes: Proteins with a grain of salt. Biophys. Chem. 2000, 86, 155–164. [Google Scholar] [CrossRef]
- Kumar, S.; Nussinov, R. Salt bridge stability in monomeric proteins. J. Mol. Biol. 1999, 293, 1241–1255. [Google Scholar] [CrossRef]

| Resolution Range | 32.78–2.49 (2.58–2.49) |
|---|---|
| Space group | P 63 |
| Unit cell | 100.13 100.13 137.43 90 90 120 |
| Total reflections, unique reflections | 539,035 (43,095), 27,088 (1673) |
| Multiplicity | 19.9 (16.5) |
| Completeness (%) | 86.53 (61.47) |
| Mean I/sigma (I) | 15.40 (0.64) |
| Wilson B-factor | 75.69 |
| R-merge, R-meas, R-pim | 0.1509 (4.187), 0.1549 (4.319), 0.03468 (1.045) |
| Correlation coefficient, CC1/2, CC | 0.999 (0.556), 1 (0.845) |
| Reflections used in refinement and for R-free | 23,592 (1672), 1230 (103) |
| Reflections used | |
| R-work, R-free | 0.2457 (0.4856), 0.3080 (0.5417) |
| CC (work), CC (free) | 0.895 (0.077), 0.855 (0.192) |
| Number of non-hydrogen atoms | 5280 |
| Macromolecules, ligands, solvent | 5275, 2, 3 |
| Protein residues | 668 |
| Root-mean square (bonds, angles) | 0.005, 0.98 |
| Ramachandran favored, allowed, outliers (%) | 92.75, 7.1, 0.15 |
| Rotamer outliers (%) | 0 |
| Clash score | 23.11 |
| Average B-factor | 100.99 |
| Macromolecules, ligands, solvent | 101.01, 93.91, 66.24 |
| Number of translation/libration/screw (TLS) groups | 1 |
| Sequence analysis | hla_bga | Psychro | Meso | Thermo | Halo | |
| Isoelectric point (pI) | 4.4 | 5.7 | 5.1 | 5.8 | 4.5 | |
| Grand average hydrophobicity | −0.55 | −0.34 | −0.35 | 0.36 | −0.5 | |
| Aliphatic amino acids, Ala, Ile, Leu, Val (%) | 27.5 | 28.6 | 28.4 | 29.4 | 27.8 | |
| Positively charged amino acids, R, K and H (%) | 11.4 | 13.1 | 12.2 | 14.3 | 11.2 | |
| Negatively charged amino acids, D and E (%) | 18.6 | 12.2 | 13.9 | 13.5 | 17.5 | |
| Small amino acids, G and A (%) | 18.9 | 16.7 | 17.0 | 16.2 | 18.6 | |
| Hydrophobic residues, F, I, L, V, M (%) | 21.4 | 25.9 | 25.5 | 27.0 | 22.0 | |
| Polar uncharged residues (%) | 14 | 18.2 | 18.2 | 14.2 | 15.7 | |
| Non-polar residues (%) | 45.6 | 46.1 | 44.3 | 45.4 | 44.8 | |
| Aspartic acids (%) | 11.7 | 5.8 | 7.6 | 5.2 | 10.1 | |
| Glutamic acids (%) | 6.9 | 6.5 | 6.3 | 8.3 | 7.4 | |
| Aromatic residues (%) | 10.4 | 10.9 | 11.3 | 12.7 | 10.8 | |
| Proline amino acids (%) | 7.1 | 5.4 | 4.6 | 6.3 | 6.2 | |
| Arginine amino acids (%) | 7.7 | 5.7 | 5.9 | 7.4 | 7.4 | |
| Structural analysis | Positive residues among total number of surface residues (%) | 14.8 | 17.3 | 20.5 | 23.7 | NA |
| Negative residues among total number of surface residues (%) | 33.6 | 17.2 | 25.6 | 24.3 | NA | |
| Polar residues among total number of surface residues (%) | 12.7 | 20.2 | 18.8 | 13.0 | NA | |
| Non-polar residues among total number of surface residues (%) | 34.9 | 38.2 | 29.8 | 33.2 | NA | |
| Aromatic residues among total number of surface residues (%) | 3.9 | 7.4 | 5.4 | 5.9 | NA | |
| Amino acids that form helices (%) | 30.4 | 36.9 | 32.3 | 33.5 | NA | |
| Amino acids that form strands (%) | 17.7 | 19.9 | 22.3 | 21.0 | NA | |
| Amino acids with bulky hydrophobic side chains (F, I, L) in the protein surface (%) | 5.5 | 11.8 | 5.0 | 9.4 | NA | |
| Hydrogen bonds (all H-bonds and inter-chain H-bonds) | 663, 18 | 771, 23 | 805, 25 | 778, 23 | NA | |
| Salt bridges (all salt bridges and inter-chain salt bridges) | 44, 4 | 40, 3 | 43, 4 | 50, 6 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karan, R.; Mathew, S.; Muhammad, R.; Bautista, D.B.; Vogler, M.; Eppinger, J.; Oliva, R.; Cavallo, L.; Arold, S.T.; Rueping, M. Understanding High-Salt and Cold Adaptation of a Polyextremophilic Enzyme. Microorganisms 2020, 8, 1594. https://doi.org/10.3390/microorganisms8101594
Karan R, Mathew S, Muhammad R, Bautista DB, Vogler M, Eppinger J, Oliva R, Cavallo L, Arold ST, Rueping M. Understanding High-Salt and Cold Adaptation of a Polyextremophilic Enzyme. Microorganisms. 2020; 8(10):1594. https://doi.org/10.3390/microorganisms8101594
Chicago/Turabian StyleKaran, Ram, Sam Mathew, Reyhan Muhammad, Didier B. Bautista, Malvina Vogler, Jorg Eppinger, Romina Oliva, Luigi Cavallo, Stefan T. Arold, and Magnus Rueping. 2020. "Understanding High-Salt and Cold Adaptation of a Polyextremophilic Enzyme" Microorganisms 8, no. 10: 1594. https://doi.org/10.3390/microorganisms8101594
APA StyleKaran, R., Mathew, S., Muhammad, R., Bautista, D. B., Vogler, M., Eppinger, J., Oliva, R., Cavallo, L., Arold, S. T., & Rueping, M. (2020). Understanding High-Salt and Cold Adaptation of a Polyextremophilic Enzyme. Microorganisms, 8(10), 1594. https://doi.org/10.3390/microorganisms8101594







