Abstract
Glucose repression has been extensively studied in Saccharomyces cerevisiae, including the regulatory systems responsible for efficient catabolism of glucose, the preferred carbon source. However, how these regulatory systems would alter central metabolism if new foreign pathways are introduced is unknown, and the regulatory networks between glycolysis and the pentose phosphate pathway, the two major pathways in central carbon metabolism, have not been systematically investigated. Here we disrupted gcr2, a key transcriptional regulator, in S. cerevisiae strain SR7 engineered to heterologously express the xylose-assimilating pathway, activating genes involved in glycolysis, and evaluated the global metabolic changes. gcr2 deletion reduced cellular growth in glucose but significantly increased growth when xylose was the sole carbon source. Global metabolite profiling revealed differential regulation of yeast metabolism in SR7-gcr2Δ, especially carbohydrate and nucleotide metabolism, depending on the carbon source. In glucose, the SR7-gcr2Δ mutant showed overall decreased abundance of metabolites, such as pyruvate and sedoheptulose-7-phosphate, associated with central carbon metabolism including glycolysis and the pentose phosphate pathway. However, SR7-gcr2Δ showed an increase in metabolites abundance (ribulose-5-phosphate, sedoheptulose-7-phosphate, and erythrose-4-phosphate) notably from the pentose phosphate pathway, as well as alteration in global metabolism when compared to SR7. These results provide insights into how the regulatory system GCR2 coordinates the transcription of glycolytic genes and associated metabolic pathways.
1. Introduction
Glucose repression, a phenomenon whereby cells grown on glucose repress the metabolism of alternate carbon sources, has been extensively studied and established in Saccharomyces cerevisiae [1]. The glucose repression pathway is a complex network of signals and regulations where native regulator systems adjust the yeast cellular metabolism and fermentation profiles depending on the available carbon source. Understanding the mechanism and regulation of glucose metabolism in yeast is critical for the development of biotechnological processes to redirect the carbon fluxes toward products of interest [2].
The S. cerevisiae Gcr2 is a transcriptional activator that regulates expression of most glycolytic genes [3]. Gcr2 protein enhances the CT box-dependent transcriptional activation of a RAP1-GCR1 complex required for the expression of glycolytic genes [4]. As a transcriptional activation complex, RAP1-GCR1 provides the specific DNA-binding and the activation of glycolytic and ribosomal genes [5,6]. Rap1 is a multifunctional DNA-binding protein that activates transcription of glycolysis genes and translational components. Rap1 acts interdependently with Gcr1, which mediates transcription of its target genes, along with Rap1 by providing an activation domain [7]. Gcr2, the other component of the activation complex, provides an axillary activation domain to the GCR1-GCR2 complex, mediating high level of glycolytic gene expression [3]. However, the mechanism by which the regulatory systems function in the presence of an alternative carbon source and the working of the system in the case of introduction of new foreign pathways, such as the heterologous xylose-assimilating genes, is currently unknown. Finally, the regulatory networks between glycolysis and the pentose phosphate pathway, the two major pathways in central carbon metabolism, have not been systematically investigated.
A global system-based approach to investigate changes in small-molecule metabolite profiles is an important tool to understand the phenotypes and fermentation outcomes in biological processes. Recently, high-resolution mass spectrometry has been used to elucidate the metabolic interactions, enabling a better understanding of the biological systems and regulations utilized under different physiological conditions [8]. In addition, the development of analytical and associated statistical methods allow high-throughput screening of hundreds of metabolites from complex biological samples, providing integrative information of the functional status of the cell in response to either genetic or environmental changes [9].
Here, we systemically investigated the regulatory mechanism of GCR2 in the S. cerevisiae SR7-gcr2Δ strain engineered for xylose metabolism at the metabolomic level. We provide extensive evidence for how global metabolic pathways are affected in the SR7-gcr2Δ strain when grown on different carbon sources, namely glucose and xylose. We found that the genetic alteration caused differential regulation of yeast metabolism, depending on the available carbon source, and the metabolic changes ultimately resulted in growth improvement via the activated pentose phosphate pathway when cells were grown on xylose as a sole carbon source. This finding suggests that native regulator systems, primarily transcriptional regulations, are highly associated with suboptimal xylose fermentation by xylose-fermenting S. cerevisiae, improving a highly efficient biotechnological process toward the generation of products of interest.
2. Materials and Methods
2.1. Strain Construction
We constructed a xylose-fermenting S. cerevisiae D452-2 strain (SR7) using the linear expression cassette of Scheffersomyces stipitis XYL1, XYL2, and XYL3 genes as described previously (Table 1) [10]. To construct the gcr2 mutant, the gcr2::KanMX4 cassette was amplified from the genomic DNA of the BY4742 gcr2 strain (clone ID: 12013) of the Yeast Knockout Collection (Thermo Fisher Scientific, Waltham, MA, USA) by polymerase chain reaction (PCR) using GCR2-specific primers (upstream: 5′-CAACCCTATGCTACAAGAGCAG-3′ and downstream: 5′-CGACACTAAACCCAGCTAACTC-3′). The PCR product was purified and genome-integrated to the SR7 strain by the LiAc transformation method [11]. The resulting deletion mutant was selected on an agar medium containing 10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose (YPD), 15 g/L agar, and 300 mg/mL G418 sulfate (GoldBio, St. Louis, MO, USA).

Table 1.
Strains used in this study.
2.2. Culture Conditions and Fermentation Experiments
Yeast cells were pre-cultured in 5 mL of YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) for 24 h at 30 °C and 250 rpm. Cells were harvested by centrifugation at 15,000 rpm for 1 min. The cells were adjusted to a concentration of OD600 1.0 (OD600 1.0 = 3.5 × 107 cells/mL) and into inoculated 20 mL of YPD (20 g/L glucose) or YPX (20 g/L xylose) in a 96-well microplate at 30 °C. Cell growth was monitored at 600 nm using a spectrophotometer (OD600). All experiments were repeated in triplicate.
2.3. Intracellular Metabolite Extraction
Metabolite extraction was conducted according to the method described previously [13]. Briefly, a colony of yeast cells was pre-cultured in 5 mL YPD for 24 h at 30 °C and 250 rpm. Cells were harvested by centrifugation at 15,000 rpm for 1 min. The cells were adjusted to a concentration of OD600 1.0 (OD600 1.0 = 3.5 × 107 cells/mL) and inoculated in 50 mL YPD (40 g/L glucose) or YPX (40 g/L xylose) in a 250 mL Erlenmeyer flask at 30 °C and 80 rpm. Five milliliters of cell culture at mid-exponential growth phase (8 h and 10 h of incubation for SR7 and SR7-gcr2Δ grown on glucose, respectively; 72 h and 48 h of incubation for SR7 and SR7-gcr2Δ grown on xylose, respectively) were quenched by quick injection into 25 mL of 60% (v/v) cold methanol (HEPES, 10 mM; pH 7.1) at −40 °C. The cells were centrifuged at 4000 rpm (3134× g) for 10 min at −20 °C and the supernatant was completely removed. Subsequently, 75% (v/v) boiling ethanol (HEPES, 10 mM; pH 7.1) was added to the quenched cell pellet. The mixture was vortexed for 30 s, incubated for 5 min at 80 °C, and immediately cooled in an ice-bath for 5 min. The cell residues were separated from the extract by centrifugation at 4000 rpm (3134× g) for 10 min at 4 °C. The supernatant was vacuum-dried for 6 h using a speed vacuum concentrator.
2.4. Derivatization of Metabolites and Metabolite Analysis Using GC/MS
Prior to GC/MS analysis, the vacuum-dried samples were derivatized by methoxyamination and trimethylsilylation. For methoxyamination, 40 µL of methoxyamine hydrochloride in pyridine (40 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) was added to the samples and incubated for 90 min at 30 °C. For trimethylsilylation, 40 µL of N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich, St. Louis, MO, USA) was added to the samples and incubated for 30 min at 37 °C.
GC/MS analysis was conducted using an Agilent 6890 GC equipped with Leco TOF. A 1 µL aliquot of derivatized samples was injected into the GC in a splitless mode and separated on an RTX-5Sil MS column (30 m × 0.25 mm, 0.25 µm film thickness; Restek, Bellefonte, PA, USA). The initial oven temperature was set at 50 °C for 1 min, and then ramped at 20 °C/min to a final temperature of 330 °C, held for 2 min. Helium was used as a carrier gas at a constant flow rate of 1.5 mL/min. The temperatures of ion source and transfer line were set at 250 °C and 280 °C, respectively. An electron impact of 70 eV was used for ionization. The mass selective detector was operated in scan mode with a mass range of 50–800 m/z.
2.5. Statistical Analysis
The metabolites were evaluated by principal component analysis (PCA) using STATISTICA software (Version 7.0, Stat Soft, Tulsa, OK, USA). The presented data were based on significance (p < 0.05) by t-test using GraphPad Prism 8 (San Diego, CA, USA). Metabolites were mapped into a biochemical network using the MetaMapp software [14]. Fold changes were mapped to node size, and direction was mapped to node color, where blue and red mean lower and higher metabolites abundance, respectively. Label font size indicates p-value by t-test. Hierarchical clustering was conducted using ClustVis (version 2.0, https://biit.cs.ut.ee/clustvis), a web tool for visualization of multivariate data clustering based on PCA and heatmap [15].
3. Results
3.1. Deletion of gcr2 Enhances Growth of the Engineered S. cerevisiae SR7-gcr2Δ on Xylose
GCR2 is a transcriptional regulator that upregulates glycolytic gene expression, especially at the energy payoff phase of glycolysis [5]. We hypothesized that SR7-gcr2Δ would reduce glycolytic activity, resulting in stimulation of the non-oxidative pentose phosphate pathway, when xylose is used as the alternative carbon source. To test this hypothesis, we characterized the phenotypic change induced by SR7-gcr2Δ by comparing it with the wild-type strain of S. cerevisiae SR7 grown using either glucose or xylose as the sole carbon source. The SR7 strain is an engineered S. cerevisiae D452-2 with heterologous expression of the xylose-assimilating pathway genes from Scheffersomyces stipitis XYL1, XYL2, and XYL3, encoding xylose reductase, xylitol dehydrogenase, and xylulokinase, respectively [10,16]. As expected, when the strains were grown on glucose, the SR7-gcr2Δ strain showed slower growth than the SR7 strain (Figure 1A, specific growth rate 1.78 ± 0.35 h−1 and 0.87 ± 0.03 h−1 for SR7 and SR7-gcr2Δ, respectively). However, when the strains were grown on xylose, the SR7-gcr2Δ mutant grew significantly faster than SR7 strain (Figure 1B, specific growth rate 0.012 ± 0.004 h−1 and 0.052 ± 0.001 h−1 for SR7 and SR7 SR7-gcr2Δ, respectively). These data suggest that gcr2Δ triggers transcriptional alteration of glycolysis and the associated pentose phosphate pathway, which directly improves xylose metabolism in the engineered S. cerevisiae strain.
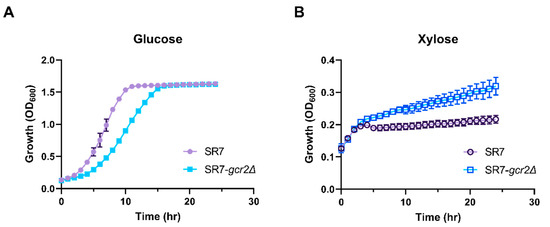
Figure 1.
Growth profiles of S. cerevisiae SR7 and SR7-gcr2Δ on glucose (A) and xylose (B) media. Cell density was measured by optical density at 600 nm. All of the experiments were performed in triplicate. The error bars represent the standard deviations. ●: S. cerevisiae SR7; and ■: S. cerevisiae SR7-gcr2Δ.
3.2. Global Metabolic Changes Are Induced in SR7-gcr2Δ
To understand the global metabolic changes in SR7-gcr2Δ, untargeted metabolite profiling was performed using GC-TOF MS platform with cultures of SR7 and the SR7-gcr2Δ mutant grown on glucose and xylose, respectively. As shown in Supplementary Table S1, a total of 110 intracellular metabolites were identified including amino acids, polyamines, organic acids, sugars, and free fatty acids. The hierarchical clustering and PCA analysis showed that the carbon source in the culture media was the primary determinant of the metabolic changes, and the metabolism of SR7-gcr2Δ mutant grown on xylose was different from SR7 (Supplementary Figure S1). The PCA score plot exhibited a good fit of R2X, 0.58, and a prediction of Q2, 0.69, based on the cumulative values up to principal component 2, showing a separation among the metabolite profiles of the four groups (Figure 2A). Especially, the PCA plot shows that the range of metabolic changes in the SR7-gcr2Δ mutant was greater than SR7 when grown on xylose. Separation of the plots in the PCA indicates significant alteration of global metabolism in the strain. The PCA result suggests that Gcr2 would affect the xylose metabolism and in turn the associated downstream metabolic pathways more than other metabolic conditions. The volcano plots comparing the detected metabolites between the two strains grown on each carbon source clearly show significantly different metabolic markers based on the statistical significance of the magnitude of the changes (Figure 2B,C). The metabolomics analysis corroborated the results from the growth analysis, showing that the deletion of gcr2 in S. cerevisiae SR7 background induces global changes in yeast metabolism.
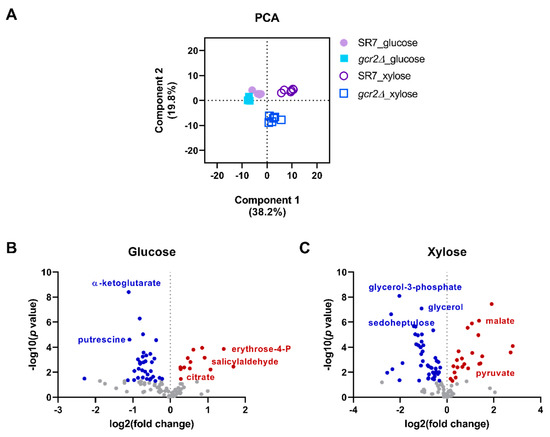
Figure 2.
(A) Principal component analysis of S. cerevisiae metabolites. Intensities of metabolites in SR7 and SR7-gcr2Δ grown on glucose and xylose media were detected by GC-TOF MS. (B,C) Volcano plot analysis of identified metabolites on glucose (B) and xylose (C). The scatter plot shows features with fold change (x) and t-test threshold (y) of 0.05, and both values were log-transformed by 2 and 10, respectively. The red or blue dots indicate significantly increased or decreased metabolites in SR7-gcr2Δ compared with SR7 (p < 0.05), respectively.
3.3. Deletion of gcr2 Differentially Alters Yeast Metabolism Depending on Carbon Sources
In addition to the multi- and univariate analyses, we generated maps to visualize the integrative metabolic network for a better understanding of the SR7-gcr2Δ effect with respect to the carbon sources. Using MetaMapp, which helps visualize metabolomics data by integrating information from biochemical pathways and chemical and mass spectral similarity, the metabolites relation network was visualized by CytoScape (Figure 3) [14]. In the depicted network, large sizes of font and circles indicate statistical significance and fold change in the strain SR7-gcr2Δ, respectively. On the map, a clear interrelationship between SR7-gcr2Δ and metabolic pathways was evident. Specifically, for both glucose and xylose media, the SR7-gcr2Δ mutant showed significantly decreased abundances of fatty acids and organic acids, including 1-monopalmitin, salicylaldehyde, and hydroxypyrimidines. However, compared to the glucose metabolism, the SR7-gcr2Δ grown in the presence of xylose showed significant changes in metabolite levels. Among the changed metabolites, the abundance of sugars and their derivatives were notably affected by the mutation. Nucleotides levels, such as guanine, hypoxanthine, inosine, and uracil, were also significantly decreased in SR7-gcr2Δ grown in xylose, while growth in glucose did not show a significant difference between the two genotypes. Taken together, the depicted network clearly showed that the relationship between SR7-gcr2Δ and associated metabolites changes, depending on the available carbon source.
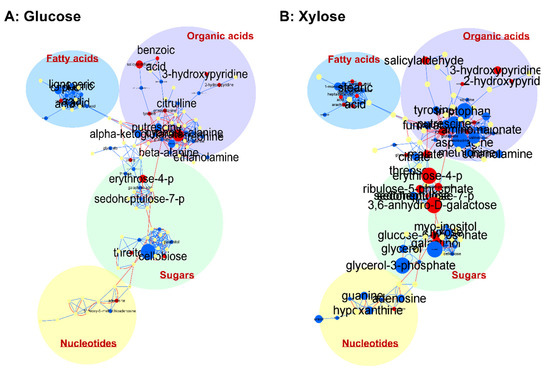
Figure 3.
Network analysis of metabolites on glucose (A) and xylose (B). Each node is a name of structurally identified metabolite. Red edges are KEGG reactant pair annotation, and blue edges are chemical similarity annotations. The graph was visualized using organic layout in Cytoscape. Red nodes are increased metabolites, and blue nodes are decreased metabolites. The size of nodes and labels reflect fold-changes (bigger nodes mean higher fold-changes) and p values assigned by t-test (bigger font sizes mean more statistically significant changes), respectively. The metabolites were clustered into fatty acids, organic acids including amino acids, sugars, and nucleotides.
3.4. Deletion of gcr2 Alters Pathways in Central Carbon Metabolism
Genetic studies have shown that the expression of most glycolytic enzymes is severely reduced in the SR7-gcr2Δ mutant and may regulate gene expression in the downstream pathways, such as the TCA cycle and respiration [17]. However, the genetic effect is unknown at the metabolic phenotype level, especially in the presence of an alternative carbon source. With respect to glycolysis, we found that the intracellular abundance of metabolites in the central carbon metabolism was not significantly different between the two strains grown on glucose medium, except for pyruvate (Figure 4). However, levels of glucose-6-phosphate, fructose-6-phosphate, and pyruvate were significantly higher in the presence of xylose. Byproducts of glycolysis, including glycerol and glycerol-3-phosphate, were reduced in the mutant strain grown in xylose. For the TCA cycle, we could detect intermediates in the pathway, such as citrate, alpha-ketoglutarate, succinate, fumarate, and malate (Figure 4B). The abundance of most of these metabolites decreased in SR7-gcr2Δ, but citrate and malate intermediates showed particularly higher abundance in glucose and xylose, respectively. Overall, the xylose culture provided a greater impact of SR7-gcr2Δ on glycolytic genes than the glucose culture, and central carbon metabolism was differentially affected by the deletion of gcr2, depending on the type of sugar available.
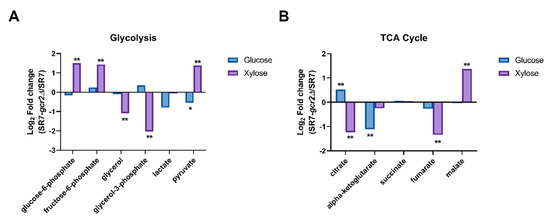
Figure 4.
Intracellular metabolite fold-changes in glycolysis (A) and TCA cycle (B) of SR7 and SR7-gcr2Δ grown on glucose (■) or xylose (■). Intracellular metabolites were detected using GC-TOF MS, and fold-changes were converted to a log 2 scale. Statistical analysis was performed using t-test, and differences were considered significant when p was < 0.01 (**) or 0.05 (*).
3.5. Deletion of gcr2 Upregulates Pentose Phosphate Pathway on Xylose
The pentose phosphate pathway is a universal metabolic pathway producing xylulose-5-phosphate as a key metabolic core intermediate, linking xylose assimilation, an oxidative phase, and a non-oxidative phase, in the pathway [18]. We could detect xylulose-5-phosphate, ribulose-5-phosphate, sedoheptulose-7-phosphate, erythrose-4-phosphate, and sedoheptulose as intermediates in the pentose phosphate pathway. All these metabolites showed higher abundance when cells were grown on xylose than on glucose (Figure 5). For xylose assimilation, there was no significant difference in xylulose-5-phosphate levels between the SR7 and SR7-gcr2Δ strains, implicating that the heterologous steps were unaffected in SR7-gcr2Δ.
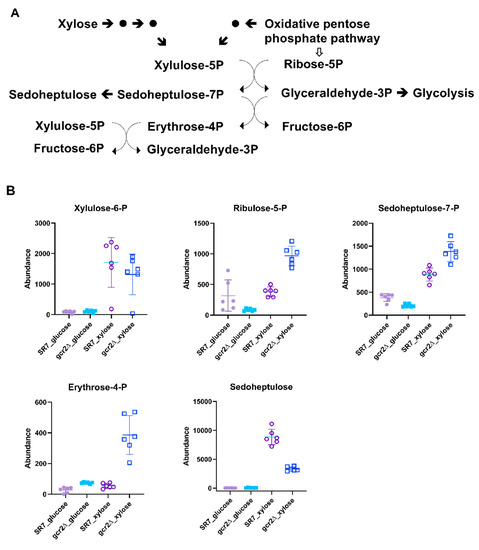
Figure 5.
(A) The pentose phosphate pathway. (B) Abundances of the intermediate metabolites in the pentose phosphate pathway of SR7 (●) and SR7-gcr2Δ (■) grown on glucose (closed symbols) or xylose (open symbols). The error bars indicate standard deviation, and all experiments were performed in six replicates.
Unlike xylulose-5-phosphate, the metabolic abundances of ribulose-5-phosphate, sedoheptulose-7-phosphate, and sedoheptulose in the oxidative and the non-oxidative phases were differentially regulated in SR7-gcr2Δ, depending on the carbon source. Higher levels of these metabolites indicated increased metabolic flow, via the pentose phosphate pathway of the SR7-gcr2Δ mutant grown in xylose. Notably, the abundance of erythrose-4-phosphate increased in SR7-gcr2Δ grown in both carbon sources, although the absolute abundance was much higher with xylose than with glucose. Overall, this study showed that the deletion of gcr2 induced the metabolic flow toward the pentose phosphate pathway and improved cellular growth when only xylose was available. In many xylose-fermenting yeast strains, the enhanced flux through the pentose phosphate pathway induces greater xylose utilization and ethanol production [10,16,18]. We speculate that the higher abundance of the intermediate results from downregulation of glycolysis, accumulating erythrose-4-phosphate in SR7-gcr2Δ. These results suggest that the glycolytic transcription activator GCR2 has a negative impact on xylose fermentation, more specifically, on the pentose phosphate pathway, thereby, ultimately providing improved xylose utilization and ethanol production in the SR7-gcr2Δ mutant.
3.6. Deletion of gcr2 Increases Cellular Defense against Oxidative Stress
Inhibition of glycolysis is known to promote flux into the pentose phosphate pathway to generate NADPH. The pentose phosphate pathway is strongly associated with cellular defense against oxidative stress by producing NADPH, which provides the reducing power fueling the antioxidant systems, and recycles oxidized glutathione [19]. We hypothesized that downregulation of glycolysis in SR7-gcr2Δ would alter the cellular redox homeostasis during oxidative stress. To test our hypothesis, we pre-cultured the two strains in YPD broth for 24 h and adjusted the optical density of the culture to one. Then, we spotted culture dilutes onto 20 g/L YPD or YPX agar plates, respectively, with supplementation of hydrogen peroxide. As shown in Figure 6, when cells were treated with hydrogen peroxide, the SR7-gcr2Δ mutant showed superior cell viability than SR7 on glucose medium. The mutant yeast strain also exhibited improved viability on xylose, however, this effect also could be due to better xylose utilization, not solely from the antioxidant activity.
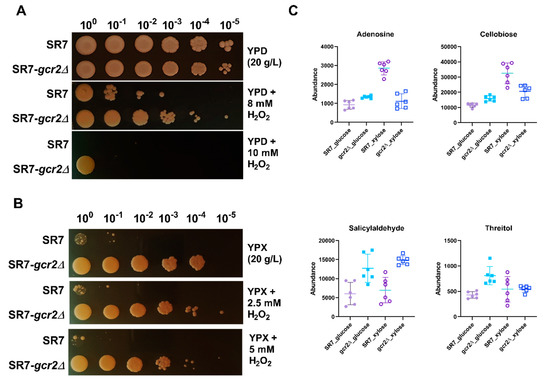
Figure 6.
Spot plate assay for SR7 and SR7-gcr2Δ grown on glucose (A) and xylose (B) under control and in the presence of hydrogen peroxide (H2O2). (C) Abundances of the metabolites associated with the cellular antioxidant activity of SR7 (●) and SR7-gcr2Δ (■) grown on glucose (closed symbols) or xylose (open symbols). The error bars indicate standard deviation, and all experiments were performed in six replicates.
In addition to the pentose phosphate pathway, we compared the abundance of other metabolites known for their antioxidant activity in yeast, such as adenosine, cellobiose, salicylaldehyde, and threitol [20,21,22,23]. All these metabolite levels were significantly higher in the mutant strain, especially in glucose medium (Figure 6C). In xylose, only the abundance of salicylaldehyde increased in SR7-gcr2Δ, while other metabolite levels were unchanged, or even decreased, suggesting the possibility of a different antioxidant mechanism based on the carbon sources. Central carbon metabolism involves a network of interrelated pathways that provide various redox cofactors and generate energy sources. The complicated metabolic network affects different cellular responses as means of the cofactors and energies. Our results showed higher antioxidant activity of SR7-gcr2Δ when compared to its wild type, which may be attributed to either the increased pentose phosphate pathway, inhibited glycolysis, or unidentified defense mechanisms against oxidative stress. Taken together, the deletion of gcr2 induces cells to adjust and reprogram their cellular responses, caused by environmental disruption beyond glycolysis.
4. Discussion
Gcr2 is a glycolytic gene transcriptional activator that regulates the expression of most glycolytic genes, in a complex with RAP1 and GCR1. Here, we report that deletion of gcr2 in xylose-fermenting S. cerevisiae improved cellular growth via upregulation of the pentose phosphate pathway. Metabolomic analysis showed global metabolic changes were induced by the deletion of gcr2, as well as altered central carbon metabolism. The SR7-gcr2Δ strain showed a clearly activated pentose phosphate pathway when grown in the presence of xylose and affected yeast cellular antioxidant activity, especially in glucose, suggesting the global effects of the transcriptional activator beyond glycolysis.
Transcription factors regulate global cellular responses and are, therefore, frequently targeted in the field of synthetic biology and metabolic engineering. To date, a few studies have succeeded to improve ethanol fermentation in yeast by deleting or over-expressing transcription factor genes, such as HAA1, WHI2, YAP1, NRM1, and CAT8 [24,25,26,27,28]. These transcription factors are involved in expression regulation of membrane proteins (Haa1), coordination of growth and stress responses (Whi2), redox homeostasis (Yap1), cell-cycle-dependent transcription (Nrm1), and gluconeogenesis (Cat8), all resulting in higher growth in xylose and enhanced ethanol yield and productivity. In the same context, when xylose was provided as a carbon source, the deletion of gcr2 effectively shifted the metabolic flux toward the pentose phosphate pathway, resulting in improved xylose utilization and cellular growth. Since GCR2 regulates glycolysis, the central cellular carbon metabolism was greatly affected. Yeast metabolic pathway will be mainly directed to the pentose phosphate pathway from xylose in the xylose culture. The intermediates in the pathway are fructose-6-phosphate and glyceraldehyde-3-phosphate shared by glycolysis, which are key components especially for the second phase of glycolysis. Therefore, we speculate that deletion of gcr2 would greatly impact the pentose phosphate pathway in SR7-gcr2Δ, resulting in subsequent changes in glycolysis and overall central carbon metabolism. Notably, we speculate that the changes in the metabolic profiles shown in the current study may have resulted from using xylose as an alternative carbon source and not from the absence of glucose in the medium. Sasaki et al. reported that the deletion of gcr2 in S. cerevisiae did not affect the cellular growth nor the expression of genes in TCA cycle and oxidative phosphorylation when cells were grown on glycerol plus lactate as alternative carbon sources of glucose [17]. Therefore, the metabolic outcomes present here would be the effect of xylose as an alternative carbon source. Additional transcript expression profiles in these pathways need to be investigated in SR7-gcr2Δ to further understand the role of GCR2 in SR7.
Although there have been a number of successful strategies to implement xylose metabolism in S. cerevisiae, their efficiency so far has not been comparable to that of glucose [10,18]. Xylose paradox is a paradoxical phenomenon in the engineered yeast that do not sense xylose as a fermentable carbon source despite being able to grow on and ferment xylose [29]. Many studies showed that high xylose concentrations would trigger cellular signals similar to those generated due to low glucose concentrations, resulting in a starvation response rather than a fermentation response [29,30]. The results of the current study support the notion of xylose paradox and associated cellular signaling pathways, involving GCR2 as a metabolic regulator. In the presence of GCR2, high concentration of xylose may trigger non-fermentative response that inhibits xylose uptake and utilization, probably due to inhibition of the pentose phosphate pathway by GCR2. However, deletion of gcr2 would result in a synergy of the stimulating effect of the pentose metabolism together with bypassing the glucose and hexokinase signaling cascades, ultimately resulting in avoiding the non-fermentative response. To further test this hypothesis, genetic modification and application of biosensor strains for GCR2 will be required.
To our knowledge, this is the first metabolic study on the roles of GCR2 in the pentose phosphate pathway in the presence of an alternative carbon source. We showed that SR7-gcr2Δ increased the metabolic flow toward the pentose phosphate pathway and cellular resistance to oxidative stress. Similarly, Lamas-Maceiras et al. reported that the deletion of the Kluyveromyces lactis GCR1 increased the pentose phosphate pathway by the activation of glucose-6-phosphate dehydrogenase (encoded by ZWF1), the first enzyme in the oxidative phase of the pentose phosphate pathway [31]. They found that there would be a direct regulation of the KlZWF1 gene expression by KlGcr1 where the KlZWF1 promoter contains a consensus sequence for Gcr1 binding. In addition, the ΔKlgcr1 strain had increased resistance to oxidative stress and glutathione species GSSG/GSH ratio, supported by other reports highlighting the importance of the pentose phosphate pathway for cellular antioxidant activity based on heterologous expressions of functional ZWF1 from various hosts [32]. These findings support the roles of GCR2 identified in this study, assuming that the two glycolytic transcriptional activators, Gcr1 and Gcr2, co-activate the transcription of genes involved in central carbon metabolism.
The field of mass spectrometry-based metabolome analysis has developed remarkably over the last two decades [33]. Datasets generated using this method have contributed to a more comprehensive understanding of cell processes and to an application of metabolic engineering-driven optimization of cell factories [34]. Our results also provide extensive knowledge on global metabolic changes induced by genetic modification of a glycolytic transcriptional activator. Based on the GC-MS instrumental platform, we could detect a broad range in the physico-chemically diverse metabolite pool, including sugars, organic acids, amino acids, fatty acids, and polyamines. The advantages of GC are high separation efficiency and reproducible retention times via the retention index using retention time markers [35]. However, the GC-based methodology has limitations on unsuitability for non-volatile and thermally unstable metabolites and on differentiation of sugar isomers [36]. Therefore, future studies, involving a combination of LC-MS and GC-MS-based methodologies, are required to identify metabolites more precisely.
Taken together, our study investigated the metabolic responses to xylose fermentation in S. cerevisiae. Based on metabolomic changes induced in the SR7-gcr2Δ mutant, we conclude that upregulation of the pentose phosphate pathway is the primary metabolic change observed in the SR7-gcr2Δ mutant when an alternative carbon source is present. These changes ultimately resulted in significantly improved xylose metabolism in the yeast strain expressing a heterologous xylose assimilation pathway. This study provides insights into how the native regulatory system GCR2 coordinates the expression of genes in central carbon metabolism and associated metabolic pathways.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/10/1499/s1, Supplementary Table S1: Metabolites list, Supplementary Figure S1: Hierarchical clustering of S. cerevisiae metabolites.
Author Contributions
S.R.K. designed the experiments. M.S. and S.R.K. carried out all experiments and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019R1F1A1062633).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lane, S.; Xu, H.; Oh, E.J.; Kim, H.; Lesmana, A.; Jeong, D.; Zhang, G.; Tsai, C.-S.; Jin, Y.-S.; Kim, S.R. Glucose repression can be alleviated by reducing glucose phosphorylation rate in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 2613. [Google Scholar] [CrossRef] [PubMed]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef] [PubMed]
- Uemura, H.; Jigami, Y. Role of GCR2 in transcriptional activation of yeast glycolytic genes. Mol. Cell. Biol. 1992, 12, 3834–3842. [Google Scholar] [CrossRef] [PubMed]
- Huie, M.A.; Scott, E.W.; Drazinic, C.M.; Lopez, M.C.; Hornstra, I.K.; Yang, T.P.; Baker, H.V. Characterization of the DNA-binding activity of GCR1: In vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 2690–2700. [Google Scholar] [CrossRef] [PubMed]
- Uemura, H.; Fraenkel, D.G. gcr2, a new mutation affecting glycolytic gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 6389–6396. [Google Scholar] [CrossRef]
- Sasaki, H.; Kishimoto, T.; Mizuno, T.; Shinzato, T.; Uemura, H. Expression of GCR1, the transcriptional activator of glycolytic enzyme genes in the yeast Saccharomyces cerevisiae, is positively autoregulated by Gcr1p. Yeast 2005, 22, 305–319. [Google Scholar] [CrossRef]
- Zeng, X.; Deminoff, S.J.; Santangelo, G.M. Specialized Rap1p/Gcr1p transcriptional activation through Gcr1p DNA contacts requires Gcr2p, as does hyperphosphorylation of Gcr1p. Genetics 1997, 147, 493–505. [Google Scholar]
- Roullier-Gall, C.; David, V.; Hemmler, D.; Schmitt-Kopplin, P.; Alexandre, H. Exploring yeast interactions through metabolic profiling. Sci. Rep. 2020, 10, 6073. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Mas, S.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef]
- Kim, S.R.; Skerker, J.M.; Kang, W.; Lesmana, A.; Wei, N.; Arkin, A.P.; Jin, Y.S. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e57048. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Ha, S.J.; Kong, I.I.; Jin, Y.S. High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Kim, S.; Yang, J.; Seo, J.H.; Kim, K.H. Intracellular metabolite profiling of Saccharomyces cerevisiae evolved under furfural. Microb. Biotechnol. 2017, 10, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Haldiya, P.K.; Wohlgemuth, G.; Kind, T.; Kothari, S.L.; Pinkerton, K.E.; Fiehn, O. MetaMapp: Mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinform. 2012, 13, 99. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Kim, S.R.; Xu, H.; Lesmana, A.; Kuzmanovic, U.; Au, M.; Florencia, C.; Oh, E.J.; Zhang, G.; Kim, K.H.; Jin, Y.S. Deletion of PHO13, encoding haloacid dehalogenase type IIA phosphatase, results in upregulation of the pentose phosphate pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015, 81, 1601–1609. [Google Scholar] [CrossRef]
- Sasaki, H.; Uemura, H. Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast 2005, 22, 111–127. [Google Scholar] [CrossRef]
- Kwak, S.; Jin, Y.S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Fact. 2017, 16, 82. [Google Scholar] [CrossRef]
- Mullarky, E.; Cantley, L.C. Diverting glycolysis to combat oxidative stress. In Innovative Medicine: Basic Research and Development; Nakao, K., Minato, N., Uemoto, S., Eds.; Springer: Tokyo, Japan, 2015; ISBN 978-4-431-55650-3. [Google Scholar]
- Maggirwar, S.B.; Dhanraj, D.N.; Somani, S.M.; Ramkumar, V. Adenosine acts as an endogenous activator of the cellular antioxidant defense system. Biochem. Biophys. Res. Commun. 1994, 201, 508–515. [Google Scholar] [CrossRef]
- Tang, W.; Harris, L.C.; Outhavong, V.; Newton, R.J. Antioxidants enhance in vitro plant regeneration by inhibiting the accumulation of peroxidase in Virginia pine (Pinus virginiana Mill.). Plant Cell Rep. 2004, 22, 871–877. [Google Scholar] [CrossRef]
- Kim, J.H.; Chan, K.L.; Faria, N.C.G.; de Martins, M.L.; Campbell, B.C. Targeting the oxidative stress response system of fungi with redox-potent chemosensitizing agents. Front. Microbiol. 2012, 3, 88. [Google Scholar] [CrossRef] [PubMed]
- Sulej, J.; Osińska-Jaroszuk, M.; Jaszek, M.; Grąz, M.; Kutkowska, J.; Pawlik, A.; Chudzik, A.; Bancerz, R. Antimicrobial and antioxidative potential of free and immobilised cellobiose dehydrogenase isolated from wood degrading fungi. Fungal Biol. 2019, 123, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Stabryla, L.; Wei, N. Improved acetic acid resistance in Saccharomyces cerevisiae by overexpression of the WHI2 gene identified through inverse metabolic engineering. Appl. Environ. Microbiol. 2016, 82, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Ruchala, J.; Kurylenko, O.O.; Soontorngun, N.; Dmytruk, K.V.; Sibirny, A.A. Transcriptional activator Cat8 is involved in regulation of xylose alcoholic fermentation in the thermotolerant yeast Ogataea (Hansenula) polymorpha. Microb. Cell. Fact. 2017, 16, 36. [Google Scholar] [CrossRef]
- Wu, G.; Xu, Z.; Jönsson, L.J. Profiling of Saccharomyces cerevisiae transcription factors for engineering the resistance of yeast to lignocellulose-derived inhibitors in biomass conversion. Microb. Cell Fact. 2017, 16, 199. [Google Scholar] [CrossRef]
- Wei, S.; Liu, Y.; Wu, M.; Ma, T.; Bai, X.; Hou, J.; Shen, Y.; Bao, X. Disruption of the transcription factors Thi2p and Nrm1p alleviates the post-glucose effect on xylose utilization in Saccharomyces cerevisiae. Biotechnol. Biofuels 2018, 11, 112. [Google Scholar] [CrossRef]
- Cunha, J.T.; Costa, C.E.; Ferraz, L.; Romaní, A.; Johansson, B.; Sá-Correia, I.; Domingues, L. HAA1 and PRS3 overexpression boosts yeast tolerance towards acetic acid improving xylose or glucose consumption: Unravelling the underlying mechanisms. Appl. Microbiol. Biotechnol. 2018, 102, 4589–4600. [Google Scholar] [CrossRef]
- Osiro, K.O.; Borgström, C.; Brink, D.P.; Fjölnisdóttir, B.L.; Gorwa-Grauslund, M.F. Exploring the xylose paradox in Saccharomyces cerevisiae through in vivo sugar signalomics of targeted deletants. Microb. Cell Fact. 2019, 18, 88. [Google Scholar] [CrossRef]
- Osiro, K.O.; Brink, D.P.; Borgström, C.; Wasserstrom, L.; Carlquist, M.; Gorwa-Grauslund, M.F. Assessing the effect of d-xylose on the sugar signaling pathways of Saccharomyces cerevisiae in strains engineered for xylose transport and assimilation. FEMS Yeast Res. 2018, 18, fox096. [Google Scholar] [CrossRef]
- Lamas-Maceiras, M.; Rodríguez-Belmonte, E.; Becerra, M.; González-Siso, M.I.; Cerdán, M.E. KlGcr1 controls glucose-6-phosphate dehydrogenase activity and responses to H2O2, cadmium and arsenate in Kluyveromyces lactis. Fungal Genet. Biol. 2015, 82, 95–103. [Google Scholar] [CrossRef]
- Heinisch, J.J.; Knuesting, J.; Scheibe, R. Investigation of heterologously expressed glucose-6-phosphate dehydrogenase genes in a yeast zwf1 deletion. Microorganisms 2020, 8, 546. [Google Scholar] [CrossRef]
- Røst, L.M.; Brekke Thorfinnsdottir, L.; Kumar, K.; Fuchino, K.; Eide Langørgen, I.; Bartosova, Z.; Kristiansen, K.A.; Bruheim, P. Absolute quantification of the central carbon metabolome in eight commonly applied prokaryotic and eukaryotic model systems. Metabolites 2020, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Kim, S.; Sorek, H.; Lee, Y.; Jeong, D.; Kim, J.; Oh, E.J.; Yun, E.J.; Wemmer, D.E.; Kim, K.H.; et al. PHO13 deletion-induced transcriptional activation prevents sedoheptulose accumulation during xylose metabolism in engineered Saccharomyces cerevisiae. Metab. Eng. 2016, 34, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Brennan, L.; Fiehn, O.; Hankemeier, T.; Kristal, B.S.; van Ommen, B.; Pujos-Guillot, E.; Verheij, E.; Wishart, D.; Wopereis, S. Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 2009, 5, 435–458. [Google Scholar] [CrossRef]
- Ren, J.-L.; Zhang, A.-H.; Kong, L.; Wang, X.J. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018, 8, 22335–22350. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).