Abstract
Two extremely halophilic archaeal strains, designated SB29T and SB3T, were isolated from the brine-seawater interface of Discovery Deep in the Red Sea. Cells of both strains were pleomorphic (irregular polyhedrals, ovals, and rods) and stained Gram-negative; colonies were pigmented pink. The sequence similarity of the 16S rRNA gene of strain SB29T with that of its most closely related validly described species (Hfx. sulfurifontis DSM 16227T) and that of strain SB3T with its closest validly described relative (Hfx. denitrificans ATCC 35960T) was 98.1% and 98.6%, respectively. The incomplete draft genomes of SB29T and SB3T are 3,871,125 bp and 3,904,985 bp in size, respectively, and their DNA G + C contents are 60.75% and 65.64%, respectively. The highest ANI values between the genomes of SB29T and SB3T and the most closely related genomes in GenBank were determined as 82.6% (Hfx. sulfurifontis ATCC BAA-897T, GenBank accession no. GCA_000337835.1) and 92.6% (Haloferax denitrificans ATCC 35960T, GenBank accession no. GCA_000337795.1), respectively. These data indicate that the two new isolates cannot be classified into any recognized species of the genus Haloferax, and, therefore, two novel species of the genus Haloferax are proposed: Haloferax profundi sp. nov. (type strain SB29T = JCM 19567T = CGMCC 1.14960T) and Haloferax marisrubri sp. nov. (type strain SB3T = JCM 19566T = CGMCC 1.14958T).
1. Introduction
Many studies indicated that deep-sea brine pools harbor distinctive microbial communities [1,2,3]. The isolation and characterization of novel extremophiles from brine environments will significantly advance our understanding of the in situ metabolisms, energy-producing processes, and environmentally adaptive activities in these harsh environments. The new strains in this study were isolated from the brine-seawater interface of Discovery Deep in the Red Sea, at a depth of 2000–2060 m, and formed two separate branches within the genus Haloferax.
Currently, there are 13 recognized species of Haloferax in the family Haloferacaceae of the order Haloferacales, according to the reclassification of the class Halobacteria into three orders and three families by Gupta [4]: Hfx. volcanii [5], Hfx. mediterranei [6], Hfx. denitrificans [7], Hfx. gibbonsii [8], Hfx. alexandrinus [9], Hfx. lucentense [10], Hfx. sulfurifontis [11], Hfx. prahovense [12], Hfx. larsenii [13], Hfx. elongans, Hfx. mucosum [14], Hfx. chudinovii [15], and Hfx. namakaokahaiae [16]. These 13 species were isolated from solar salterns, salt lakes, Permian potassium salt deposits, microbial mats from Hamelin Pool (Shark Bay, Australia), and sulfide- and sulfur-rich springs. No species of Haloferax had been previously isolated from deep-sea brine pool environments. Here, we describe the physiological and genomic properties of two novel strains within the genus Haloferax isolated from a brine-seawater interface, and propose to delineate them as two novel species.
2. Materials and Methods
2.1. Isolation and Growth of Strains SB29T and SB3T
Strains SB29T and SB3T were isolated using procedures described by Zhang et al. [17], and the two strains were grown aerobically in a modified R2A (MR2A) medium [18] for all physiological-chemical tests. The media contained the following ingredients (L−1): 0.5 g casamino acids (Difco), 0.5 g yeast extract (Difco), 0.5 g sodium pyruvate, 0.5 g peptone, 0.5 g glucose, 3.0 g trisodium citrate, 2.0 g KCl, 0.3 g K2HPO4, 0.5 g CaCl2, 20.0 g MgSO4∙7H2O, 200.0 g NaCl, and the pH was adjusted to 7.0–7.2.
2.2. Selection, Morphology and Phylogeny of Strains
In our cultivation experiment, we isolated 15 strains that formed two groups based on 16S rRNA gene sequences and colony and cell morphologies. Eight strains from the Discovery interface 1 (Di1), designated strains SB1 to SB8, formed small (1 mm), pink, translucent, convex colonies on an agar medium. This group of strains all consisted of pleomorphic rods (Supplementary Figures S1 and S2), and an initial analysis of their partial 16S rRNA gene sequences indicated they were most closely related to Hfx. denitrificans ATCC 35960T (98.6% similarity). Strain SB3T was selected as a representative strain for further taxonomic characterization. Seven strains isolated from the Discovery interface 2 (Di2), designated SB22–25 and SB28–30, formed pinkish-red, opaque, domed, mucoid colonies, 3–5 mm in diameter, containing pleomorphic cells. Analyses of their partial 16S rRNA gene sequences indicated they were most closely related to Hfx. sulfurifontis (98.1% similarity). Strain SB29T was selected as a representative strain for further taxonomic characterization.
2.3. Phenotypic Tests
Phenotypic tests of SB29T and SB3T were performed according to the proposed minimum standards for the description of novel taxa in the order Halobacteriales [19]. Of the 13 published type strains, 12 were selected as reference strains for physiological tests (Table 1). Hfx. namakaokahaiae was described after our experiments were already completed. As Hfx. namakaokahaiae did not represent one of the closest relatives of either novel strain (Figure 1), we amended Table 1 with data from McDuff et al. [16]. Cell morphology and motility in exponentially growing cultures in the liquid MR2A medium were examined using a Leica microscope equipped with phase-contrast optics (Leica DM 6000 B). The minimal salt concentration required to prevent cell lysis was tested by suspending washed cells in serial sterile saline solutions containing NaCl ranging from 0 to 15.0% (w/v), and the stability of the cells was observed by light microscopy. Sample preparation for transmission electron microscopy (TEM) was based on previously published methods [20,21] and the cells were analyzed on a Technai 12 transmission electron microscope (FEI Company, Hillsboro, OR, USA). Gram-staining was performed using the method outlined by Dussault [22]. Most biochemical and nutritional tests were performed as described by Allen et al. [14], Xu et al. [13], and Goh et al. [23]. In brief, anaerobic archaeal growth and gas formation with nitrate as an electron acceptor were measured in 10 mL stoppered tubes filled with 4 mL media containing NaNO3 (5.0 g L−1), L-arginine (5.0 g L−1), and DMSO (5.0 g L−1). An inverted Durham tube was added to capture the gas production; nitrite formation was monitored colorimetrically. The optimal growth temperature was determined after incubation on MR2A agar and shaking in MR2A liquid medium at 10, 15, 20, 25, 28, 30, 33, 37, 40, 45, and 50 °C. Growth was measured as an increase in turbidity at 660 nm, determined in a spectrophotometer. Tolerance to NaCl concentration was tested on both MR2A agar and liquid media with 0.5, 1.0, 1.2, 1.5, 2.0, 3.0, 4.0, 4.3, 4.5, 5.0, 5.5, 5.8, and 6.0 M NaCl added. Similarly, tolerance to MgCl2 was tested on both MR2A agar liquid media containing 0, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.7, 0.75, and 1.0 M MgCl2. pH tolerance of the strains SB29T and SB3T was tested in an MR2A liquid medium, adjusted to pH 5.5 to 10.0 with 20 mM of the following buffer systems: MES (pH 5.5 and 6.0), PIPES (pH 6.5 and 7.0), HEPES (pH 7.5 and 8.0), Tricine (pH 8.5), and CAPSO (pH 9.0, 9.5 and 10.0). Oxidase activity was tested using the oxidase reagent kit (bioMérieux) according to the manufacturer’s instructions. Catalase activity was determined by pouring 3.0% H2O2 solution onto bacterial colonies and observing bubble production. Esterase activity was detected as outlined by Gutíérrez and González [24].

Table 1.
Phenotypic and other characteristics that distinguish the Discovery Deep brine-seawater interface isolates SB29T and SB3T from other Haloferax species. Taxa: 1, SB29T; 2, SB3T; 3, Hfx. elongans JCM 14791T [14]; 4, Hfx. mucosum JCM 14792T [14]; 5, Hfx. mediterranei JCM 8866T [6]; 6, Hfx. volcanii JCM 8879T [5]; 7, Hfx. lucentense DSM 14919T [10]; 8, Hfx. denitrificans JCM 8864T [7]; 9, Hfx. gibbonsii JCM 8863T [8]; 10, Hfx. alexandrinus JCM 10717T [9]; 11, Hfx. sulfurifontis DSM 16227T [11]; 12, Hfx. prahovense DSM 18310T [12]; 13, Hfx. larsenii JCM 13917T [13]; 14, Hfx. chudinovii DSM 26526T [15]; 15, Hfx. namakaokahaiae DSM 29988T [16]. +, positive; −, negative; ±, variable; w, weakly positive; Hfx., Haloferax.
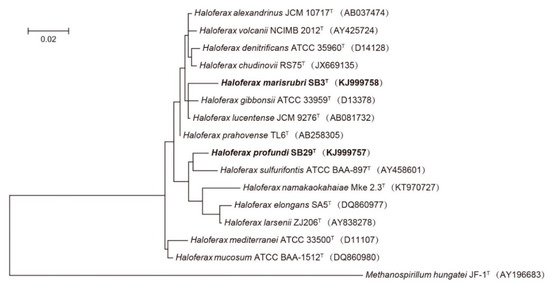
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on 16S rRNA gene sequences showing the relationships between strain SB3T and SB29T and closely related taxa. Scale bar indicates 0.01 substitutions per nucleotide position. Methanospirillum hungatei JF-1T (AY196683) was used as outgroup. GenBank accession numbers are listed for each sequence in parentheses.
Isolates and reference strains were grown in tubes containing an MR2A liquid medium supplemented with 0.5% (w/v) Na2S2O3, and H2S production was analyzed using a C2V-200 micro gas chromatograph from Thermo Scientific. Acid production was determined using the API 50 CHB system (bioMérieux), where the API 50 CHB medium was supplemented with (L−1): 2.0 g KCl, 0.3 g K2HPO4, 0.5 g CaCl2, 20.0 g MgSO4∙7H2O, and 180.0 g NaCl. The pH was maintained between 7.0–7.2. The neutral oligotrophic haloarchaeal medium contained the following ingredients (L−1): 0.05 g yeast extract, 5.4 g KCl, 0.3 g K2HPO4, 0.25 g CaCl2, 0.25 g NH4Cl, 26.8 g MgSO4 ∙ 7H2O, 23.0 g MgCl2∙ 6H2O, and 180.0 g NaCl (pH adjusted to 7.0–7.2 with a 1.0 M NaOH solution). This medium was combined with single carbon sources (5.0 g L−1) to test the hydrolysis of casein, starch, gelatin, and Tween 80 [25,26]. Biochemical activities and the use of organic substrates as sole carbon and energy sources were also evaluated using API 20E and API 20NE kits (bioMérieux) and Biolog GEN III MicroPlates (bioMérieux), according to the manufacturers’ instructions, with the exception of a salinity adjustment to 20% (w/v) NaCl and supplementing the media with (L−1) 2.0 g KCl, 0.3 g K2HPO4, 0.5 g CaCl2, and 20.0 g MgSO4∙7H2O (pH between 7.0–7.2). Susceptibility to antibiotics was assessed on MR2A agar using the Kirby–Bauer disc diffusion method according to Gutíérrez et al. [11] and Du et al. [27]. Generation times for SB29T and SB3T were determined at 33 °C and 37 °C, respectively, by the method of Robinson et al. [28]. The NaCl, pH, and temperature ranges for growth, sensitivity to antibiotics, and use of carbon sources are presented in the species descriptions. The characteristics that distinguish strains SB29T and SB3T from other validly described Haloferax species are shown in Table 1. Whole-cell protein profiles determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [29] were used as a rapid method for distinguishing between bacterial species [30]. Cells of strain SB29T and SB3T were grown in an MR2A liquid medium at 37 °C and were harvested at the late exponential growth phase for characterization of polar lipids. Polar lipids were extracted from 200 mg of freeze-dried cell material using a chloroform/methanol (0.3%, w/v)/aqueous NaCl mixture (1:2:0.8, v/v/v), modified after Bligh and Dyer [31], and were recovered into the chloroform phase by adjusting the mixture to a ratio of 1:1:0.9 (v/v/v) and subsequently separated by two-dimensional silica gel thin-layer chromatography (silica gel GF254, 0.25-mm thick, Haiyang Chemical Co., Qingdao, China). The first direction was developed in a chloroform/methanol/water (65:25:4, v/v/v) mixture and the second was developed in a chloroform/methanol/acetic acid/water (80:12:15:4, v/v/v/v) mixture. Total lipid material was detected using staining reagents (phosphomolybdic acid, molybdenum blue, ninhydrin, and α-naphthol) specific for defined functional groups [32]. Polar lipid analysis was performed by the identification services at the China Center of Industrial Culture Collection in Beijing, China. The analysis of respiratory quinones in strain SB29T and its closest relative, Hfx. sulfurifontis DSM 16227T, as well as strain SB3T and its closest relative, Hfx. denitrificans ATCC 35960T, was performed by the identification service of the DSMZ, Braunschweig, Germany.
2.4. Genomic Analyses
Genomic DNA was extracted using a commercial kit (TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit V 3.0) and subsequently sequenced on the Illumina HiSeq 2000 platform at the Bioscience Core Lab at the King Abdullah University of Science and Technology (KAUST, Saudi Arabia). Filtering and trimming of the raw data was performed using PRINSEQ v0.20.4 [33]. SOAPdenovo v1.05 [34,35] with default parameters was used to assemble the trimmed reads. The size of the genome was estimated by k-mer analysis [36] and genome completeness was determined with CheckM v1.0.3 [37]. Calculation of the DNA G + C content and prediction of protein-coding open reading frames (ORFs) was done in Glimmer v3.02 [38]. We used RNAmmer v1.2 to predict rRNAs [39] and tRNAscan-SE v1.21 to predict tRNAs [40]. Genome completeness was determined by CheckM v1.0.3.
The DNA–DNA relatedness value is a standard method for defining individual species, with 70% being the recommended minimum relatedness value for the DNA of strains of the same species [41,42]. Digital DNA–DNA hybridization (DDH) values were obtained by means of genome-to-genome sequence comparison via GGDC 2.0 using Formula (2) as implemented in the software [43]. Furthermore, as a complement to DNA–DNA hybridization [44], average nucleotide identity (ANI) values of the total genomic sequences shared between the genomic sequences of strain SB29T or SB3T with closely related genomic sequences from GenBank were determined according to Goris et al. [45]. Whole-genome sequences in a pairwise comparison were split into consecutive 1000 bp windows. Sequences were aligned with nucmer in MUMmer v3.23 [46] and ANI values were calculated using JSpeciesWS [47].
2.5. Phylogenetic Analyses
Multiple sequence alignments of 16S rRNA gene sequences of strains SB29T and SB3T with those of their most closely related taxa were performed using CLUSTALX v1.81 [48]. A phylogenetic tree was constructed with the maximum likelihood (ML) method using MEGA v6.0 [49], based on recommendations by Minegishi et al. (2012) [50], and sequences of strains SB29T and SB3T were placed within the genus Haloferax (Figure 1). Maximum parsimony (MP) and neighbor-joining (NJ) methods were additionally used to analyze and verify the taxonomic positions of the novel isolates and reference strains (Supplementary Figures S5 and S6). All available genome sequences of validly described Haloferax species (11 genomes) were retrieved from NCBI and re-annotated using PROKKA [51]. We used Roary to identify the core genes of these type strains [52] and a multiple sequence alignment of 31 single-copy core genes was obtained using MAFFT [53]. A phylogenetic tree based on the concatenation of these single-copy core genes was inferred using the neighbor-joining (NJ) method and rooted with sequences retrieved from Methanospirillum hungatei JF-1T (Supplementary Figure S7).
2.6. Data Availability
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA genes of strains SB29T and SB3T are KJ999757 and KJ999758, respectively. The whole genome shotgun projects were deposited at DDBJ/EMBL/GenBank under the accession identifications LOPV00000000 and LOPW00000000 for strains SB29T and SB3T, respectively. Phase-contrast micrographs of the late exponential phase of strains SB29T and SB3T cells, sodium dodecyl sulfate polyacrylamide gel electrophoresis patterns of the whole-cell proteins, thin-layer chromatograms of the polar lipids extracted from strains SB29T and SB3T, additional phylogenetic trees, and a table of average nucleotide identity values with closely related strains are available as Supplementary Materials.
3. Results
All phylogenetic analyses are in agreement and show that both strains branch separately from the existing validly described species of the genus Haloferax, indicating that these strains represent novel species (Figure 1, Figures S6 and S7). Based on the results of the genomic assembly, both genomes are estimated to be 99.0% complete. The draft genome of SB29T is 3,871,125 bp in size with 771 contigs, 57 scaffolds, and a 136-fold coverage; the draft genome of strain SB3T is 3904,985 bp in size with 66 contigs, 26 scaffolds, and a 154-fold coverage. A total of 4053 ORFs, 47 tRNAs, and 2 rRNAs are predicted for strain SB29T, and a total of 3824 ORFs, 51 tRNAs, and 2 rRNAs are predicted for strain SB3T. The DNA G + C contents for strain SB29T and SB3T are 60.75% and 65.64%, respectively, which are very similar to those reported for other Haloferax species (Table 1). The genomic data suggests that both strains SB29T (1473 bp) and SB3T (1446 bp) only possess one copy of the 16S rRNA gene.
Strains SB29T and SB3T had low DNA–DNA relatedness and DDH with each other and with all recognized Haloferax species (Table 1, Tables S1 and S2). An ANI boundary of 95–96% is recommended to taxonomically circumscribe prokaryotic species [54,55]. The highest ANI values between the genomes of SB29T and SB3T and their closely related genomes in GenBank were determined as 82.6% (Hfx. sulfurifontis ATCC BAA-897T, GenBank accession no. GCA_000337835.1) and 92.6% (Haloferax denitrificans ATCC 35960T, GenBank accession no. GCA_000337795.1), respectively (Supplementary Tables S1 and S2). This further indicates that strains SB29T and SB3T represent two different novel species.
Phylogenetic, phenotypic, and chemotaxonomic data indicate that strains SB29T and SB3T are members of the genus Haloferax. The DNA–DNA hybridization data, whole-cell protein profiles, and phenotypic characteristics (Table 1) justify the creation of two novel species within the genus Haloferax to accommodate these strains, for which the names Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., respectively, are proposed. The major respiratory quinones of strain SB29T were MK8 (69%) and MK8 (VIII-H2, 31%), while those of SB3T were MK8 (90%) and MK8 (VIII-H2, 10%), which differ from other closely related species of the same genus Haloferax, i.e., Hfx. sulfurifontis DSM 16227T (MK8, 74%, and MK8 (VIII-H2, 26%)) and Hfx. denitrificans ATCC 35960T (MK8, 81%, and MK8 (VIII-H2, 19%)).
Whole-cell protein analysis of strains SB29T and SB3T with various other Haloferax strains using SDS-PAGE (Supplementary Figure S3) showed complex band patterns, but clear differences between the novel strains reported here and validly described Haloferax species.
Both strains contained phosphatidyl glycerol (PG), phosphatidyl glycerorophosphate-methyl ester (PGP-Me), sulfated mannosyl glucosyl diether (S-DGD-1), and mannosyl glucosyldiether (DGD-1), but lacked phosphatidyl glycerosulfate (PGS), which is a characteristic of other species of the genus Haloferax [56] (Supplementary Figure S4).
4. Discussion
Based on the phenotypic, phylogenetic, and genetic differences between strains SB29T and SB3T and their most closely related species, we propose to delineate them as two novel species, Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov.
4.1. Description of Haloferax profundi sp. nov.
Haloferax profundi (pro.fun’di. L. gen. n. profundi, from the depths of the sea). Cells are Gram-negative, motile, and pleomorphic, shaped as irregular polyhedra, ovals, and rods (average size of 0.3–0.6 × 1.0–6.8 μm); under optimal conditions, cells undergo exponential growth and elongate up to 13 μm (Supplementary Figures S1 and S2). Colonies on a complex agar medium formed pinkish-red, opaque, domed colonies, with a diameter of 3–5 mm. Growth occurs in 1.0–5.5 M NaCl (optimum between 3.5–4.5 M NaCl), at 15–45 °C (optimum at 33 °C), and at pH 5.5–9.0 (optimum between pH 7.0–7.5). Growth occurs in media containing between 0.05–0.7 M Mg2+ (optimum at 0.2 M Mg2+), and cell lysis occurs in solutions below 0.9 M NaCl. The generation time at 33 °C is 36 ± 4 min. Cells are chemoorganotrophic, aerobic, oxidase, and catalase positive, and esterase negative. Nitrate is not reduced to nitrite, nor is any gas produced from nitrate. Indole is produced from tryptophan, but H2S is not produced from thiosulfate. No anaerobic growth is observed with nitrate, DMSO, or L-arginine. Gelatin, casein, and starch are hydrolyzed, unlike Tween 80. The following substrates are used as carbon sources: D-glucose, D-mannitol, D-maltose, malate, citrate, glycerol, sucrose, and trehalose. Acid is produced from D-glucose, glycerol, D-maltose, sucrose, and trehalose. The following substrates are not used as carbon sources: D-sorbitol, d-arabitol, ethanol, fumarate, D-mannose, D-fructose, D-galactose, L-rhamnose, xylose, glycine, lactose, raffinose, L-histidine, L-lysine, or L-ornithine. Strain SB29T contains phosphatidyl glycerol (PG), phosphatidyl glycerol phosphate-methylester (PGP-Me), sulfated mannosyl glucosyldiether (S-DGD-1), mannosyl glucosyldiether (DGD-1), two unidentified phospholipids (UPL1 and UPL2), and glycolipid (GL), but phosphatidyl glycerol sulfate (PGS) was absent. The major respiratory quinones of strain SB29T are MK8 (69%) and MK8 (VIII-H2, 31%). The genome draft as presented here has a size of 3,871,125 bp and a DNA G + C content of 60.75 mol%. The type strain is SB29T (=JCM 19567T = CGMCC 1.14960T), its genome accession number is GCA_001469865.1, and it was isolated from the Discovery Deep brine-seawater (Di2) in the Red Sea.
4.2. Description of Haloferax marisrubri sp. nov.
Haloferax marisrubri (ma.ris.ru’bri. L. n. mare, maris, the sea; L. adj. ruber, red; N.L. gen. n. marisrubri, of the Red Sea). Cells are Gram-negative, motile, and pleomorphic, with irregular polyhedra, ovals, and rods (average, 0.4–0.7 × 1.0–5.3 μm). Under optimal conditions, growth is exponential and cells elongate to up to 11 μm (Supplementary Figures S1 and S2). Colonies on a complex agar medium are pink, translucent, convex, and about 1.0 mm in diameter. Growth occurs between 1.5–5.8 M NaCl (optimum between 4.5–5.0 M NaCl), at 20–50 °C (optimum at 37 °C), and between pH 6.5–9.0 (optimum between pH 7.5–8.0). Growth occurs in media containing 0.05–1.0 M Mg2+ (optimum at 0.35 M Mg2+), and cell lysis occurs in NaCl solutions below 0.95 M. The generation time at 37 °C is 47 ± 5 min. Cells are chemoorganotrophic, aerobic, oxidase, and catalase positive, and esterase negative. Nitrate is not reduced to nitrite, and no gas is produced from nitrate. Indole is not produced from tryptophan and H2S is not produced from thiosulfate. Anaerobic growth is not observed with nitrate, DMSO, or L-arginine. Gelatin and starch are hydrolyzed, unlike casein and Tween 80. The following substrates are used as carbon sources: D-glucose, D-mannitol, D-maltose, citrate, glycerol, and trehalose. Acid is produced from D-glucose, glycerol, D-maltose, and trehalose. The following substrates are not used as carbon sources: D-mannose, D-fructose, sucrose, D-sorbitol, D-arabitol, ethanol, malate, fumarate, D-galactose, L-rhamnose, xylose, glycine, lactose, raffinose, L-histidine, L-lysine, or L-ornithine. Strain SB3T contains phosphatidyl glycerol (PG), phosphatidyl glycerol phosphate-methylester (PGP-Me), sulfated mannosyl glucosyldiether (S-DGD-1), mannosyl glucosyldiether (DGD-1), two unidentified phospholipids (UPL1 and UPL2), and glycolipid (GL), but phosphatidyl glycerol sulfate (PGS) is absent. The major respiratory quinones of SB3T are MK8 (90.0%) and MK8 (VIII-H2, 10.0%). The genome draft, as presented here, has a size of 3,904,985 bp and a DNA G + C content of 65.64 mol%. The type strain is SB3T (= JCM 19567T = CGMCC 1.14960T), its genome accession number is GCA_001469875.2, and it was isolated from the Discovery Deep brine-seawater (Di1) in the Red Sea.
5. Conclusions
In this study, we isolated and described two novel strains of extremely halophilic Archaea from the brine-seawater interface of Discovery Deep in the Red Sea. Phylogenetic, phenotypic, and genetic analyses indicated that these strains represent two novel species of the genus Haloferax, for which we propose the names Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/10/1475/s1, Figure S1. Phase-contrast micrograph of strain SB29T (a) and SB3T (b) after 72 h growth in liquid MR2A medium at 37 °C, illustrating the pleomorphic nature of cells of strains SB29T and SB3T. Scale bars are shown on the images, Figure S2. Scanning Electron Microscope (SEM) micrograph of strain SB29T (a) and SB3T (b) after 72 h of growth in liquid MR2A medium at 37 °C, scale bar = 1 μm for panel a, scale bar = 2 μm for panel b, Figure S3. Whole-cell proteins from various halophilic archaeal strains and isolates SB29T and SB3T, following separation by SDS-PAGE. Following cell lysis, approximately 15 µg of protein was applied to each lane. Proteins were stained with Coomassie brilliant blue. Lanes: 1, molecular mass marker; 2, Strain SB29T; 3, Strain SB3T; 4, Hfx. alexandrinus JCM 10717T; 5, Hfx. larsenii JCM 13917T; 6, Hfx. lucentense DSM14919T; 7, Hfx. prahovense DSM 18310T; 8, Hfx. sulfurifontis DSM 16227T; 9. Hfx. elongans JCM 14791T, Figure S4. Polar lipids profiles for strain SB3T (a) and SB29T (b) include PG, PGP-Me, S-DGD-1, DGD-1, UPL1 and UPL2; and an unidentified GL, which support the placement of both strains into the genus Haloferax. The TLC plates were stained with 10% phosphomolybdic acid in 95% ethanol for detection of total lipids, 0.2 % ninhydrin in 1-butanol for aminolipids, molybdenum blue reagent (Sigma) for phospholipids, and a-naphthol staining reagent (Sigma) for cholines, Figure S5. Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationships between strain SB3T and SB29T (bold) and related taxa. Scale bar indicates 0.01 substitutions per nucleotide position. Methanospirillum hungatei JF-1T (AY196683) was used as outgroup. GenBank accession numbers are indicated for each strain in parentheses, Figure S6. Maximum parsimony phylogenetic tree based on 16S rRNA gene sequences showing the relationships between strain SB3T and SB29T (both in bold) and related taxa. Methanospirillum hungatei JF-1T (AY196683) was used as outgroup. GenBank accession numbers are indicated for each strain in parentheses, Figure S7. Phylogenomic tree based on the concatenation of the 31 single-copy core genes from all 11 available genomes of type strains for Haloferax spp. were inferred with Neighbor-Joining (NJ) method and rooted by Methanospirillum hungatei JF-1T. The genes were katG2, top6A, ntpB, atpA, ftsZ_2, aroB, fusA, uvrA, Ion, gatA, ndk, rplB, ftsZ_1, pdxS, nikR_2, guaA_2, rpoB_1, rpsK, rpsJ_1, tufB_1, rps12P, rps28e, DUF2073, rps2P, rpoK, rpl40e, rps3p, Mcm2 and three hypothetical proteins (PRK00939, PRK04016, PRK04243; https://www.ncbi.nlm.nih.gov/cdd) as determined in Roary (see methods section), Table S1. Average nucleotide identity (ANIm) and digital DNA-DNA hybridization (dDDH) values (%) between the genomes of strain SB3T (GenBank accession: LOPW00000000), and closely related genomes in GenBank, Table S2. Average nucleotide identity (ANIm) and digital DNA-DNA hybridization (dDDH) values (%) between the genomes of strain SB29T (GenBank accession no.: LOPV00000000), and closely related genomes in GenBank.
Author Contributions
Conceptualization, U.S.; Formal analysis, X.D., Y.S. and J.W.; Funding acquisition, U.S.; Investigation, G.Z.; Methodology, A.A., T.H. and M.F.H.; Supervision, U.S.; Validation, U.S.; Writing—review & editing, A.A. and U.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by baseline support from the King Abdullah University of Science and Technology (KAUST) and the Saudi Economic and Development Company Research Excellence Award to U.S.
Acknowledgments
We would like to thank Aharon Oren for the very valuable help in naming the organisms.
Conflicts of Interest
All of the authors of the manuscript entitled "Haloferax profundi sp. nov. and Haloferax marisrubri sp. nov., isolated from the Discovery Deep brine-seawater interface in the Red Sea" declare that there is no conflict of interest to this work. We declare that we do not have any commercial or any other associative interest that represents a conflict of interest in connection with the work submitted.
References
- Bougouffa, S.; Yang, J.K.; Lee, O.O.; Wang, Y.; Batang, Z.; Al-Suwailem, A.; Qian, P.Y. Distinctive Microbial Community Structure in Highly Stratified Deep-Sea Brine Water Columns. Appl. Environ. Microbiol. 2013, 79, 3425–3437. [Google Scholar] [CrossRef] [PubMed]
- Eder, W.; Jahnke, L.L.; Schmidt, M.; Huber, R. Microbial Diversity of the Brine-Seawater Interface of the Kebrit Deep, Red Sea, Studied via 16S rRNA Gene Sequences and Cultivation Methods. Appl. Environ. Microbiol. 2001, 67, 3077–3085. [Google Scholar] [CrossRef]
- Daffonchio, D.; Borin, S.; Brusa, T.; Brusetti, L.; van der Wielen, P.W.J.J.; Bolhuis, H.; Yakimov, M.M.; D’Auria, G.; Giuliano, L.; Marty, D.; et al. Stratified prokaryote network in the oxic–anoxic transition of a deep-sea halocline. Nature 2006, 440, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Naushad, S.; Baker, S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: A proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 1050–1069. [Google Scholar] [PubMed]
- Mullakhanbhai, M.F.; Larsen, H. Halobacteriumvolcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 1975, 104, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Valera, F.; Juez, G.; Kushner, D.J. Halobacterium mediterranei spec, nov., a New Carbohydrate-Utilizing Extreme Halophile. Syst. Appl. Microbiol. 1983, 4, 369–381. [Google Scholar] [CrossRef]
- Ga, T.; Li, J.; Li, H. Halobacterium denitrificans sp. nov., an extremely halophilic denitrifying bacterium. Int. J. Syst. Bacteriol. 1986, 36, 66–70. [Google Scholar]
- Juez, G.; Rodriguez-Valera, F.; Ventosa, A.; Kushner, D.J. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic archaebacteria. Syst. Appl. Microbiol. 1986, 8, 75–79. [Google Scholar] [CrossRef]
- Asker, D.; Ohta, Y. Haloferax alexandrinus sp. nov., an extremely halophilic canthaxanthin-producing archaeon from a solar saltern in Alexandria (Egypt). Int. J. Syst. Evol. Microbiol. 2002, 52, 729–738. [Google Scholar]
- Gutierrez, M.C.; Kamekura, M.; Holmes, M.L.; Dyall-Smith, M.L.; Ventosa, A. Taxonomic characterization of Haloferax sp. (“H. alicantei”) strain Aa 2.2: Description of Haloferax lucentensis sp. nov. Extremophiles 2002, 6, 479–483. [Google Scholar] [CrossRef]
- Elshahed, M.S.; Savage, K.N.; Oren, A.; Gutierrez, M.C.; Ventosa, A.; Krumholz, L.R. Haloferax sulfurifontis sp. nov., a halophilic archaeon isolated from a sulfide- and sulfur-rich spring. Int. J. Syst. Evol. Microbiol. 2004, 54, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Enache, M.; Itoh, T.; Kamekura, M.; Teodosiu, G.; Dumitru, L. Haloferax prahovense sp. nov., an extremely halophilic archaeon isolated from a Romanian salt lake. Int. J. Syst. Evol. Microbiol. 2007, 57, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Wu, Y.H.; Wang, C.S.; Oren, A.; Zhou, P.J.; Wu, M. Haloferax larsenii sp. nov., an extremely halophilic archaeon from a solar saltern. Int. J. Syst. Evol. Microbiol. 2007, 57, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.A.; Goh, F.; Leuko, S.; Echigo, A.; Mizuki, T.; Usami, R.; Kamekura, M.; Neilan, A.B.; Burns, B.P. Haloferax elongans sp. nov. and Haloferax mucosum sp. nov., isolated from microbial mats from Hamelin Pool, Shark Bay, Australia. Int. J. Syst. Evol. Microbiol. 2008, 58, 798–802. [Google Scholar] [CrossRef]
- Saralov, A.I.; Baslerov, R.V.; Kuznetsov, B.B. Haloferax chudinovii sp. nov., a halophilic archaeon from Permian potassium salt deposits. Extremophiles 2013, 17, 499–504. [Google Scholar] [CrossRef]
- Mcduff, S.; King, G.M.; Neupane, S.; Myers, M.R. Isolation and characterization of extremely halophilic CO-oxidizing Euryarchaeota from hypersaline cinders, sediments and soils and description of a novel CO oxidizer, Haloferax namakaokahaiae Mke2.3T, sp. nov. FEMS Microbiol. Ecol. 2016, 92, fiw028. [Google Scholar] [CrossRef][Green Version]
- Zhang, G.; Gu, J.; Zhang, R.; Rashid, M.; Haroon, M.F.; Xun, W.; Ruan, Z.; Dong, X.; Stingl, U. Haloprofundus marisrubri gen. nov., sp. nov., an extremely halophilic archaeon isolated from a brine-seawater interface. Int. J. Syst. Evol. Microbiol. 2017, 67, 9–16. [Google Scholar] [CrossRef]
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef]
- Oren, A.; Ventosa, A.; Grant, W.D. Proposed minimal standards for description of new taxa in the order Halobacteriales. Int. J. Syst. Bacteriol. 1997, 47, 233–238. [Google Scholar] [CrossRef]
- Rieger, G.; Müller, K.; Hermann, R.; Stetter, K.O.; Rachel, R. Cultivation of hyperthermophilic archaea in capillary tubes resulting in improved preservation of fine structures. Arch. Microbiol. 1997, 168, 373–379. [Google Scholar] [CrossRef]
- Huber, H.; Burggraf, S.; Mayer, T.; Wyschkony, I.; Rachel, R.; Stetter, K.O. Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov and Ignicoccus pacificus sp. nov and Ignicoccus pacificus sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50. [Google Scholar] [CrossRef] [PubMed]
- Dussault, H.P. An improved technique for staining red halophilic bacteria. J. Bacteriol. 1955, 70, 484–485. [Google Scholar] [CrossRef] [PubMed]
- Goh, F.; Leuko, S.; Allen, M.A.; Bowman, J.P.; Kamekura, M.; Neilan, B.A.; Burns, B.P. Halococcus hamelinensis sp. nov., a novel halophilic archaeon isolated from stromatolites in Shark Bay, Australia. Int. J. Syst. Evol. Microbiol. 2006, 56, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, C.; González, C. Method for Simultaneous Detection of Proteinase and Esterase Activities in Extremely Halophilic Bacteria. Appl. Microbiol. 1972, 24, 516–517. [Google Scholar] [CrossRef]
- Cui, H.L.; Gao, X.; Sun, F.F.; Dong, Y.; Xu, X.W.; Zhou, Y.-G.; Liu, H.-C.; Oren, O.; Zhou, P.-J. Halogranum rubrum gen. nov., sp. nov., a halophilic archaeon isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2010, 60, 1366–1371. [Google Scholar] [CrossRef]
- Dyall-Smith, M.L. The Halohandbook: Protocols for Haloarchaeal Genetics. 2008. Available online: http://www.haloarchaea.com/resources/halohandbook/ (accessed on 7 July 2020).
- Du, Z.J.; Wang, Y.; Dunlap, C.; Rooney, A.P.; Chen, G.J. Draconibacterium orientale gen. nov., sp. nov., isolated from two distinct marine environments, and proposal of Draconibacteriaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 1690–1696. [Google Scholar] [CrossRef]
- Robinson, J.L.; Pyzyna, B.; Atrasz, R.G.; Henderson, C.A.; Morrill, K.L.; Anna Mae, B.; Desoucy, E.; Fogleman, R.E., 3rd; Naylor, J.B.; Steele, S.M.; et al. Growth kinetics of extremely halophilic Archaea (family Halobacteriaceae) as revealed by Arrhenius plots. J. Bacteriol. 2005, 187, 923–929. [Google Scholar] [CrossRef]
- Stan-Lotter, H.; Pfaffenhuemer, M.; Legat, A.; Busse, H.J.; Radax, C.; Gruber, C. Halococcus dombrowskii sp. nov., an archaeal isolate from a Permian alpine salt deposit. Int. J. Syst. Evol. Microbiol. 2002, 52, 1807–1814. [Google Scholar]
- Jackman, P.J.H. Microbial systematics based on electrophoretic whole-cell protein patterns. Methods Microbiol. 1987, 19, 209–225. [Google Scholar]
- Bligh, E.G. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Tindall, B.J.; Sikorski, J.; Smibert, R.M.; Krieg, N.R. Phenotypic characterization and the principles of comparative systematics. In Methods for General and Molecular Microbiology, 3rd ed.; Reddy, C.A., Beveridge, T.J., Breznak, J.A., Marzluf, G., Marzluf, T.M., Schmidt, L.R., Eds.; Snyder ASM Press: Washington, DC, USA, 2007; pp. 330–393. [Google Scholar]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shi, Y.; Yuan, J.; Hu, X.; Zhang, H.; Liu, N.; Li, Z.; Chen, Y.; Mu, D.; Fan, W. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv 2013, arXiv:1308.2012. [Google Scholar]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.; Morre, L.H.; Morre, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Graham, P.H.; Sadowsky, M.J.; Keyser, H.H.; Barnet, Y.M.; Bradley, R.S.; Cooper, J.E.; de Ley, D.J.; Jarvis, B.D.W.; Roslycky, E.B.; Strijdom, B.W.; et al. Proposed minimal standards for the description of new genera and species of root- and stem-nodulating bacteria. Int. J. Syst. Bacteriol. 1991, 41, 582–587. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. SpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Minegishi, H.; Kamekura, M.; Kitajima-Ihara, T.; Nakasone, K.; Echigo, A.; Shimane, Y.; Usami, R.; Itoh, T.; Ihara, K. Gene orders in the upstream of 16S rRNA genes divide genera of the family Halobacteriaceae into two groups. Int. J. Syst. Evol. Microbiol. 2012, 62, 188–195. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Kamekura, M.; Mizuki, T.; Usami, R.; Yoshida, Y.; Horikoshi, K.; Vreeland, V.H. The potential use of signature bases from 16S rRNA gene sequences to aid the assignment of microbial strains to genera of halobacteria. In Halophilic Microorganisms; Ventosa, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 77–100. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).