Whole Genome Sequencing for Studying Bacillus anthracis from an Outbreak in the Abruzzo Region of Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Outbreak Scenario
2.2. DNA, CanSNPs, and WGS
2.3. NGS Data Analysis
3. Results
3.1. Genomic Features of the Abruzzo Outbreak Strain
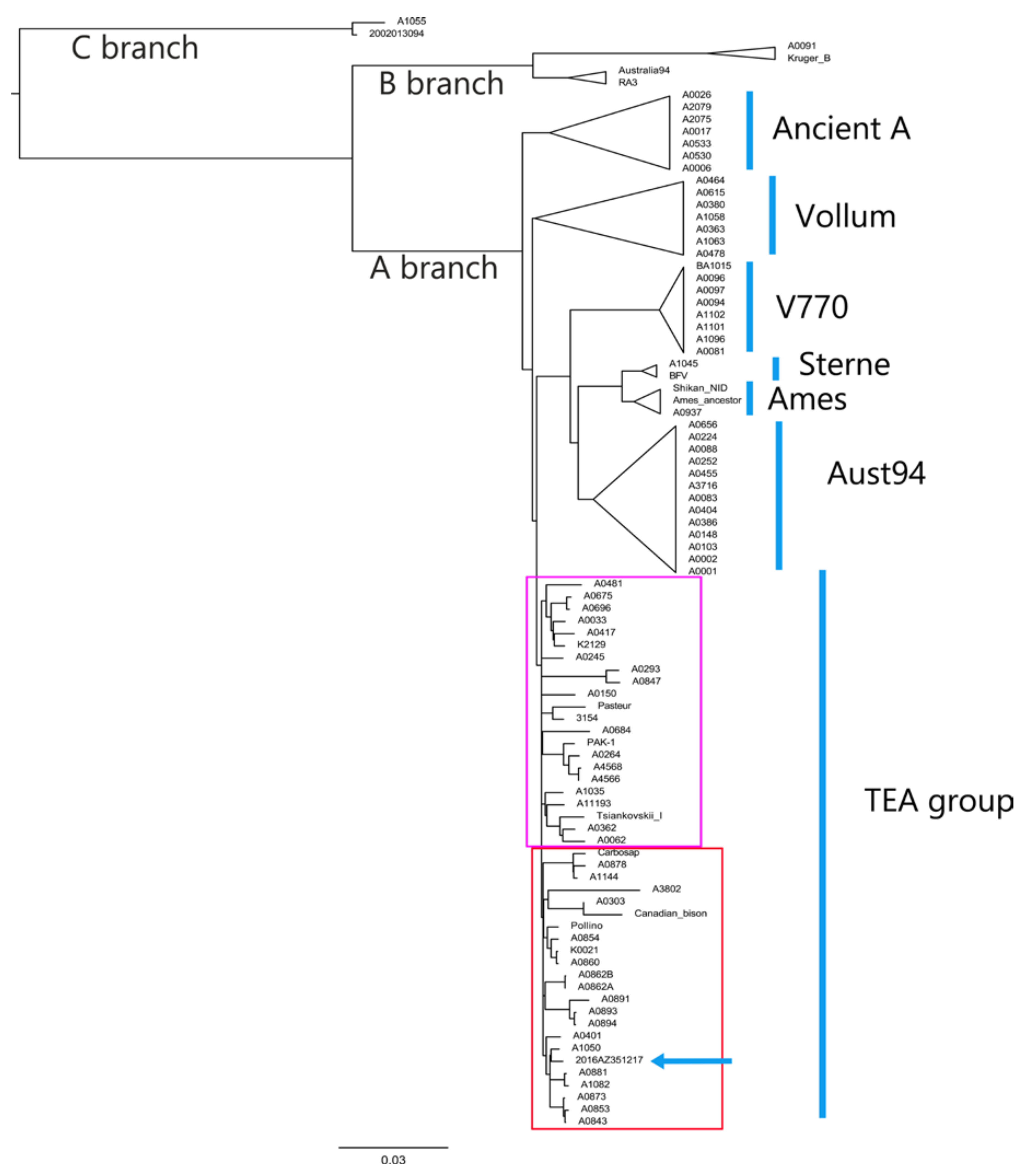
3.2. Worldwide Phylogenetic Positioning of the Abruzzo Outbreak Strain
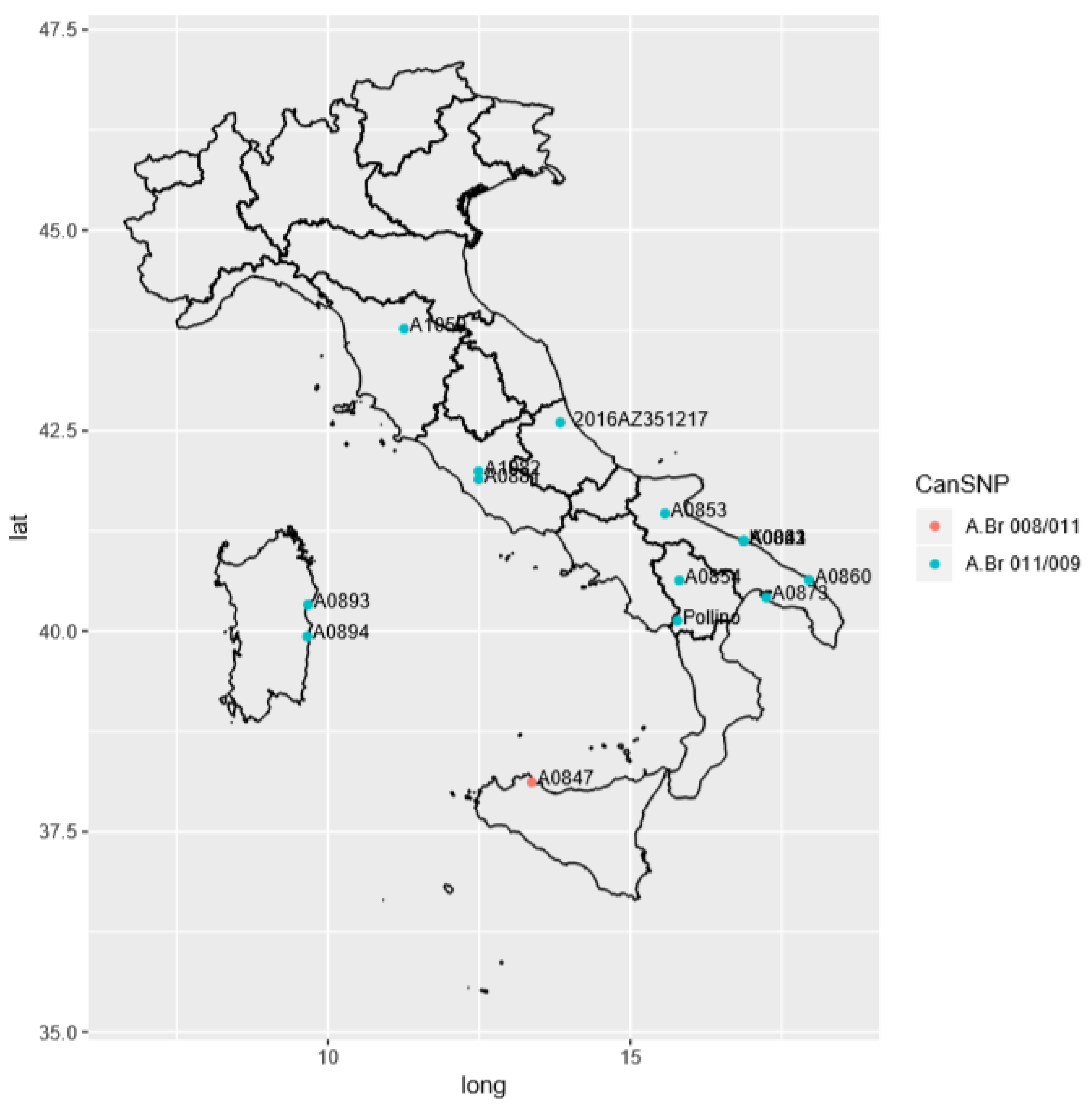
3.3. Detailed Italian Genetic Positioning of the Abruzzo Outbreak Strain
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fasanella, A.; Serrecchia, L.; Chiaverini, A.; Garofolo, G.; Muuka, G.M.; Mwambazi, L. Use of Canonical Single Nucleotide Polymorphism (CanSNPs) to characterize. PeerJ 2018, 6, e5270. [Google Scholar] [CrossRef]
- Glassman, H.N. World incidence of anthrax in man. Public Health Rep. 1958, 73, 22–24. [Google Scholar] [CrossRef]
- Hugh-Jones, M. 1996-97 Global Anthrax Report. J. Appl. Microbiol. 1999, 87, 189–191. [Google Scholar] [CrossRef]
- Braun, P.; Grass, G.; Aceti, A.; Serrecchia, L.; Affuso, A.; Marino, L.; Grimaldi, S.; Pagano, S.; Hanczaruk, M.; Georgi, E.; et al. Microevolution of Anthrax from a Young Ancestor (M.A.Y.A.) Suggests a Soil-Borne Life Cycle of Bacillus anthracis. PLoS ONE 2015, 10, e0135346. [Google Scholar] [CrossRef]
- Fasanella, A.; Van Ert, M.; Altamura, S.A.; Garofolo, G.; Buonavoglia, C.; Leori, G.; Huynh, L.; Zanecki, S.; Keim, P. Molecular diversity of Bacillus anthracis in Italy. J. Clin. Microbiol. 2005, 43, 3398–3401. [Google Scholar] [CrossRef]
- Fasanella, A.; Palazzo, L.; Petrella, A.; Quaranta, V.; Romanelli, B.; Garofolo, G. Anthrax in red deer (Cervus elaphus), Italy. Emerg. Infect. Dis. 2007, 13, 1118–1119. [Google Scholar] [CrossRef]
- Palazzo, L. Recent Epidemic-Like Anthrax Outbreaks in Italy: What Are the Probable Causes? Open J. Vet. Med. 2012, 2, 74–76. [Google Scholar] [CrossRef]
- Van Ert, M.N.; Easterday, W.R.; Huynh, L.Y.; Okinaka, R.T.; Hugh-Jones, M.E.; Ravel, J.; Zanecki, S.R.; Pearson, T.; Simonson, T.S.; U’Ren, J.M.; et al. Global genetic population structure of Bacillus anthracis. PLoS ONE 2007, 2, e461. [Google Scholar] [CrossRef]
- Marston, C.K.; Allen, C.A.; Beaudry, J.; Price, E.P.; Wolken, S.R.; Pearson, T.; Keim, P.; Hoffmaster, A.R. Molecular epidemiology of anthrax cases associated with recreational use of animal hides and yarn in the United States. PLoS ONE 2011, 6, e28274. [Google Scholar] [CrossRef]
- Pisarenko, S.V.; Eremenko, E.I.; Ryazanova, A.G.; Kovalev, D.A.; Buravtseva, N.P.; Aksenova, L.Y.; Dugarzhapova, Z.F.; Evchenko, A.Y.; Kravets, E.V.; Semenova, O.V.; et al. Phylogenetic analysis of Bacillus anthracis strains from Western Siberia reveals a new genetic cluster in the global population of the species. BMC Genom. 2019, 20, 692. [Google Scholar] [CrossRef]
- Keim, P.; Price, L.B.; Klevytska, A.M.; Smith, K.L.; Schupp, J.M.; Okinaka, R.; Jackson, P.J.; Hugh-Jones, M.E. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 2000, 182, 2928–2936. [Google Scholar] [CrossRef]
- Sahl, J.W.; Pearson, T.; Okinaka, R.; Schupp, J.M.; Gillece, J.D.; Heaton, H.; Birdsell, D.; Hepp, C.; Fofanov, V.; Noseda, R.; et al. A Bacillus anthracis Genome Sequence from the Sverdlovsk 1979 Autopsy Specimens. MBio 2016, 7, e01501–e01516. [Google Scholar] [CrossRef]
- Pilo, P.; Frey, J. Pathogenicity, population genetics and dissemination of Bacillus anthracis. Infect. Genet. Evol. 2018, 64, 115–125. [Google Scholar] [CrossRef]
- Girault, G.; Blouin, Y.; Vergnaud, G.; Derzelle, S. High-throughput sequencing of Bacillus anthracis in France: Investigating genome diversity and population structure using whole-genome SNP discovery. BMC Genom. 2014, 15, 288. [Google Scholar] [CrossRef]
- Derzelle, S.; Thierry, S. Genetic diversity of Bacillus anthracis in Europe: Genotyping methods in forensic and epidemiologic investigations. Biosecur. Bioterror. 2013, 11, S166–S176. [Google Scholar] [CrossRef]
- Pisarenko, S.V.; Eremenko, E.I.; Ryazanova, A.G.; Kovalev, D.A.; Buravtseva, N.P.; Aksenova, L.Y.; Evchenko, A.Y.; Semenova, O.V.; Bobrisheva, O.V.; Kuznetsova, I.V.; et al. Genotyping and phylogenetic location of one clinical isolate of Bacillus anthracis isolated from a human in Russia. BMC Microbiol. 2019, 19, 165. [Google Scholar] [CrossRef]
- Price, E.P.; Seymour, M.L.; Sarovich, D.S.; Latham, J.; Wolken, S.R.; Mason, J.; Vincent, G.; Drees, K.P.; Beckstrom-Sternberg, S.M.; Phillippy, A.M.; et al. Molecular epidemiologic investigation of an anthrax outbreak among heroin users, Europe. Emerg. Infect. Dis. 2012, 18, 1307–1313. [Google Scholar] [CrossRef]
- Girault, G.; Thierry, S.; Cherchame, E.; Derzelle, S. Application of High-Throughput Sequencing: Discovery of Informative SNPs to Subtype Bacillus anthracis. Adv. Biosci. Biotechnol. 2014, 5, 669–677. [Google Scholar] [CrossRef]
- GitLab. WGSBAC. Available online: https://gitlab.com/FLI_Bioinfo/WGSBAC.git (accessed on 12 November 2019).
- Köster, J.; Rahmann, S. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics 2018, 34, 3600. [Google Scholar] [CrossRef]
- Grüning, B.; Dale, R.; Sjödin, A.; Chapman, B.A.; Rowe, J.; Tomkins-Tinch, C.H.; Valieris, R.; Köster, J.; Team, B. Bioconda: Sustainable and comprehensive software distribution for the life sciences. Nat. Methods 2018, 15, 475–476. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, 46. [Google Scholar] [CrossRef]
- Seeman, T. Shovill. Available online: https://github.com/tseemann/shovill (accessed on 12 November 2019).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seeman, T. ABRicate. Available online: https://github.com/tseemann/abricate (accessed on 12 November 2019).
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Seeman, T. Snippy. Available online: https://github.com/tseeman/snippy (accessed on 12 November 2019).
- FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 12 November 2019).
- Bionumerics. Bionumerics. Available online: http://www.applied-maths.com/ (accessed on 12 November 2019).
- Liu, Y.; Lai, Q.; Göker, M.; Meier-Kolthoff, J.P.; Wang, M.; Sun, Y.; Wang, L.; Shao, Z. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 2015, 5, 14082. [Google Scholar] [CrossRef]
- Fasanella, A.; Galante, D.; Garofolo, G.; Jones, M.H. Anthrax undervalued zoonosis. Vet. Microbiol. 2010, 140, 318–331. [Google Scholar] [CrossRef]
- Schuch, R.; Fischetti, V.A. The secret life of the anthrax agent Bacillus anthracis: Bacteriophage-mediated ecological adaptations. PLoS ONE 2009, 4, e6532. [Google Scholar] [CrossRef]
- Cavallo, J.D.; Ramisse, F.; Girardet, M.; Vaissaire, J.; Mock, M.; Hernandez, E. Antibiotic susceptibilities of 96 isolates of Bacillus anthracis isolated in France between 1994 and 2000. Antimicrob. Agents Chemother. 2002, 46, 2307–2309. [Google Scholar] [CrossRef]
- Muscillo, M.; La Rosa, G.; Sali, M.; De Carolis, E.; Adone, R.; Ciuchini, F.; Fasanella, A. Validation of a pXO2-A PCR assay to explore diversity among Italian isolates of Bacillus anthracis strains closely related to the live, attenuated Carbosap vaccine. J. Clin. Microbiol. 2005, 43, 4758–4765. [Google Scholar] [CrossRef]
- Garofolo, G.; Serrecchia, L.; Corrò, M.; Fasanella, A. Anthrax phylogenetic structure in Northern Italy. BMC Res. Notes 2011, 4, 273. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. ECDC Strategic Framework for the Integration of Molecular and Genomic Typing into European Surveillance and Multi Country Outbreak Investigations 2019–2021; ECDC: Stockholm, Sweden, 2019. [Google Scholar]
| Parameter | Characteristics of the Outbreak Strain |
| Total number of reads | 4,564,206 |
| Total number of nucleotides | 615,149,873 |
| G + C content | 36.0 |
| Average read length | 134 (35–151) |
| Depth of coverage | 109x |
| Genome assembly results | |
| Total number of contigs | 35 |
| Total genome length | 5,450,929 bp |
| Average contig size | 155,740 bp (524–1,052,202 bp) |
| N50 | 514,228 bp |
| Genome annotation results | |
| Total number of CDS | 5718 |
| Total number of rRNA | 7 |
| Total number of tRNA | 80 |
| Total number of tmRNA | 1 |
| Virulence genes | nheA; non-hemolytic enterotoxin A—NP_978284 |
| nhe; non-hemolytic enterotoxin B—NP_978285 | |
| nhe; non-hemolytic enterotoxin C—NP_978286 | |
| BAS3109; thiol-activated cytolysin—YP_029366 | |
| inhA; immune inhibitor A metalloprotease—YP_026915 | |
| capE; CapE involved in Poly-gamma-glutamate synthesis—YP_016572 | |
| dep/capD; gamma-glutamyltranspeptidase required for polyglutamate anchoring to peptidoglycan—AAF13660 | |
| capA; CapA required for Poly-gamma-glutamate transport—AAF13661 | |
| capC; CapC involved in Poly-gamma-glutamate synthesis—AAF13662 | |
| capB; CapB involved in Poly-gamma-glutamate synthesis—AAF13663 | |
| cya; calmodulin sensitive adenylate cyclase edema factor—AAD32426 | |
| pagA; anthrax toxin moiety protective antigen—AAD32414 | |
| lef; anthrax toxin lethal factor precursor—AAD32411 | |
| Resistance genes | fosB; FosB/FosD family fosfomycin resistance bacillithiol transferase—A7J11_05167 |
| bla; class A beta-lactamase Bla1—A7J11_05168 | |
| lsa(B); ABC-F type ribosomal protection protein Lsa(B)—A7J11_01190 | |
| fosB2; fosfomycin resistance bacillithiol transferase FosB2—A7J11_00634 | |
| bla2; BcII family subclass B1 metallo-beta-lactamase—A7J11_00039 | |
| mph(B); Mph(B) family macrolide 2’-phosphotransferase—A7J11_05208 | |
| vanZ-F; glycopeptide resistance protein VanZ-F—A7J11_00543 | |
| vanR; VanM-type vancomycin resistance DNA-binding response regulator VanR—A7J11_02292 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiaverini, A.; Abdel-Glil, M.Y.; Linde, J.; Galante, D.; Rondinone, V.; Fasanella, A.; Cammà, C.; D’Alterio, N.; Garofolo, G.; Tomaso, H. Whole Genome Sequencing for Studying Bacillus anthracis from an Outbreak in the Abruzzo Region of Italy. Microorganisms 2020, 8, 87. https://doi.org/10.3390/microorganisms8010087
Chiaverini A, Abdel-Glil MY, Linde J, Galante D, Rondinone V, Fasanella A, Cammà C, D’Alterio N, Garofolo G, Tomaso H. Whole Genome Sequencing for Studying Bacillus anthracis from an Outbreak in the Abruzzo Region of Italy. Microorganisms. 2020; 8(1):87. https://doi.org/10.3390/microorganisms8010087
Chicago/Turabian StyleChiaverini, Alexandra, Mostafa Y. Abdel-Glil, Jörg Linde, Domenico Galante, Valeria Rondinone, Antonio Fasanella, Cesare Cammà, Nicola D’Alterio, Giuliano Garofolo, and Herbert Tomaso. 2020. "Whole Genome Sequencing for Studying Bacillus anthracis from an Outbreak in the Abruzzo Region of Italy" Microorganisms 8, no. 1: 87. https://doi.org/10.3390/microorganisms8010087
APA StyleChiaverini, A., Abdel-Glil, M. Y., Linde, J., Galante, D., Rondinone, V., Fasanella, A., Cammà, C., D’Alterio, N., Garofolo, G., & Tomaso, H. (2020). Whole Genome Sequencing for Studying Bacillus anthracis from an Outbreak in the Abruzzo Region of Italy. Microorganisms, 8(1), 87. https://doi.org/10.3390/microorganisms8010087







