Experimental Challenges for Reduced Genomes: The Cell Model Escherichia coli
Abstract
1. Introduction
2. Genome Reduction in Synthetic Microbiology
2.1. E. coli as a Representative Cell Model
2.2. Genome Reduction Provides Ideal Cellular Conditions for Synthetic Design
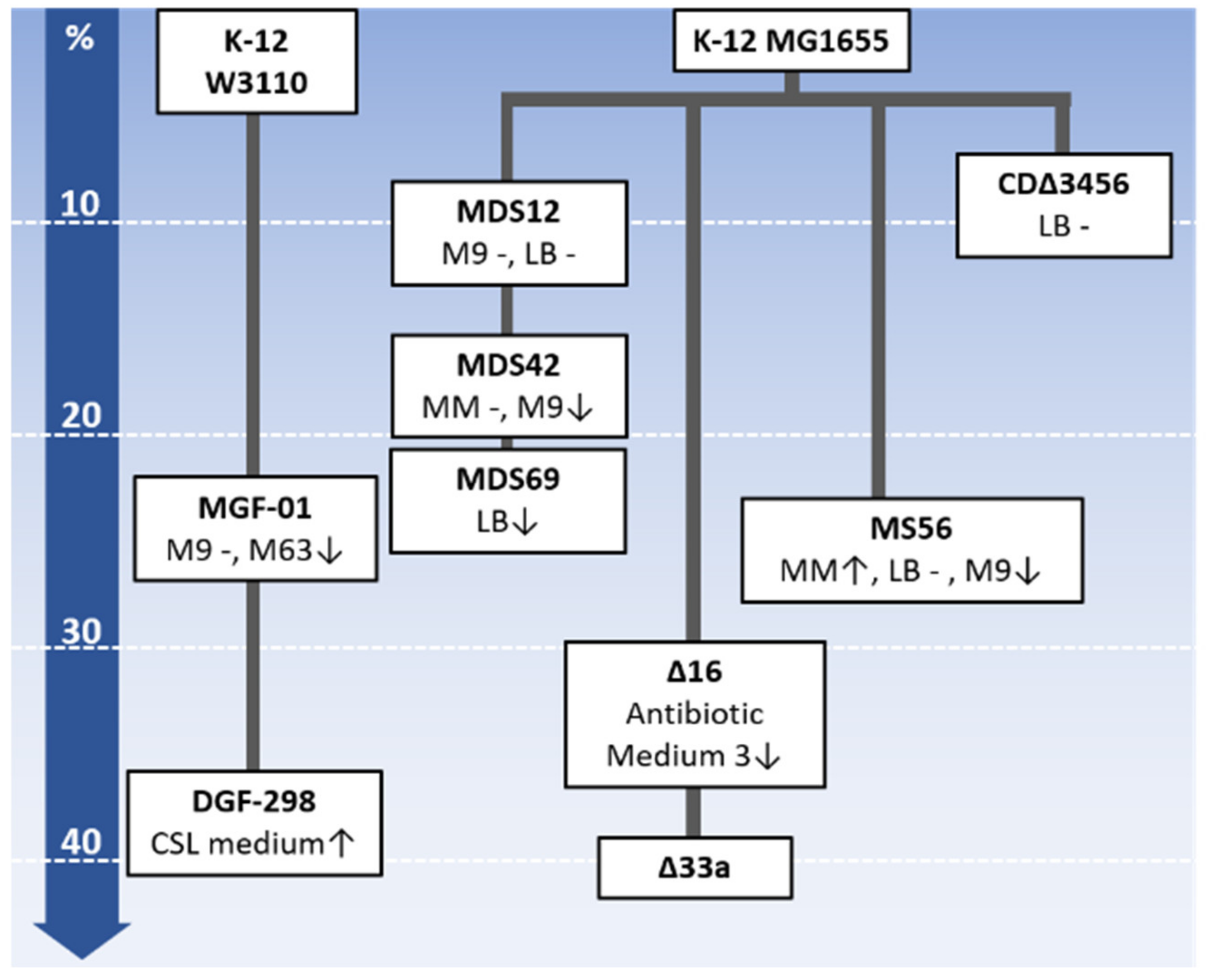
3. Collections of Reduced Genomes
3.1. Minimum Genome Factory (MGF)
3.1.1. Design Principles
3.1.2. Growth
3.2. Multiple-Deletion Series (MDS)
3.2.1. Design Principles
3.2.2. Growth
3.3. Δ16
3.3.1. Design Principles
3.3.2. Growth
3.4. MS56
3.4.1. Design Principles
3.4.2. Growth
3.5. CDΔ3456
3.5.1. Design Principles
3.5.2. Growth
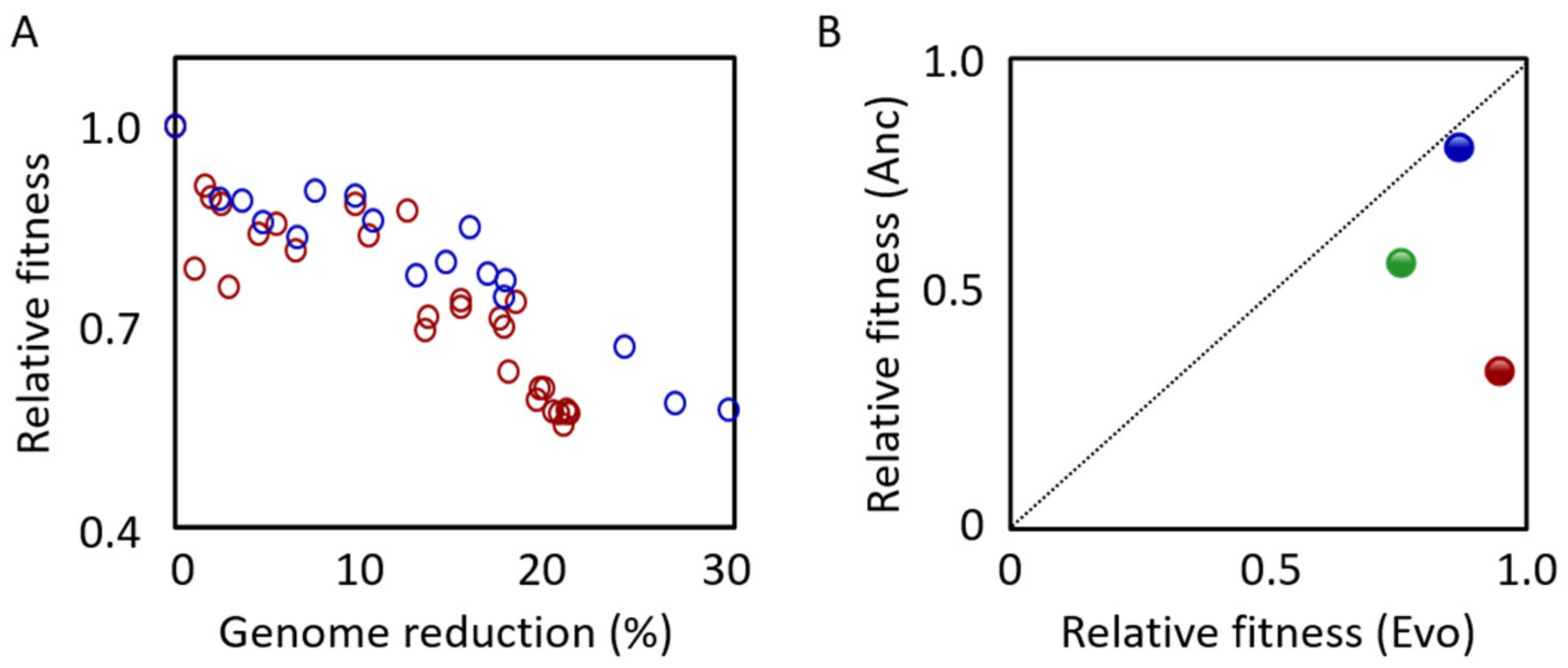
4. Aims and Additional Characteristics of Genome-Reduced Strains
4.1. Genome Stability
4.2. Genome Minimization
5. Proposals for Further Challenges
5.1. Concerns for the Environmental Factors
5.2. Approaches for Experimental Evolution
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ostrov, N.; Landon, M.; Guell, M.; Kuznetsov, G.; Teramoto, J.; Cervantes, N.; Zhou, M.; Singh, K.; Napolitano, M.G.; Moosburner, M.; et al. Design, synthesis, and testing toward a 57-codon genome. Science 2016, 353, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Yuan, S.; Xiong, B.; Sun, H.; Ye, L.; Li, J.; Zhang, X.; Bi, C. Development of a fast and easy method for Escherichia coli genome editing with CRISPR/Cas9. Microb. Cell Fact. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Chuang, R.Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351. [Google Scholar] [CrossRef] [PubMed]
- Blattner, F.R.; Iii, G.P.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- King, Z.A.; Lloyd, C.J.; Feist, A.M.; Palsson, B.O. Next-generation genome-scale models for metabolic engineering. Curr. Opin. Biotechnol. 2015, 35, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Choe, D.; Cho, S.; Kim, S.C.; Cho, B.K. Minimal genome: Worthwhile or worthless efforts toward being smaller? Biotechnol. J. 2016, 11, 199–211. [Google Scholar] [CrossRef]
- Kurokawa, M.; Seno, S.; Matsuda, H. Correlation between genome reduction and bacterial growth. DNA Res. 2016, 23, 517–525. [Google Scholar] [CrossRef]
- Hashimoto, M.; Ichimura, T.; Mizoguchi, H.; Tanaka, K.; Fujimitsu, K.; Keyamura, K.; Ote, T.; Yamakawa, T.; Yamazaki, Y.; Mori, H.; et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005, 55, 137–149. [Google Scholar] [CrossRef]
- Marr, A.G. Growth rate of Escherichia coli. Microbiol. Rev. 1991, 55, 316–333. [Google Scholar]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Linn, B.Y.S.; Arber, W. Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form. Proc. Natl. Acad. Sci. USA 1968, 59, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Dower, W.J.; Miller, J.F.; Ragsdale, C.W.; Group, M.B.; Laboratories, B.; Medical, S. High efficiency transformation of E.coli by high voltage electroporation. Nucleic Acids Res. 1988, 16, 6127–6146. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Harbor, C.S. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Morse, M.L.; Esther, M.L.; Joshua, L. Transduction in Escherichia Coli K-12. Genetics 1956, 41, 142–156. [Google Scholar] [PubMed]
- Cohen, S.N.; Chang, A.C.Y.; Boyer, H.W.; Helling, R.B. Construction of Biologically Functional Bacterial Plasmids in Vitro. Proc. Natl. Acad. Sci. USA 1973, 70, 3240–3244. [Google Scholar] [CrossRef]
- Itakura, K.; Hirose, T.; Crea, R.; Riggs, A.D.; Heyneker, H.L.; Bolivar, F.; Boyer, H.W. Expression in Escherichia coli of a Chemically Synthesized Gene for the Hormone Somatostatin. Science 1977, 198, 1056–1063. [Google Scholar] [CrossRef]
- Collins, D. Expression of Naphthalene Oxidation Genes in Escherichia coli Results in the Biosynthesis of Indigo. Science 1983, 222, 167–170. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 1–17. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef]
- Rosano, G.L.; Morales, E.S.; Ceccarelli, E.A. New tools for recombinant protein production in Escherichia coli: A 5-year update. Protein Sci. 2019, 28, 1412–1422. [Google Scholar] [CrossRef]
- Hayashi, K.; Morooka, N.; Yamamoto, Y.; Fujita, K.; Isono, K.; Choi, S.; Ohtsubo, E.; Baba, T.; Wanner, B.L.; Mori, H.; et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.; Abe, T.; Arnaud, M.B.; Berlyn, M.K.B.; Blattner, F.R.; Chaudhuri, R.R.; Glasner, J.D.; Horiuchi, T.; Keseler, I.M.; Kosuge, T.; et al. Escherichia coli K-12: A cooperatively developed annotation snapshot-2005. Nucleic Acids Res. 2006, 34, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kothari, A.; Krummenacker, M.; Latendresse, M.; Peter, E. The EcoCyc Database. EcoSal. Plus 2019, 8, 1–34. [Google Scholar]
- De Smet, R.; Marchal, K. Advantages and limitations of current network inference methods. Nat. Rev. Microbiol. 2010, 8, 717–729. [Google Scholar] [CrossRef]
- Shen-Orr, S.S.; Milo, R.; Mangan, S.; Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002, 31, 64–68. [Google Scholar] [CrossRef]
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014, 12, 381–390. [Google Scholar] [CrossRef]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef]
- Jullesson, D.; David, F.; Pfleger, B.; Nielsen, J. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol. Adv. 2015, 33, 1395–1402. [Google Scholar] [CrossRef]
- Cardinale, S.; Arkin, A.P. Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol. J. 2012, 7, 856–866. [Google Scholar] [CrossRef]
- Purnick, P.E.M.; Weiss, R. The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 2009, 10, 410–422. [Google Scholar] [CrossRef]
- Pál, C.; Papp, B.; Pósfai, G. The dawn of evolutionary genome engineering. Nat. Rev. Genet. 2014, 15, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Reuß, D.R.; Altenbuchner, J.; Mäder, U.; Rath, H.; Ischebeck, T.; Sappa, P.K.; Thürmer, A.; Guérin, C.; Nicolas, P.; Steil, L.; et al. Large-scale reduction of the Bacillus subtilis genome: Consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 2016, 27, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Simon, U.; M, B.; Andreas, R.; Marius, H.; Daniel, S.; Natalie, B.; Anna, B.; Michael, B.; Wolfgang, W.; Kay, M.; et al. Chassis organism from Corynebacterium glutamicum–a top-down approach to identify and delete irrelevant gene clusters. Biotechnol. J. 2015, 10, 290–301. [Google Scholar]
- Lieder, S.; Nikel, P.I.; Lorenzo, V.D.; Takors, R. Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microb. Cell Fact. 2015, 14, 1–14. [Google Scholar] [CrossRef]
- Juhas, M.; Reuß, D.R.; Zhu, B.; Commichau, F.M. Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiol 2014, 160, 2341–2351. [Google Scholar] [CrossRef]
- Zhu, B.; Stülke, J. SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 2018, 46, D743–D748. [Google Scholar] [CrossRef]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science 2010, 329, 52–57. [Google Scholar] [CrossRef]
- Fredens, J.; Wang, K.; Torre, D.D.; Funke, L.F.H.; Robertson, W.E.; Christova, Y.; Chia, T.; Schmied, W.H.; Dunkelmann, D.L.; Beránek, V.; et al. Total synthesis of Escherichia coli with a recoded genome. Nature 2019, 569, 514–518. [Google Scholar] [CrossRef]
- Juhas, M. On the road to synthetic life: The minimal cell and genome-scale engineering. Crit. Rev. Biotechnol. 2015, 36, 416–423. [Google Scholar] [CrossRef]
- Chiang, S.M.; Schellhorn, H.E. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 2012, 525, 161–169. [Google Scholar] [CrossRef]
- Weber, H.; Polen, T.; Heuveling, J.; Wendisch, V.F.; Hengge, R.; Ju, F.; Al, W.E.T. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005, 187, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; File, J. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006, 9, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Babakhani, S. Transposons: The agents of antibiotic resistance in bacteria. J. Basic Microbiol. 2018, 58, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Harms, H.; Bley, T.; Mu, S. Origin and analysis of microbial population heterogeneity in bioprocesses. Curr. Opin. Biotechnol. 2010, 21, 100–113. [Google Scholar]
- Rugbjerg, P.; Myling-petersen, N.; Porse, A.; Sarup-lytzen, K.; Sommer, M.O.A. Diverse genetic error modes constrain large-scale bio-based production. Nat. Commun. 2018, 9, 787. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Tripp, H.J.; Givan, S.; Podar, M.; Vergin, K.L.; Baptista, D.; Bibbs, L.; Eads, J.; Richardson, T.H.; Noordewier, M.; et al. Genome Streamlining in a Cosmopolitan Oceanic Bacterium. Science 2005, 309, 1242–1246. [Google Scholar] [CrossRef]
- Martínez-Cano, D.J.; Reyes-Prieto, M.; Martínez-Romero, E.; Partida-Martínez, L.P.; Latorre, A.; Moya, A.; Delaye, L. Evolution of small prokaryotic genomes. Front. Microbiol. 2015, 6, 1–23. [Google Scholar] [CrossRef]
- Kafri, M.; Metzl-raz, E.; Jona, G.; Barkai, N. Article the Cost of Protein Production. Cell Reports 2016, 14, 22–31. [Google Scholar] [CrossRef]
- Mizoguchi, H.; Mori, H.; Fujio, T. Escherichia coli minimum genome factory. Biotechnol. Appl. Biochem. 2007, 46, 157–167. [Google Scholar]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef]
- Hirokawa, Y.; Kawano, H.; Tanaka-Masuda, K.; Nakamura, N.; Nakagawa, A.; Ito, M.; Mori, H.; Oshima, T.; Ogasawara, N. Genetic manipulations restored the growth fitness of reduced-genome Escherichia coli. J. Biosci. Bioeng. 2013, 116, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; Volume 1, ISBN 9781936113415. [Google Scholar]
- Mizoguchi, H.; Sawano, Y.; Kato, J.; Mori, H. Superpositioning of deletions promotes growth of Escherichia coli with a reduced genome. DNA Res. 2008, 15, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, A.; Sakurai, T.; Tsuru, S.; Ying, B.; Mori, K.; Yomo, T. Construction of Escherichia coli gene expression level perturbation collection. Metab. Eng. 2009, 11, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Choe, D.; Lee, J.H.; Yoo, M.; Hwang, S.; Sung, B.H.; Cho, S.; Palsson, B.; Kim, S.C.; Cho, B. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, I.; Kurokawa, M.; Liu, L.; Ying, B.W. Coordinated Changes in Mutation and Growth Rates Induced by Genome Reduction. MBio 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Yuan, X.; Couto, J.M.; Glidle, A.; Song, Y.; Sloan, W.; Yin, H. Single-Cell Microfluidics to Study the Effects of Genome Deletion on Bacterial Growth Behavior. ACS Synth. Biol. 2017, 6, 2219–2227. [Google Scholar] [CrossRef]
- Kolisnychenko, V.; Iii, G.P.; Herring, C.D.; Fehér, T.; Pósfai, J.; Blattner, F.R.; Pósfai, G. Engineering a Reduced Escherichia coli Genome. Genome Res. 2002, 12, 640–647. [Google Scholar] [CrossRef]
- Pósfai, G.; Umenhoffer, K.; Kolisnychenko, V.; Stahl, B.; Sharma, S.S. Emergent Properties of Reduced-Genome Escherichia coli. Science 2006, 312, 1044–1047. [Google Scholar] [CrossRef]
- Karcagi, I.; Draskovits, G.; Umenhoffer, K.; Fekete, G.; Kovács, K.; Méhi, O.; Balikó, G.; Szappanos, B.; Györfy, Z.; Fehér, T.; et al. Indispensability of Horizontally Transferred Genes and Its Impact on Bacterial Genome Streamlining. Mol. Biol. Evol. 2016, 33, 1257–1269. [Google Scholar] [CrossRef]
- Neidhardt, F.C.; Bloch, P.L.; Smith, D.F. Culture Medium for Enterobacteria. J. Bacteriol. 1974, 119, 736–747. [Google Scholar]
- Lee, J.H.; Sung, B.H.; Kim, M.S.; Blattner, F.R.; Yoon, B.H.; Kim, J.H.; Kim, S.C. Metabolic engineering of a reduced-genome strain of Escherichia coli for L-threonine production. Microb. Cell Fact. 2009, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Ying, B.W.; Seno, S.; Kaneko, F.; Matsuda, H.; Yomo, T. Multilevel comparative analysis of the contributions of genome reduction and heat shock to the Escherichia coli transcriptome. BMC Genom. 2013, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Ying, B.W.; Yama, K. Gene Expression Order Attributed to Genome Reduction and the Steady Cellular State in Escherichia coli. Front. Microbiol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Lee, S.H.; Yang, K.S. Enhancing recombinant protein production with an Escherichia coli host strain lacking insertion sequences. Appl. Microbiol. Biotechnol. 2014, 98, 6701–6713. [Google Scholar] [CrossRef]
- Yu, B.J.; Sung, B.H.; Koob, M.D.; Lee, C.H.; Lee, J.H.; Lee, W.S.; Kim, M.S.; Kim, S.C. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 2002, 20, 1018–1023. [Google Scholar] [CrossRef]
- Hall, B.G. Activation of the bgl Operon by Adaptive Mutation. Mol. Biol. Evol. 1988, 15, 1–5. [Google Scholar] [CrossRef]
- Akeno, Y.; Ying, B.-W.; Tsuru, S.; Yomo, T. A reduced genome decreases the host carrying capacity for foreign DNA. Microb. Cell Fact. 2014, 13, 49. [Google Scholar] [CrossRef]
- Couto, J.M.; Mcgarrity, A.; Russell, J.; Sloan, W.T. The effect of metabolic stress on genome stability of a synthetic biology chassis Escherichia coli K12 strain. Microb. Cell Fact. 2018, 1–10. [Google Scholar] [CrossRef]
- Me, O.; Fekete, G. Competition between Transposable Elements and Mutator Genes in Bacteria. Mol. Biol. Evol. 2012, 29, 3153–3159. [Google Scholar]
- Mushegian, A.R.; Koonin, E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 1996, 93, 10268–10273. [Google Scholar] [CrossRef]
- Martínez-Carranza, E.; Barajas, H.; Alcaraz, L.D.; Servín-González, L.; Ponce-Soto, G.Y.; Soberón-Chávez, G. Variability of bacterial essential genes among closely related bacteria: The case of Escherichia coli. Front. Microbiol. 2018, 9, 1–7. [Google Scholar]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Baba, T.; Yokoyama, K.; Takeuchi, R.; Nomura, W.; Makishi, K.; Otsuka, Y.; Dose, H.; Wanner, B.L. Identification of essential genes and synthetic lethal gene combinations in Escherichia coli K-12. Methods Mol. Biol. 2015, 1279, 45–65. [Google Scholar] [PubMed]
- Goodall, E.C.A.; Robinson, A.; Johnston, I.G.; Jabbari, S.; Turner, K.A.; Cunningham, A.F.; Lund, P.A.; Cole, J.A.; Henderson, I.R.; Kline, K.A. The Essential Genome of Escherichia coli K-12. MBio 2018, 9, e02096-17. [Google Scholar] [CrossRef]
- Henry, C.S.; Overbeek, R.; Stevens, R.L. Building the blueprint of life. Biotechnol. J. 2010, 5, 695–704. [Google Scholar] [CrossRef]
- Iwadate, Y.; Honda, H.; Sato, H.; Hashimoto, M.; Kato, J.I. Oxidative stress sensitivity of engineered Escherichia coli cells with a reduced genome. FEMS Microbiol. Lett. 2011, 322, 25–33. [Google Scholar] [CrossRef]
- Wang, L.; Maranas, C.D. MinGenome: An in Silico Top-Down Approach for the Synthesis of Minimized Genomes. ACS Synth. Biol. 2018, 7, 462–473. [Google Scholar] [CrossRef]
- Peebo, K.; Neubauer, P. Application of Continuous Culture Methods to Recombinant Protein Production in Microorganisms. Microorganisms 2018, 6, 56. [Google Scholar] [CrossRef]
- Moran, N.A.; Mclaughlin, H.J.; Sorek, R. The Dynamics and Time Scale of Ongoing Genomic Erosion in Symbiotic Bacteria. Science 2009, 323, 379–382. [Google Scholar] [CrossRef]
- Nutman, A.P.; Bennett, V.C.; Friend, C.R.L.; Van Kranendonk, M.J.; Chivas, A.R. Rapid emergence of life shown by discovery of 3,700-million-year-old microbial structures. Nature 2016, 537, 535–538. [Google Scholar] [CrossRef]
- Good, B.H.; McDonald, M.J.; Barrick, J.E.; Lenski, R.E.; Desai, M.M. The dynamics of molecular evolution over 60,000 generations. Nature 2017, 551, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Jahn, L.J.; Munck, C.; Ellabaan, M.M.H.; Sommer, M.O.A. Adaptive Laboratory Evolution of Antibiotic Resistance Using Different Selection Regimes Lead to Similar Phenotypes and Genotypes. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, T.; Suzuki, S.; Hirasawa, T.; Ono, N.; Yomo, T.; Shimizu, H.; Furusawa, C. Phenotypic convergence in bacterial adaptive evolution to ethanol stress. BMC Evol. Biol. 2015, 15, 180. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, D.E.; Kepner, J.L.; Bennett, A.F.; Lenski, R.E.; Barrick, J.E. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. USA 2017, 114, E1904–E1912. [Google Scholar] [CrossRef]
- Kishimoto, T.; Iijima, L.; Tatsumi, M.; Ono, N.; Oyake, A.; Hashimoto, T.; Matsuo, M.; Okubo, M.; Suzuki, S.; Mori, K.; et al. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 2010, 6, e1001164. [Google Scholar] [CrossRef]
- Tenaillon, O.; Rodríguez-Verdugo, A.; Gaut, R.L.; McDonald, P.; Bennett, A.F.; Long, A.D.; Gaut, B.S. The molecular diversity of adaptive convergence. Science 2012, 335, 457–461. [Google Scholar] [CrossRef]
- Hamdallah, I.; Torok, N.; Bischof, K.M.; Majdalani, N.; Chadalavada, S.; Mdluli, N. crossm Experimental Evolution of Escherichia coli K-12 at High pH and with RpoS Induction. Appl. Environ. Microbiol. 2018, 84, 1–17. [Google Scholar] [CrossRef]
- Palsson, B.O.; Feist, M. Use of Adaptive Laboratory Evolution To Discover Key Mutations Enabling Rapid Growth of Escherichia coli K-12 MG1655 on Glucose Minimal Medium. Appl. Environ. Microbiol. 2015, 81, 17–30. [Google Scholar]
- Conrad, T.M.; Lewis, N.E.; Palsson, B.O. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2014, 7, 509. [Google Scholar] [CrossRef]
- Toprak, E.; Veres, A.; Michel, J.; Chait, R.; Hartl, D.L.; Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2011, 44, 101–105. [Google Scholar] [CrossRef] [PubMed]
- McCloskey, D.; Xu, S.; Sandberg, T.E.; Brunk, E.; Hefner, Y.; Szubin, R.; Feist, A.M.; Palsson, B.O. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun. 2018, 9, 3796. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.; Zhang, J. Evolutionary adaptations to new environments generally reverse plastic phenotypic changes. Nat. Commun. 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurokawa, M.; Ying, B.-W. Experimental Challenges for Reduced Genomes: The Cell Model Escherichia coli. Microorganisms 2020, 8, 3. https://doi.org/10.3390/microorganisms8010003
Kurokawa M, Ying B-W. Experimental Challenges for Reduced Genomes: The Cell Model Escherichia coli. Microorganisms. 2020; 8(1):3. https://doi.org/10.3390/microorganisms8010003
Chicago/Turabian StyleKurokawa, Masaomi, and Bei-Wen Ying. 2020. "Experimental Challenges for Reduced Genomes: The Cell Model Escherichia coli" Microorganisms 8, no. 1: 3. https://doi.org/10.3390/microorganisms8010003
APA StyleKurokawa, M., & Ying, B.-W. (2020). Experimental Challenges for Reduced Genomes: The Cell Model Escherichia coli. Microorganisms, 8(1), 3. https://doi.org/10.3390/microorganisms8010003