The Structure and Species Co-Occurrence Networks of Soil Denitrifying Bacterial Communities Differ Between A Coniferous and A Broadleaved Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Soil Sampling and Analyses
2.3. Soil DNA Extraction and Quantification of nirK, nirS, and nosZ Genes
2.4. Functional Gene Amplicon Sequencing and Processing
2.5. Network Analysis
2.6. Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties
3.2. Abundance, Diversity, and Composition of Denitrifying Bacterial Communities
3.3. Relating the Denitrifying Bacterial Communities to Soil Properties
3.4. Denitrifying Bacteria Community Networks and the Keystone Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malmer, A.; Grip, H. Converting tropical rainforest to forest plantation in sabah, malaysia. PartII.Effects on nutrient dynamics and net losses in streamwater. Hydrol. Process. 2010, 8, 195–209. [Google Scholar] [CrossRef]
- Gatti, R.C.; Castaldi, S.; Lindsell, J.A.; Coomes, D.A.; Marchetti, M.; Maesano, M.; Paola, A.D.; Paparella, F.; Valentini, R. The impact of selective logging and clearcutting on forest structure, tree diversity and above-ground biomass of African tropical forests. Ecol. Res. 2015, 30, 119–132. [Google Scholar] [CrossRef]
- Li, Z.A.; Peng, S.L.; Rae, D.J.; Zhou, G.Y. Litter decomposition and nitrogen mineralization of soils in subtropical plantation forests of southern china, with special attention to comparisons between legumes and non-legumes. Plant. Soil 2001, 229, 105–116. [Google Scholar] [CrossRef]
- Burton, J.; Chen, C.; Xu, Z.; Ghadiri, H. Gross nitrogen transformations in adjacent native and plantation forests of subtropical Australia. Soil Biol. Biochem. 2007, 39, 426–433. [Google Scholar] [CrossRef]
- Henri, C.J. Soil-site productivity of Gmelina arborea, Eucalyptus urophylla, and Eucalyptus grandis, forest plantations in western Venezuela. For. Ecol. Manag. 2001, 144, 255–264. [Google Scholar] [CrossRef]
- Ma, Y.; Geng, Y.; Huang, Y.; Shi, Y.; Niklaus, P.A.; Schmid, B.; He, J.S. Effect of clear-cutting silviculture on soil respiration in a subtropical forest of China. J. Plant. Ecol. 2013, 6, 335–348. [Google Scholar] [CrossRef]
- Wan, X.; Huang, Z.; He, Z.; Yu, Z.; Wang, M.; Davis, M.R.; Yang, Y. Soil C:N ratio is the major determinant of soil microbial community structure in subtropical coniferous and broadleaf forest plantations. Plant. Soil 2015, 387, 103–116. [Google Scholar] [CrossRef]
- Burton, J.; Chen, C.; Xu, Z.; Ghadiri, H. Soil microbial biomass, activity and community composition in adjacent native and plantation forests of subtropical Australia. J. Soil Sediment. 2010, 10, 1267–1277. [Google Scholar] [CrossRef]
- Song, P.; Ren, H.; Jia, Q.; Guo, J.; Zhang, N.; Ma, K. Effects of historical logging on soil microbial communities in a subtropical forest in southern china. Plant. Soil 2015, 397, 1–12. [Google Scholar] [CrossRef]
- Liu, J.; Dang, P.; Gao, Y.; Zhu, H.; Zhu, H.; Zhao, F.; Zhao, Z. Effects of tree species and soil properties on the composition and diversity of the soil bacterial community following afforestation. For. Ecol. Manag. 2018, 427, 342–349. [Google Scholar] [CrossRef]
- Cao, Y.S.; Fu, S.L.; Zou, X.M.; Cao, H.L.; Shao, Y.H.; Zhou, L.X. Soil microbial community composition under eucalyptus plantations of different age in subtropical china. Eur. J. Soil Biol. 2010, 46, 128–135. [Google Scholar] [CrossRef]
- Wang, C.; Lu, H.; Dong, D.; Deng, H.; Strong, P.J.; Wang, H.; Wu, W.X. Insight into the effects of biochar on manure composting: Evidence supporting the relationship between N2O emission and denitrifying community. Env. Sci. Technol. 2013, 47, 7341–7349. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Kölbl, A.; Ebli, M.; Buegger, F.; Schloter, M.; Fiedler, S. Field-scale pattern of denitrifying microorganisms and N2O emission rates indiate a high potential for complete denitrification in an agriculturally used organic soil. Microb. Ecol. 2017, 74, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Keil, D.; Niklaus, P.A.; Von Riedmatten, L.R.; Boeddinghaus, R.S.; Dormann, C.F.; Scherer-Lorenzen, M.; Kandeler, E.; Marhan, S. Effects of warming and drought on potential N2O emissions and denitrifying bacteria abundance in grasslands with different land-use. Fems Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Domeignoz-horta, L.A.; Philippot, L.; Peyrard, C.; Bru, D.; Breuil, M.C.; Bizouard, F.; Justes, E.; Mary, B.; Léonard, J.; Spor, A. Peaks of in situ N2O emissions are influenced by N2O producing and reducing microbial communities across arable soils. Glob. Chang. Biol. 2017, 24. [Google Scholar] [CrossRef]
- Pérez, T.; Trumbore, S.E.; Tyler, S.C.; Davidson, E.A.; Keller, M.; Camargo, P.B.D. Isotopic variability of N2O emissions from tropical forest soils. Glob. Biogeochem. Cycles 2000, 14, 525–535. [Google Scholar] [CrossRef]
- Liu, S.; Reiners, W.A.; Keller, M.; Schimel, D.S. Model simulation of changes in n2o and no emissions with conversion of tropical rain forests to pastures in the Costa Rican Atlantic zone. Glob. Biogeochem. Cycles 1999, 13, 663–677. [Google Scholar] [CrossRef]
- Verchot, L.V.; Hutabarat, L.; Hairiah, K.; Noordwijk, M.V. Nitrogen availability and soil N2O emissions following conversion of forests to coffee in southern Sumatra. Glob. Biogeochem. Cycles 2006, 20. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, Y.D.; Wu, S.Y. Methane and nitrous oxide fluxes in temperate secondary forest and larch plantation in northeastern china. Acta Ecol. Sin. 2013, 33, 5320–5328. [Google Scholar]
- van Lent, J.; Hergoualc’h, K.; Verchot, L.V. Soil N2O and no emissions from land use and land-use change in the tropics and subtropics: A meta-analysis. Biogeosciences 2015, 12, 12783–12821. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Prescott, C.E.; Grayston, S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 2014, 75, 11–25. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosz gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72. [Google Scholar] [CrossRef]
- Demanèche, S.; Philippot, L.; David, M.M.; Navarro, E.; Vogel, T.M.; Simonet, P. Characterization of denitrification gene clusters of soil bacteria via a metagenomic approach. Appl. Environ. Microbiol. 2009, 75. [Google Scholar] [CrossRef]
- Bartossek, R.; Nicol, G.W.; Lanzen, A.; Klenk, H.P.; Schleper, C. Homologues of nitrite reductases in ammonia-oxidizing archaea: Diversity and genomic context. Environ. Microbiol. 2010, 12, 1075–1088. [Google Scholar] [CrossRef]
- Bárta, J.; Melichová, T.; VaněK, D.; Picek, T.; Šantrůčková, H. Effect of pH and dissolved organic matter on the abundance of nirK and nirS denitrifiers in spruce forest soil. Biogeochemistry 2010, 101, 123–132. [Google Scholar] [CrossRef]
- Morales, S.E.; Cosart, T.; Holben, W.E. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 2010, 4, 799–808. [Google Scholar] [CrossRef]
- Cabello, P.; Roldán, M.D.; Moreno-Vivián, C. Nitrate reduction and the nitrogen cycle in archaea. Microbiology 2004, 150, 3527–3546. [Google Scholar] [CrossRef]
- Szukics, U.; Abell, G.; Hödl, V.; Mitter, B.; Sessitsch, A.; Hackl, E.; Zechmeister-Boltenstern, S. Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol. Ecol. 2010, 72, 395–406. [Google Scholar] [CrossRef]
- Bergaust, L.; Mao, Y.; Bakken, L.R.; Frostegård, A. Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous [corrected] oxide reductase in paracoccus denitrificans. Appl. Environ. Microbiol. 2010, 76, 6387–6396. [Google Scholar] [CrossRef]
- Kandeler, E.; Deighlmayr, K.; Tscherko, D.; Bru, D.; Philippot, L. Abundance of narG, nirS, nirK, and nosZ genes ofdenitrifying bacteria during primary successions of aglacier foreland. Appl. Environ. Microbiol. 2006, 72, 5957–5959. [Google Scholar] [CrossRef]
- Rasche, F.; Knapp, D.; Kaiser, C.; Koranda, M.; Kitzler, B.; Zechmeister-boltenstern, S.; Richte, A.; Sessitsch, A. Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 2011, 5, 389–402. [Google Scholar] [CrossRef]
- Graf, D.R.; Jones, C.M.; Hallin, S. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Rösch, C.; Mergel, A.; Bothe, H. Biodiversity of denitrifying and dinitrogen fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 2002, 68, 3818–3829. [Google Scholar] [CrossRef]
- Petersen, D.G.; Blazewicz, S.J.; Firestone, M.; Herman, D.J.; Turetsky, M.; Waldrop, M. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Env. Microbiol. 2012, 14, 993–1008. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Yang, Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. Mbio 2011, 2, e00122-11. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Feng, L.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13. Available online: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-13-113 (accessed on 23 August 2019). [CrossRef]
- Lu, L.; Yin, S.; Liu, X.; Zhang, W.; Gu, T.; Shen, Q.; Qiu, H. Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, B.; Li, H.; Liu, M.; Chen, L.; Zhou, S. Aggregate-related changes in network patterns of nematodes and ammonia oxidizers in an acidic soil. Soil Biol. Biochem. 2015, 88, 101–109. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.Z.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.M.; Brookes, P.C.; Xu, J.M.; Gilbert, J.A. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef]
- Tian, J.; He, N.; Kong, W.; Deng, Y.; Feng, K.; Green, S.M.; Wang, X.B.; Zhou, J.Z.; Kuzyakov, Y.; Yu, G.R. Deforestation decreases spatial turnover and alters the network interactions in soil bacterial communities. Soil Biol. Biochem. 2018, 123, 80–86. [Google Scholar] [CrossRef]
- He, D.; Shen, W.; Eberwein, J.; Zhao, Q.; Ren, L.; Wu, Q.L. Diversity and co-occurrence network of soil fungi are more responsive than those of bacteria to shifts in precipitation seasonality in a subtropical forest. Soil Biol. Biochem. 2017, 115, 499–510. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; Van, M.D.H. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Liu, G.S.; Jiang, N.H.; Zhang, L.D.; Liu, Z.L. Soil Physical and Chemical Analysis and Description of Soil Profiles; China Standards Press: Beijing, China, 1996; Volume 7-5066-1218, pp. 1–265. [Google Scholar]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F. QIIME allows integration and analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef]
- ter Braak, C.J.F.; Smilauer, P. CANOCO Reference Manual and CanoDraw for Windows User’s Guide: Software for Canonical Community Ordination (version 4.5); Microcomputer Power: Ithaca, NY, USA, 2012; Available online: http://www.canodraw.com/ (accessed on 23 August 2019).
- Wrage, N.; Velthof, G.L.; Beusichem, M.L.V.; Oenema, O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 2001, 33, 1723–1732. [Google Scholar] [CrossRef]
- Meng, H.; Wu, R.; Wang, Y.F.; Gu, J.D. A comparison of denitrifying bacterial community structures and abundance in acidic soils between natural forest and re-vegetated forest of nanling nature reserve in southern China. J. Environ. Manag. 2017, 198, 41–49. [Google Scholar] [CrossRef]
- Koike, I.; Hattori, A. Energy yield of denitrification: An estimate from growth yield in continuous cultures of pseudomonas denitrificans under nitrate-, nitrite- and oxide-limited conditions. J. Gen. Appl. Microbiol. 1975, 88, 11–19. [Google Scholar] [CrossRef]
- Macarthur, R. Fluctuations of animal populations and a measure of community stability. Ecology 1955, 36, 533–536. [Google Scholar] [CrossRef]
- Hatakka, A. Lignin-modifying enzymes from selected white-rot fungi: Production and role from in lignin degradation. FEMS Microbiol. Rev. 2010, 13, 125–135. [Google Scholar] [CrossRef]
- Miltner, A.; Bombach, P.; Schmidt-Brücken, B.; Kästner, M. SOM genesis: Microbial biomass as a significant source. Biogeochemistry 2012, 111, 41–55. [Google Scholar] [CrossRef]
- Bru, D.; Ramette, A.; Saby, N.P.; Dequiedt, S.; Ranjard, L.; Jolivet, C.A.; Arrouays, D.; Philippot, L. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2011, 5, 532–542. [Google Scholar] [CrossRef] [PubMed]


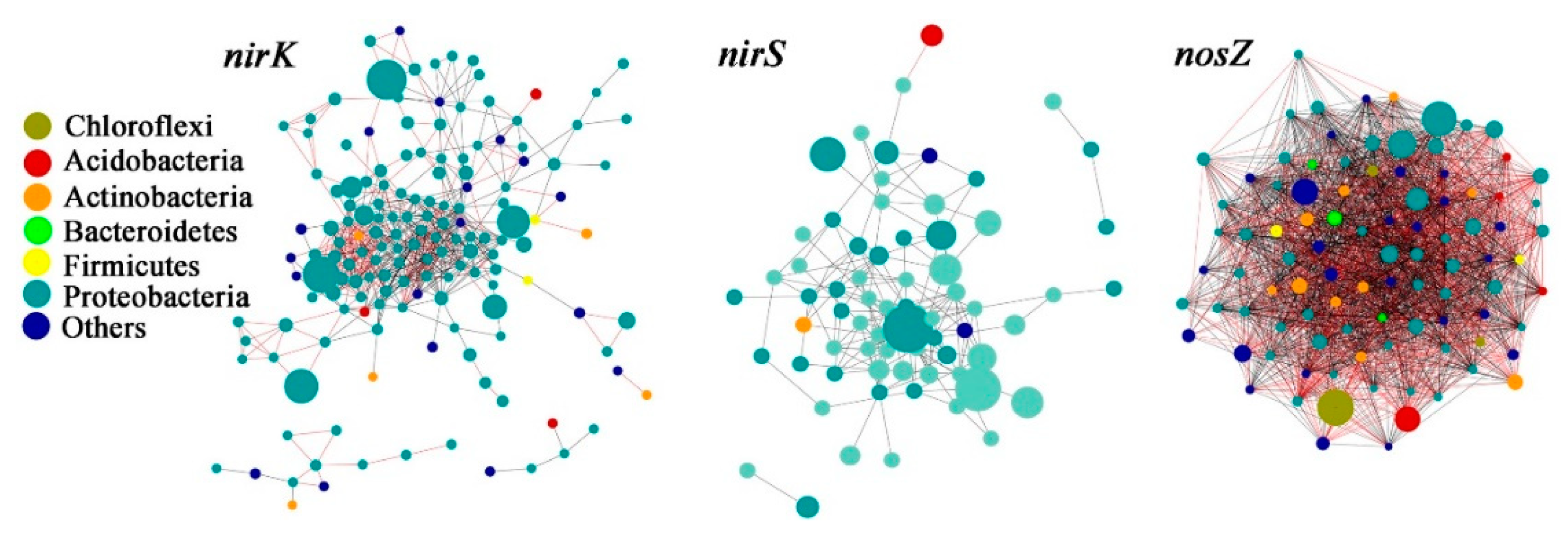
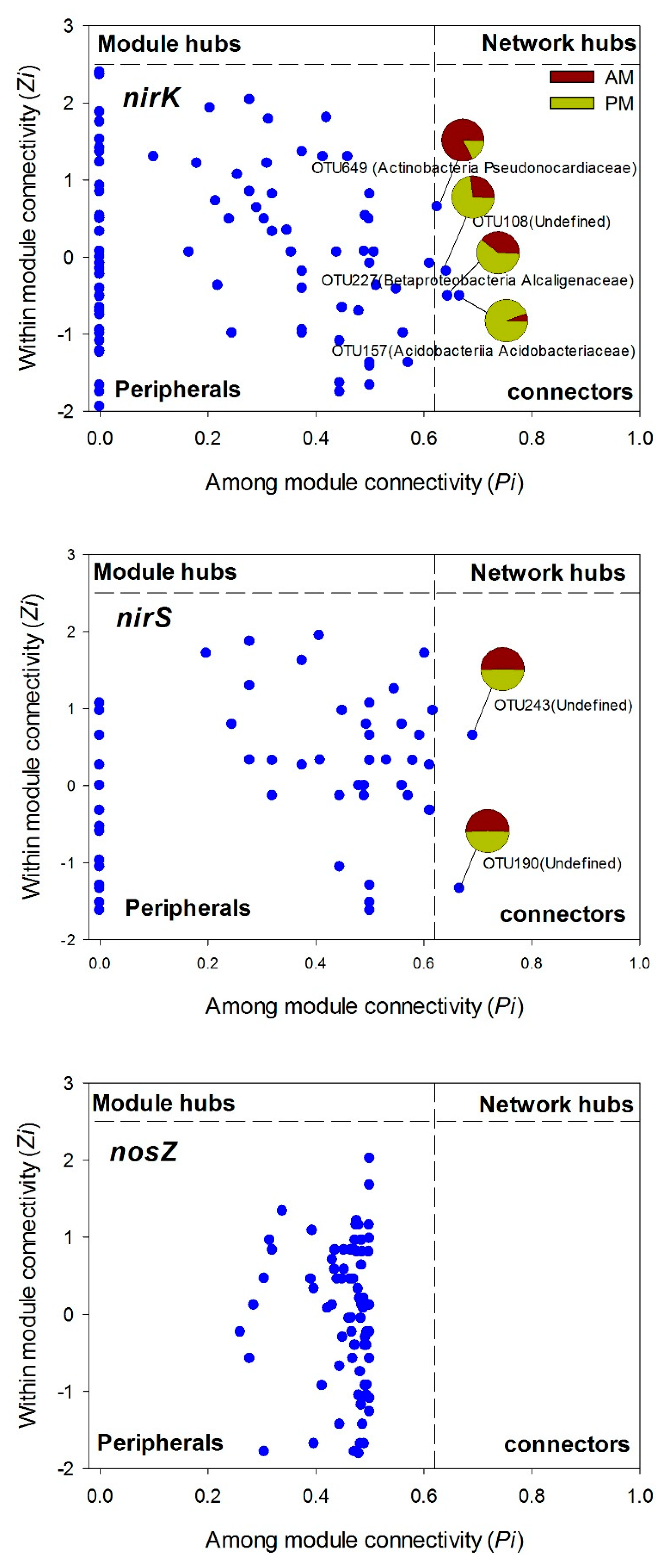
| Sites | SOM | DOC | MBC | MBN | NH4+ | NO3− | SWC | pH | DNA |
|---|---|---|---|---|---|---|---|---|---|
| AM | 111.82a (6.59) | 219.81a (17.84) | 367.42a (42.22) | 71.23a (9.08) | 12.17a (3.41) | 13.75a (1.71) | 35.37a (1.07) | 3.42a (0.03) | 11.95a (1.48) |
| PM | 70.65b (5.65) | 195.63a (9.73) | 213.15b (23.01) | 39.61b (4.29) | 2.22b (0.81) | 10.83a (1.53) | 23.15b (1.21) | 3.55a (0.03) | 5.47b (1.79) |
| Sites | Abundance | Shannon-Weiner Index | Simpson Index | ||||||
|---|---|---|---|---|---|---|---|---|---|
| nirK | nirS | nosZ | nirK | nirS | nosZ | nirK | nirS | nosZ | |
| AM | 6.28 × 105aA (2.12 × 105) | 1.83 × 106aB (6.04 × 105) | 2.01 × 107aC (2.76 × 106) | 2.33aAB (0.32) | 1.57bA (0.14) | 2.83bB (0.27) | 0.75aA (0.08) | 0.65aA (0.04) | 0.81aA (0.07) |
| PM | 3.48 × 105aA (1.70 × 105) | 9.83 × 106aB (3.95 × 106) | 5.40 × 106aB (2.31 × 106) | 2.51aA (0.30) | 2.40aA (0.20) | 3.78aB (0.11) | 0.78aA (0.07) | 0.87aAB (0.03) | 0.95aB (0.01) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Li, J.; Shen, W.; Xu, H.; Li, Y.; Luo, T. The Structure and Species Co-Occurrence Networks of Soil Denitrifying Bacterial Communities Differ Between A Coniferous and A Broadleaved Forests. Microorganisms 2019, 7, 361. https://doi.org/10.3390/microorganisms7090361
Chen J, Li J, Shen W, Xu H, Li Y, Luo T. The Structure and Species Co-Occurrence Networks of Soil Denitrifying Bacterial Communities Differ Between A Coniferous and A Broadleaved Forests. Microorganisms. 2019; 7(9):361. https://doi.org/10.3390/microorganisms7090361
Chicago/Turabian StyleChen, Jie, Jiajia Li, Weijun Shen, Han Xu, Yide Li, and Tushou Luo. 2019. "The Structure and Species Co-Occurrence Networks of Soil Denitrifying Bacterial Communities Differ Between A Coniferous and A Broadleaved Forests" Microorganisms 7, no. 9: 361. https://doi.org/10.3390/microorganisms7090361
APA StyleChen, J., Li, J., Shen, W., Xu, H., Li, Y., & Luo, T. (2019). The Structure and Species Co-Occurrence Networks of Soil Denitrifying Bacterial Communities Differ Between A Coniferous and A Broadleaved Forests. Microorganisms, 7(9), 361. https://doi.org/10.3390/microorganisms7090361




