Response of Microbial Communities and Their Metabolic Functions to Drying–Rewetting Stress in a Temperate Forest Soil
Abstract
1. Introduction
2. Material and Methods
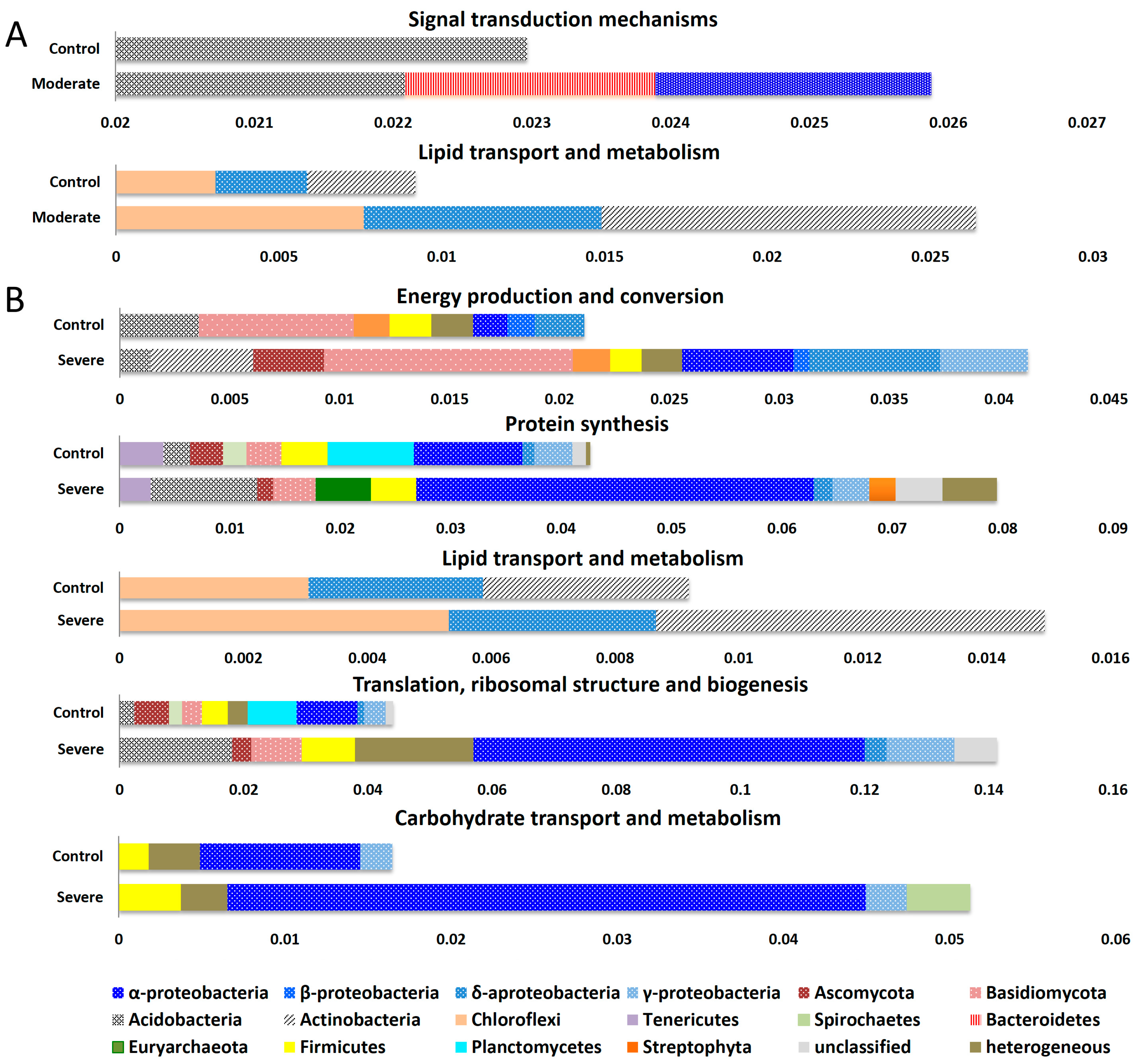
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneva, Switzerland, 2014; ISBN 9789291691432. [Google Scholar]
- Evans, S.E.; Wallenstein, M.D. Climate change alters ecological strategies of soil bacteria. Ecol. Lett. 2014, 17, 155–164. [Google Scholar] [CrossRef]
- Leitner, S.; Minixhofer, P.; Inselsbacher, E.; Keiblinger, K.M.; Zimmermann, M.; Zechmeister-Boltenstern, S. Short-term soil mineral and organic nitrogen fluxes during moderate and severe drying–rewetting events. Appl. Soil Ecol. 2017, 114, 28–33. [Google Scholar] [CrossRef]
- Fuchslueger, L.; Bahn, M.; Fritz, K.; Hasibeder, R.; Richter, A. Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow. New Phytol. 2014, 201, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, S.; Schimel, J.P.; Porporato, A. Responses of soil microbial communities to water stress: Results from a meta-analysis. Ecology 2012, 93, 930–938. [Google Scholar] [CrossRef]
- Schimel, J.; Balser, T.C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef]
- Birch, H.F. The effect of soil drying on humus decomposition and nitrogen availability. Plant Soil 1958, 10, 9–31. [Google Scholar] [CrossRef]
- Borken, W.; Matzner, E. Reappraisal of drying and wetting effects on C and N mineralization and fluxes in soils. Glob. Chang. Biol. 2009, 15, 808–824. [Google Scholar] [CrossRef]
- Inglima, I.; Alberti, G.; Bertolini, T.; Vaccari, F.P.; Gioli, B.; Miglietta, F.; Cotrufo, M.F.; Peressotti, A. Precipitation pulses enhance respiration of Mediterranean ecosystems: The balance between organic and inorganic components of increased soil CO2 efflux. Glob. Chang. Biol. 2009, 15, 1289–1301. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P. A proposed mechanism for the pulse in carbon dioxide production commonly observed following the rapid rewetting of a dry Soil. Soil Sci. Soc. Am. J. 2003, 67, 798–805. [Google Scholar] [CrossRef]
- Manzoni, S.; Schaeffer, S.M.; Katul, G.; Porporato, A.; Schimel, J.P. A theoretical analysis of microbial eco-physiological and diffusion limitations to carbon cycling in drying soils. Soil Biol. Biochem. 2014, 73, 69–83. [Google Scholar] [CrossRef]
- Lawrence, C.R.; Neff, J.C.; Schimel, J.P. Does adding microbial mechanisms of decomposition improve soil organic matter models? A comparison of four models using data from a pulsed rewetting experiment. Soil Biol. Biochem. 2009, 41, 1923–1934. [Google Scholar] [CrossRef]
- Leitner, S.; Homyak, P.M.; Blankinship, J.C.; Eberwein, J.; Jenerette, G.D.; Zechmeister-Boltenstern, S.; Schimel, J.P. Linking NO and N2O emission pulses with the mobilization of mineral and organic N upon rewetting dry soils. Soil Biol. Biochem. 2017, 115, 461–466. [Google Scholar] [CrossRef]
- Evans, S.; Dieckmann, U.; Franklin, O.; Kaiser, C. Synergistic effects of diffusion and microbial physiology reproduce the Birch effect in a micro-scale model. Soil Biol. Biochem. 2016, 93, 28–37. [Google Scholar] [CrossRef]
- Bouskill, N.J.; Lim, H.C.; Borglin, S.; Salve, R.; Wood, T.E.; Silver, W.L.; Brodie, E.L. Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J. 2013, 7, 384–394. [Google Scholar] [CrossRef]
- Botton, S.; Van Heusden, M.; Parsons, J.R.; Smidt, H.; Van Straalen, N. Resilience of microbial systems towards disturbances. Crit. Rev. Microbiol. 2006, 32, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Fornara, D.; Ikenaga, M.; Akagi, I.; Zhang, R.; Jia, Z. The resilience of microbial community under drying and rewetting cycles of three forest soils. Front. Microbiol. 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.; Freeman, C.; Golyshin, P.N.; Ackermann, G.; Fenner, N.; McDonald, J.E.; Ehbair, A.; Jones, T.G.; Murphy, L.M.; Creer, S. Subtle shifts in microbial communities occur alongside the release of carbon induced by drought and rewetting in contrasting peatland ecosystems. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Uhlírová, E.; Elhottová, D.; Tríska, J.; Santrůcková, H. Physiology and microbial community structure in soil at extreme water content. Folia Microbiol. (Praha) 2005, 50, 161–166. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; Hall, E.K. A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 2012, 109, 35–47. [Google Scholar] [CrossRef]
- Meisner, A.; Jacquiod, S.; Snoek, B.L.; Ten Hooven, F.C.; van der Putten, W.H. Drought legacy effects on the composition of soil fungal and prokaryote communities. Front. Microbiol. 2018, 9, 294. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Singh, B.K.; Maestre, F.T. Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 2017, 20, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Luo, G.; Wang, T.; Li, K.; Ling, N.; Shen, Q.; Zhang, J.; Li, L. Historical-nitrogen deposition and straw addition facilitate the resistance of soil multifunctionality to drying-wetting cycles. Appl. Environ. Microbiol. 2019. [Google Scholar] [CrossRef]
- Maron, P.A.; Ranjard, L.; Mougel, C.; Lemanceau, P. Metaproteomics: A new approach for studying functional microbial ecology. Microb. Ecol. 2007, 53, 486–493. [Google Scholar] [CrossRef]
- Hettich, R.L.; Pan, C.; Chourey, K.; Giannone, R.J. Metaproteomics: Harnessing the power of high performance mass spectrometry to identify the suite of proteins that control metabolic activities in microbial communities. Anal. Chem. 2013, 85, 4203–4214. [Google Scholar] [CrossRef]
- Pulleman, M.; Tietema, A. Microbial C and N transformations during drying and rewetting of coniferous forest floor material. Soil Biol. Biochem. 1999, 31, 275–285. [Google Scholar] [CrossRef]
- Jumpponen, A.; Trappe, J.M. Dark septate endophytes: A review of facultative biotrophic root-colonizing fungi. New Phytol. 1998, 140, 295–310. [Google Scholar] [CrossRef]
- Leitner, S.; Sae-Tun, O.; Kranzinger, L.; Zechmeister-Boltenstern, S.; Zimmermann, M. Contribution of litter layer to soil greenhouse gas emissions in a temperate beech forest. Plant Soil 2016, 403, 455–469. [Google Scholar] [CrossRef]
- IUSS. World Reference Base for Soil Resources; FAQ: Rome, Italy, 2007. [Google Scholar]
- Ross, D.J. Measurements of microbial biomass c and n in grassland soils by fumigation-incubation procedures: influence of inoculum size and the control. Soil Biol. Biochem. 1990, 22, 289–294. [Google Scholar] [CrossRef]
- Saiya-Cork, K.R.; Sinsabaugh, R.L.; Zak, D.R. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 2002, 34, 1309–1315. [Google Scholar] [CrossRef]
- German, D.P.; Weintraub, M.N.; Grandy, A.S.; Lauber, C.L.; Rinkes, Z.L.; Allison, S.D. Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol. Biochem. 2011, 43, 1387–1397. [Google Scholar] [CrossRef]
- Kaiser, C.; Koranda, M.; Kitzler, B.; Fuchslueger, L.; Schnecker, J.; Schweiger, P.; Rasche, F.; Zechmeister-Boltenstern, S.; Sessitsch, A.; Richter, A. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010, 187, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Keiblinger, K.M.; Wilhartitz, I.C.; Schneider, T.; Roschitzki, B.; Schmid, E.; Eberl, L.; Riedel, K.; Zechmeister-Boltenstern, S. Soil metaproteomics—Comparative evaluation of protein extraction protocols. Soil Biol. Biochem. 2012, 54, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Zybailov, B.; Mosley, A.L.; Sardiu, M.E.; Coleman, M.K.; Florens, L.; Washburn, M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006, 5, 2339–2347. [Google Scholar] [CrossRef]
- Liu, D.; Keiblinger, K.M.; Schindlbacher, A.; Wegner, U.; Sun, H.; Fuchs, S.; Lassek, C.; Riedel, K.; Zechmeister-Boltenstern, S. Microbial functionality as affected by experimental warming of a temperate mountain forest soil—A metaproteomics survey. Appl. Soil Ecol. 2017, 117–118, 196–202. [Google Scholar] [CrossRef]
- Schwen, A.; Zimmermann, M.; Leitner, S.; Woche, S. Soil water repellency and its impact on hydraulic characteristics in a beech forest under simulated climate change. Vadose Zone J. 2015, 14, 1–11. [Google Scholar] [CrossRef]
- Černohlávková, J.; Jarkovský, J.; Nešporová, M.; Hofman, J. Variability of soil microbial properties: Effects of sampling, handling and storage. Ecotoxicol. Environ. Saf. 2009, 72, 2102–2108. [Google Scholar] [CrossRef]
- Inselsbacher, E. Recovery of individual soil nitrogen forms after sieving and extraction. Soil Biol. Biochem. 2014, 71, 76–86. [Google Scholar] [CrossRef]
- Rousk, J.; Jones, D.L. Loss of low molecular weight dissolved organic carbon (DOC) and nitrogen (DON) in H2O and 0.5M K2SO4 soil extracts. Soil Biol. Biochem. 2010, 42, 2331–2335. [Google Scholar] [CrossRef]
- Chodak, M.; Gołębiewski, M.; Morawska-Płoskonka, J.; Kuduk, K.; Niklińska, M. Soil chemical properties affect the reaction of forest soil bacteria to drought and rewetting stress. Ann. Microbiol. 2015, 65, 1627–1637. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Hillekens, R.H.E.; de Hollander, M.; van der Heijden, M.G.A.; van den Berg, M.; van Straalen, N.M.; Kowalchuk, G.A. Structural and functional variation in soil fungal communities associated with litter bags containing maize leaf. FEMS Microbiol. Ecol. 2013, 84, 519–531. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Pold, G.; Topçuoglu, B.D.; van Diepen, L.T.A.; Varney, R.M.; Blanchard, J.L.; Melillo, J.; Frey, S.D. Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 2015, 6, 1–13. [Google Scholar]
- Starke, R.; Kermer, R.; Ullmann-Zeunert, L.; Baldwin, I.T.; Seifert, J.; Bastida, F.; von Bergen, M.; Jehmlich, N. Bacteria dominate the short-term assimilation of plant-derived N in soil. Soil Biol. Biochem. 2016, 96, 30–38. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Breznak, J.A.; Schmidt, T.M. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 2007, 73, 2708–2717. [Google Scholar] [CrossRef]
- Pankratov, T.A.; Dedysh, S.N. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 2010, 62 Pt 1, 2097–2106. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef]
- Hao, Y.; Cui, X.; Qian, X.; Liu, W.; Zhang, H.; Zhao, H.; Kang, X.; Wang, Y. Changes in soil microbial community response to precipitation events in a semi-arid steppe of the Xilin River Basin, China. J. Arid Land 2018, 11, 97–110. [Google Scholar]
- Blazewicz, S.J.; Schwartz, E.; Firestone, M.K. Growth and death of bacteria and fungi underlie rainfall-induced carbon dioxide pulses from seasonally dried soil. Ecology 2014, 95, 1162–1172. [Google Scholar] [CrossRef]
- Landesman, W.J.; Dighton, J. Shifts in Microbial Biomass and the Bacteria: Fungi Ratio Occur Under Field Conditions within 3 h After Rainfall. Microb. Ecol. 2011, 62, 228–236. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J. 2015, 9, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Kaisermann, A.; Maron, P.A.; Beaumelle, L.; Lata, J.C. Fungal communities are more sensitive indicators to non-extreme soil moisture variations than bacterial communities. Appl. Soil Ecol. 2015, 86, 158–164. [Google Scholar] [CrossRef]
- Cosentino, D.; Chenu, C.; Le Bissonnais, Y. Aggregate stability and microbial community dynamics under drying-wetting cycles in a silt loam soil. Soil Biol. Biochem. 2006, 38, 2053–2062. [Google Scholar] [CrossRef]
- Yuste, J.C.; Peñuelas, J.; Estiarte, M.; Garcia-Mas, J.; Mattana, S.; Ogaya, R.; Pujol, M.; Sardans, J. Drought-resistant fungi control soil organic matter decomposition and its response to temperature. Glob. Chang. Biol. 2011, 17, 1475–1486. [Google Scholar] [CrossRef]
- Bastida, F.; Torres, I.F.; Andrés-Abellán, M.; Baldrian, P.; López-Mondéjar, R.; Větrovský, T.; Richnow, H.H.; Starke, R.; Ondoño, S.; García, C.; et al. Differential sensitivity of total and active soil microbial communities to drought and forest management. Glob. Chang. Biol. 2017, 23, 4185–4203. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, U.N.; Ball, B.A. Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob. Chang. Biol. 2015, 21, 1407–1421. [Google Scholar] [CrossRef]
- Placella, S.A.; Brodie, E.L.; Firestone, M.K. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. USA 2012, 109, 10931–10936. [Google Scholar] [CrossRef]
- Bastida, F.; Jehmlich, N. It’s all about functionality: How can metaproteomics help us to discuss the attributes of ecological relevance in soil? J. Proteom. 2016, 144, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, K.C.; Karaoz, U.; Hanson, C.A.; Santee, C.A.; Bradford, M.A.; Treseder, K.K.; Wallenstein, M.D.; Brodie, E.L. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2011, 2, 94. [Google Scholar] [CrossRef]
- Bastida, F.; Selevsek, N.; Torres, I.F.; Hernández, T.; García, C. Soil restoration with organic amendments: linking cellular functionality and ecosystem processes. Sci. Rep. 2015, 5, 15550. [Google Scholar] [CrossRef] [PubMed]

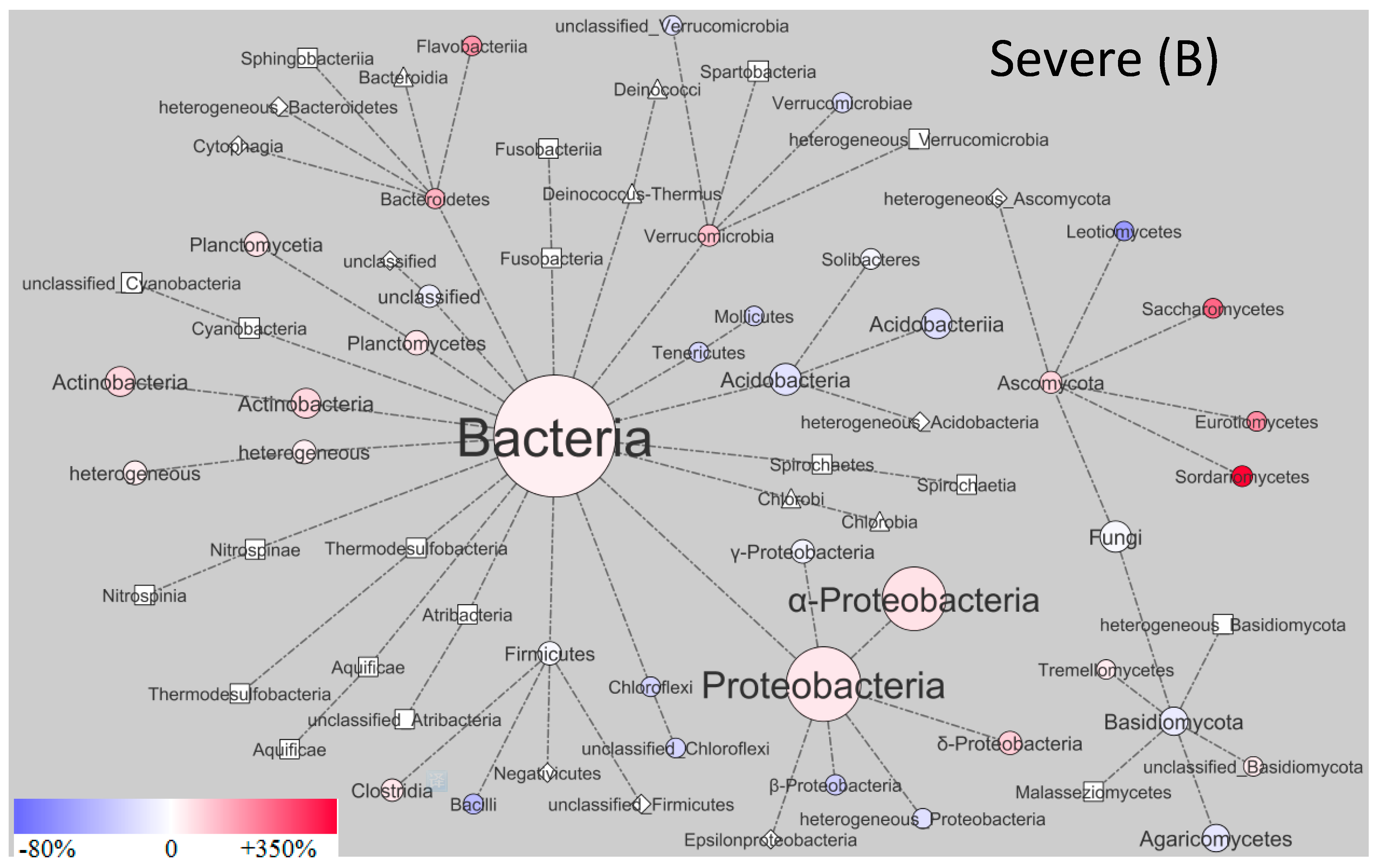
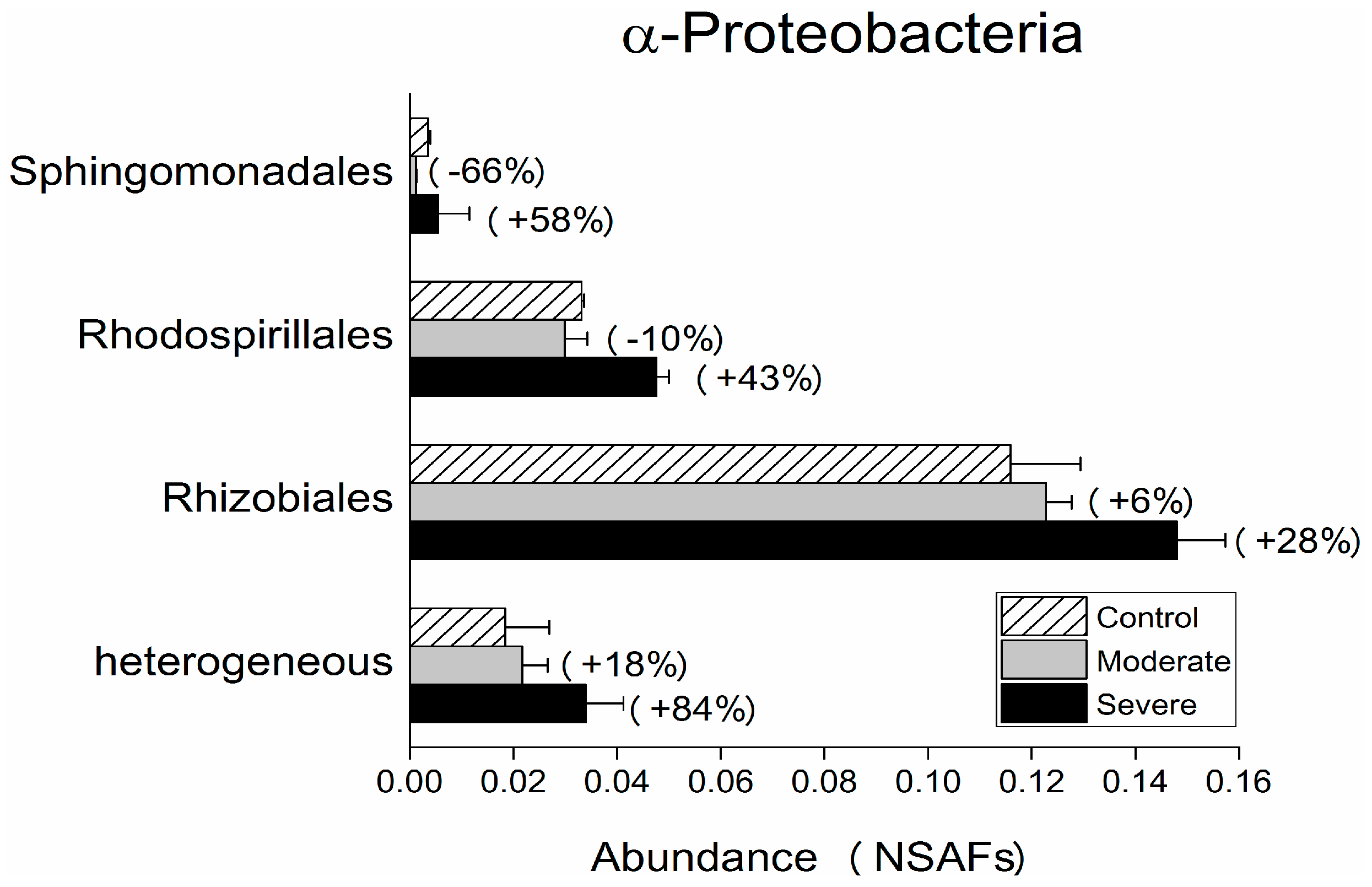
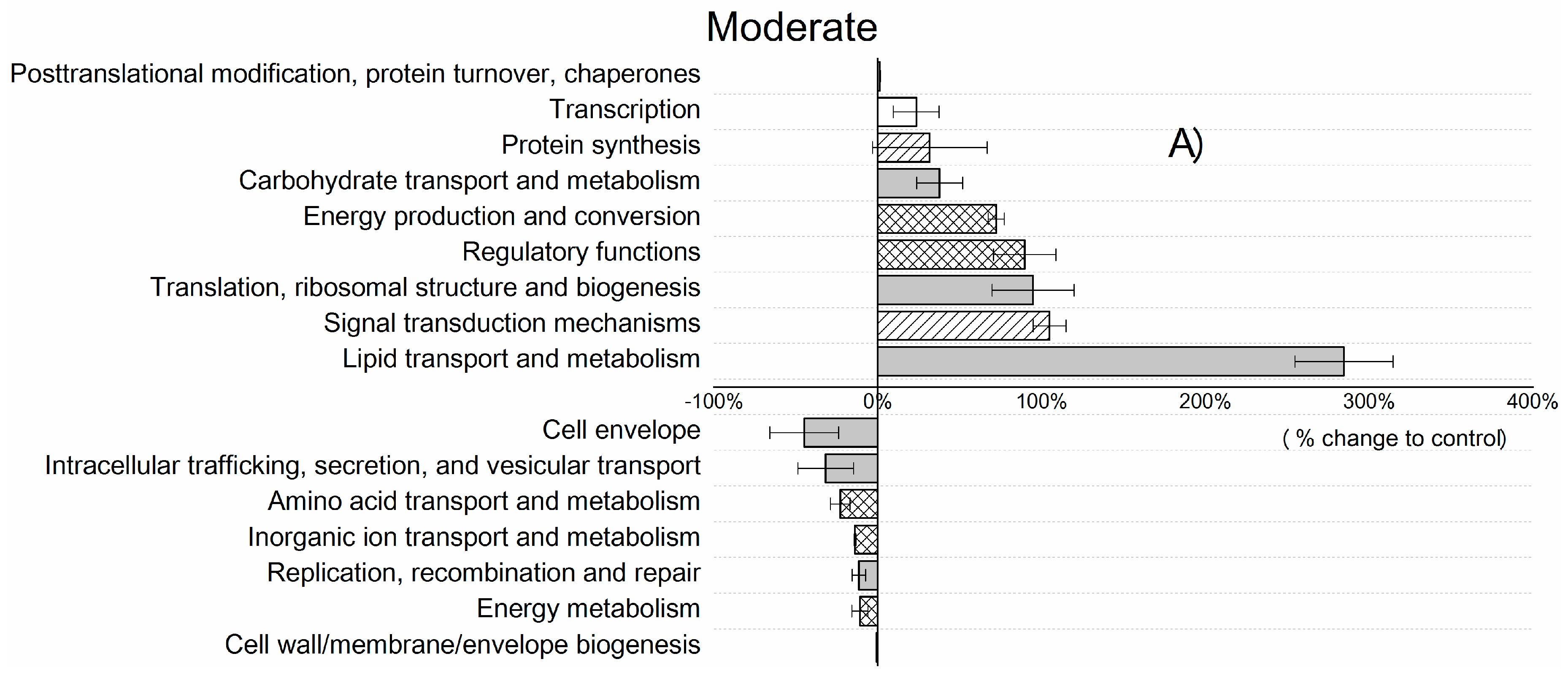
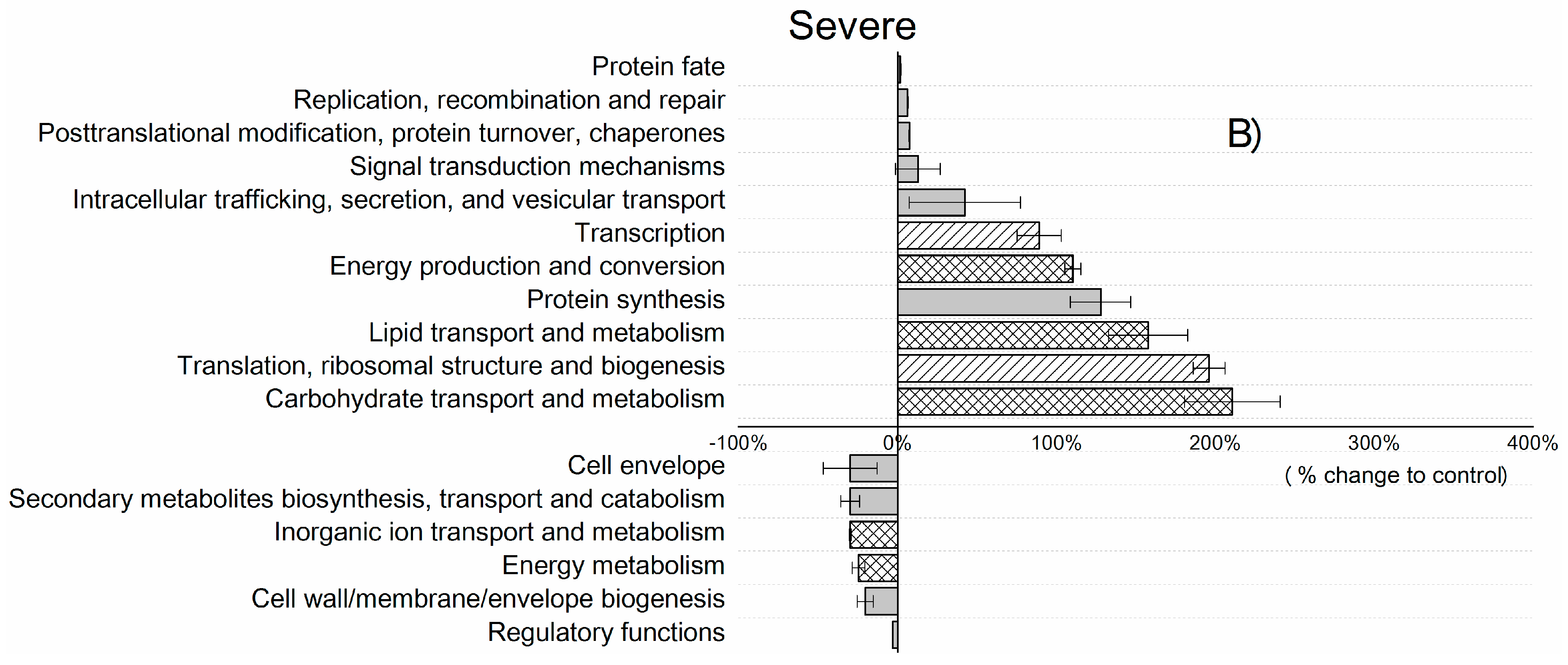
| Control | Moderate | Severe | |
|---|---|---|---|
| * Water content %w/w | 30.9 ± 2.6 a | 21.3 ± 1.7 b | 17.7 ± 2.1 b |
| NH4+–N (µg g−1) | 2.72 ± 0.27 | 2.54 ± 0.26 | 2.48 ± 0.33 |
| NO3−–N (µg g−1) | 4.32 ± 0.44 | 4.11 ± 0.74 | 3.63 ± 0.25 |
| PO43−–P (µg g−1) | 1.56 ± 0.55 | 1.31 ± 0.36 | 1.10 ± 0.49 |
| Dissolved organic carbon (µg g−1) | 197 ± 18 | 187 ± 53 | 212 ± 61 |
| Total dissolved N (µg g-1) | 25.7 ±1.9 | 25.8 ± 6.35 | 26.7 ± 9.5 |
| Microbial biomass carbon (mg g−1) | 1.21 ±0.24 | 0.99 ± 0.10 | 0.96 ± 0.19 |
| Microbial biomass nitrogen (mg g−1) | 0.12 ± 0.04 | 0.10 ± 0.01 | 0.09 ± 0.02 |
| Cmic/Nmic ratio | 10.3 ± 1.5 | 10.1 ± 1.38 | 10.2 ± 0.8 |
| * Cellobiohydrolase (nmol g−1 min−1) | 4.0 ± 2.1 b | 3.4 ± 1.6 b | 7.1 ± 1.1 a |
| * Chitinase (nmol g−1 min−1) | 13.1 ± 1.8 a | 9.4 ± 2.4 b | 8.9 ± 3.2 b |
| Phosphatase (nmol g−1 min−1) | 54.8 ± 10.3 | 35.2 ± 6.2 | 42.5 ± 11.7 |
| Protease (nmol g−1 min−1) | 2.8 ± 0.2 | 2.1 ± 0.1 | 2.1 ± 0.2 |
| Phenoloxidase (nmol g−1 min−1) | 14.6 ± 7.1 | 8.9 ± 3.1 | 10.8 ± 4.4 |
| Peroxidase (nmol g−1 min−1) | 84.6 ± 46.4 | 123.4 ± 55.9 | 204.6 ± 118.8 |
| Archaeal proteins (NSAFs) | 0.0017 | 0.0030 | 0.0045 |
| Expressed ribosomal proteins | 7 | 38 | 23 |
| Ascomycota/Basidiomycota (NSAFs ratio) | 0.16 | 0.34 | 0.31 |
| Shannon a-taxa diversity b | 2.73 | 2.79 | 2.51 |
| Shannon-functional diversity c | 5.39 | 5.71 | 5.62 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Keiblinger, K.M.; Leitner, S.; Wegner, U.; Zimmermann, M.; Fuchs, S.; Lassek, C.; Riedel, K.; Zechmeister-Boltenstern, S. Response of Microbial Communities and Their Metabolic Functions to Drying–Rewetting Stress in a Temperate Forest Soil. Microorganisms 2019, 7, 129. https://doi.org/10.3390/microorganisms7050129
Liu D, Keiblinger KM, Leitner S, Wegner U, Zimmermann M, Fuchs S, Lassek C, Riedel K, Zechmeister-Boltenstern S. Response of Microbial Communities and Their Metabolic Functions to Drying–Rewetting Stress in a Temperate Forest Soil. Microorganisms. 2019; 7(5):129. https://doi.org/10.3390/microorganisms7050129
Chicago/Turabian StyleLiu, Dong, Katharina M. Keiblinger, Sonja Leitner, Uwe Wegner, Michael Zimmermann, Stephan Fuchs, Christian Lassek, Katharina Riedel, and Sophie Zechmeister-Boltenstern. 2019. "Response of Microbial Communities and Their Metabolic Functions to Drying–Rewetting Stress in a Temperate Forest Soil" Microorganisms 7, no. 5: 129. https://doi.org/10.3390/microorganisms7050129
APA StyleLiu, D., Keiblinger, K. M., Leitner, S., Wegner, U., Zimmermann, M., Fuchs, S., Lassek, C., Riedel, K., & Zechmeister-Boltenstern, S. (2019). Response of Microbial Communities and Their Metabolic Functions to Drying–Rewetting Stress in a Temperate Forest Soil. Microorganisms, 7(5), 129. https://doi.org/10.3390/microorganisms7050129







