Occurrence and Function of the Na+-Translocating NADH:Quinone Oxidoreductase in Prevotella spp.
Abstract
:1. Introduction
2. Materials and Methods
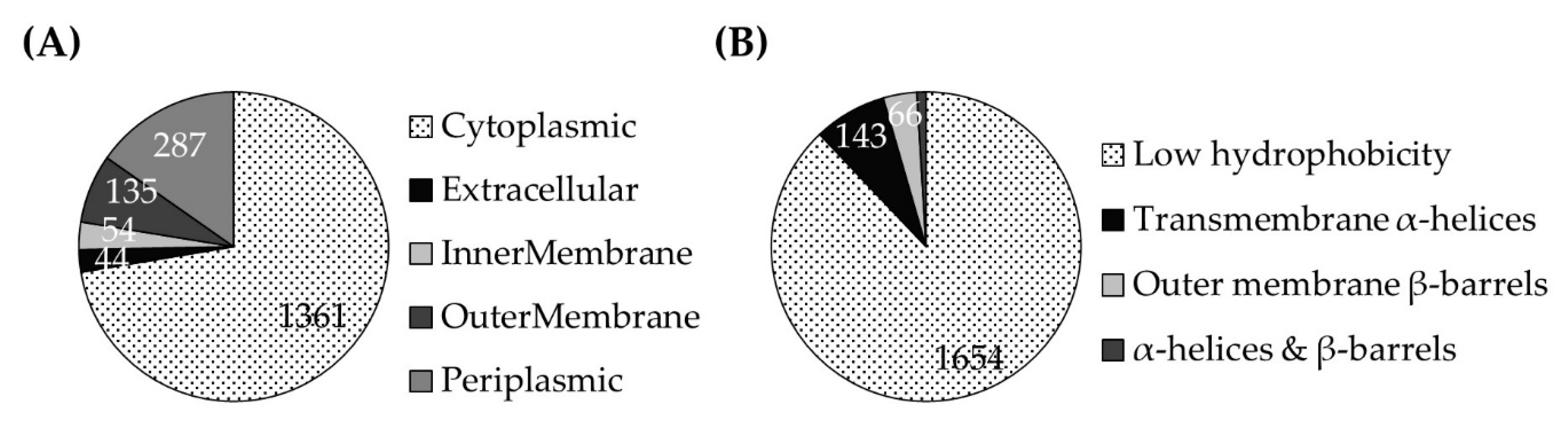
2.1. Analysis of Prevotellaceae-Derived Proteins from the Rumen
2.2. Organism and Growth Conditions
2.3. Proteome Extraction, Fractionation and Sodium Dodecyl Sulfate PolyAcrylamide Gel Electrophoresis (SDS-PAGE)
2.4. Isolation of Membranes, Solubilization and Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
2.5. Trypsin Digestion, Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS) Measurements and Bioinformatic Processing
2.6. Detection of NADH:Quinone Oxidoreductase (NQR) Homologues by In-Gel Fluorescence
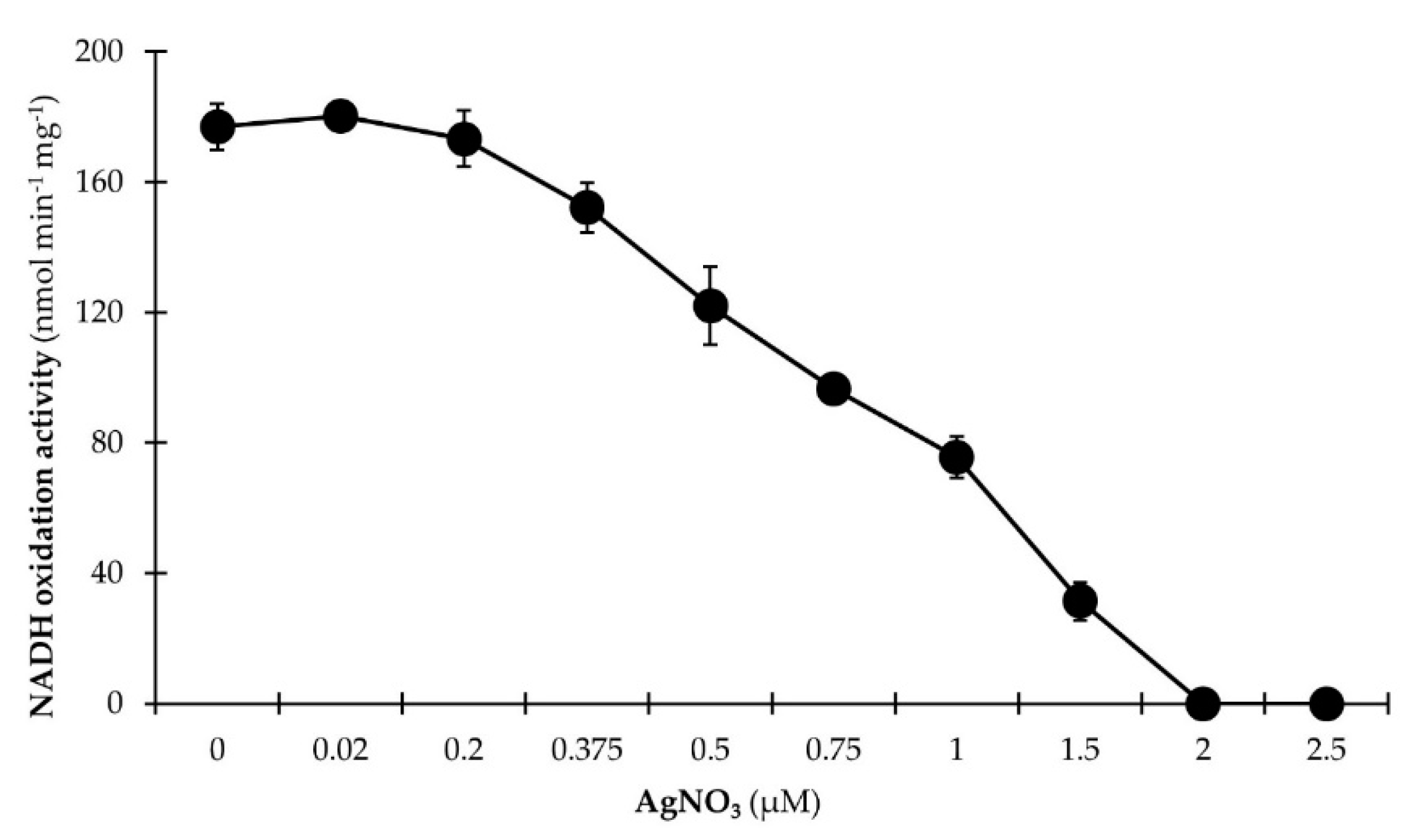
2.7. Inhibition of the NADH:Quinone Oxidoreduction Activity by Ag+
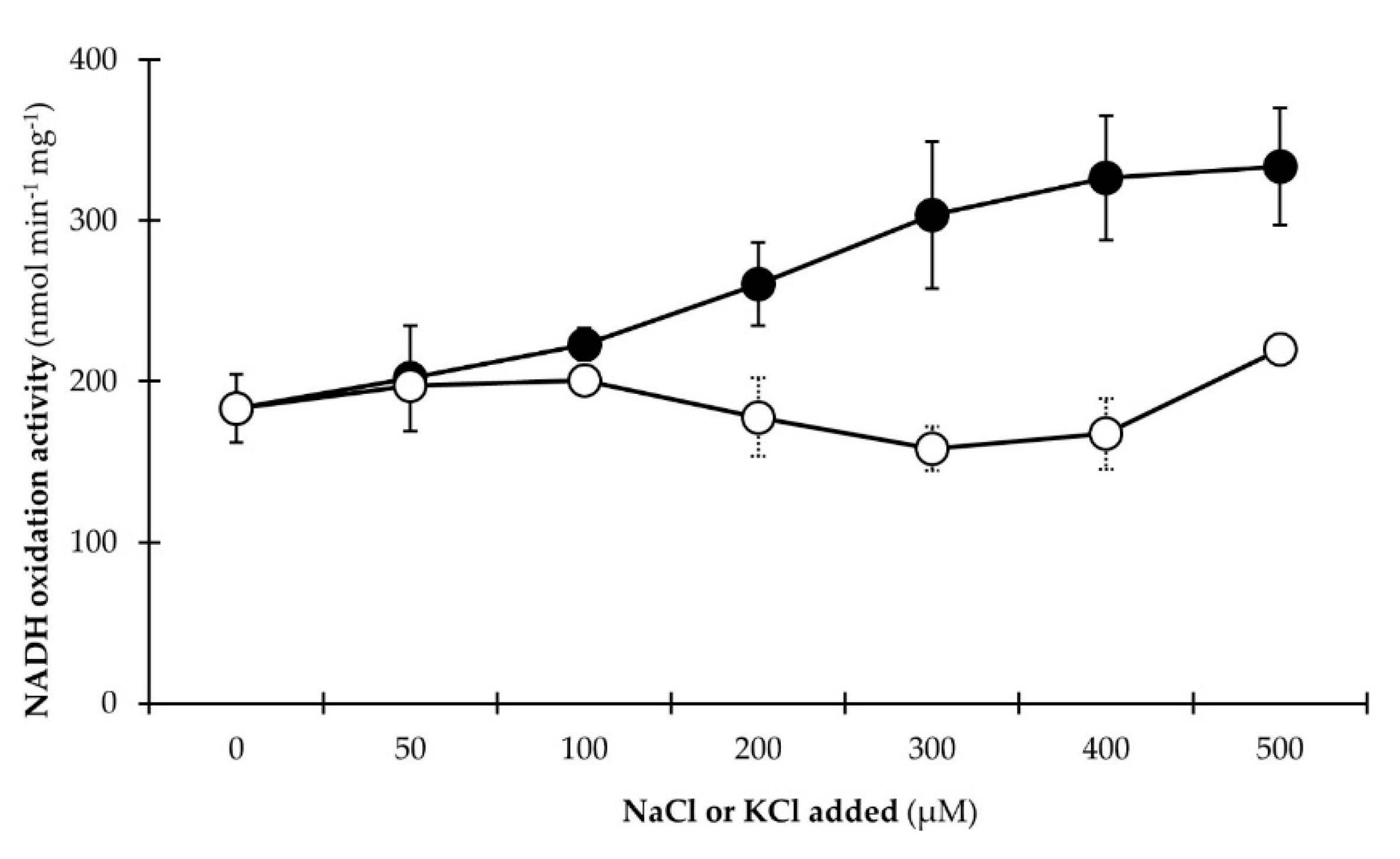
2.8. Stimulation of the NADH:Quinone Oxidoreduction Activity by Na+
3. Results and Discussion
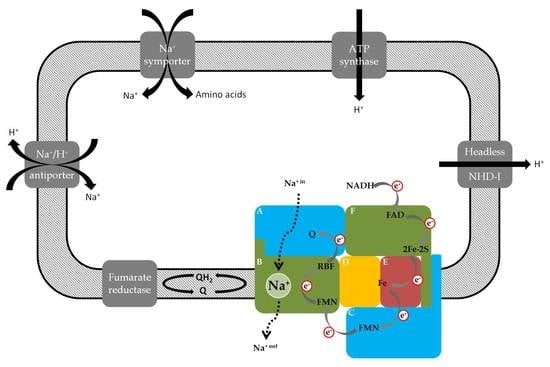
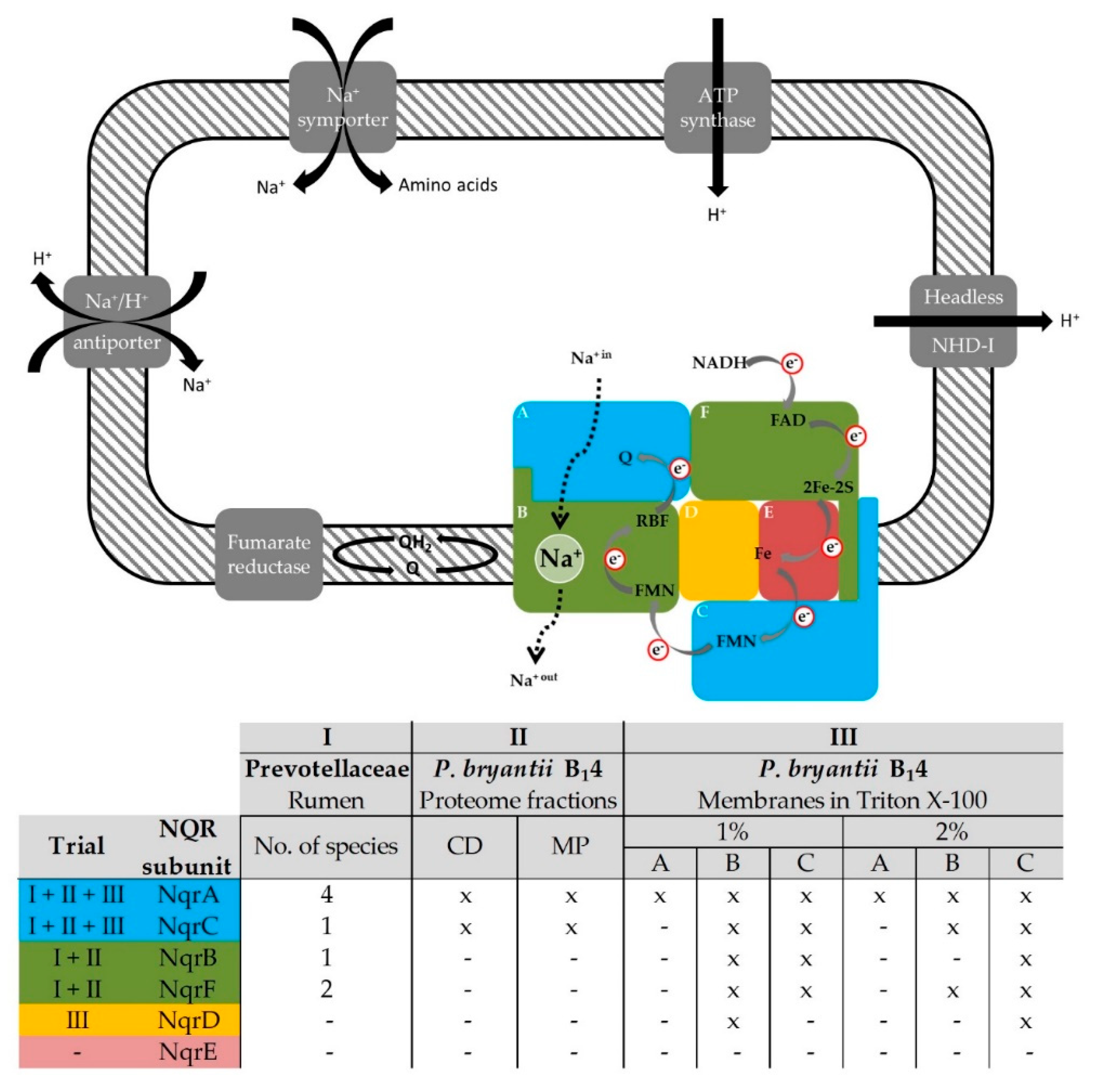
3.1. Identification of Na+-Translocating NADH:Quinone Oxidoreductase Subunits by LC–MS/MS
3.1.1. Ruminal Prevotellaceae Proteins Include Subunits of the NQR
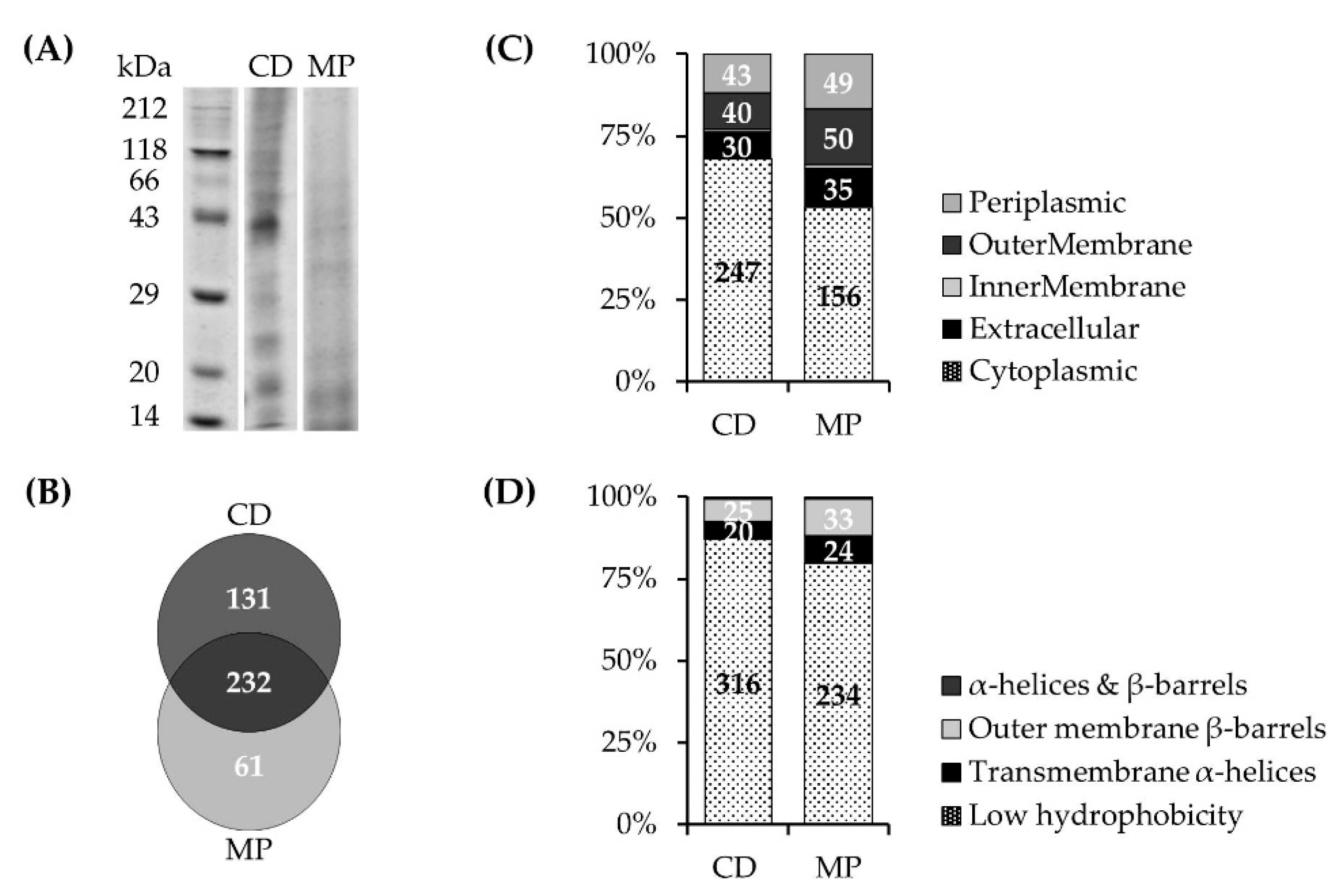
3.1.2. NQR Subunits Detected in Fractions of the Prevotella bryantii B14 Proteome
3.1.3. NQR Subunits Identified in Solubilized Membranes of Prevotella bryantii B14
3.2. Prevotella bryantii B14 Contains a Functional NQR
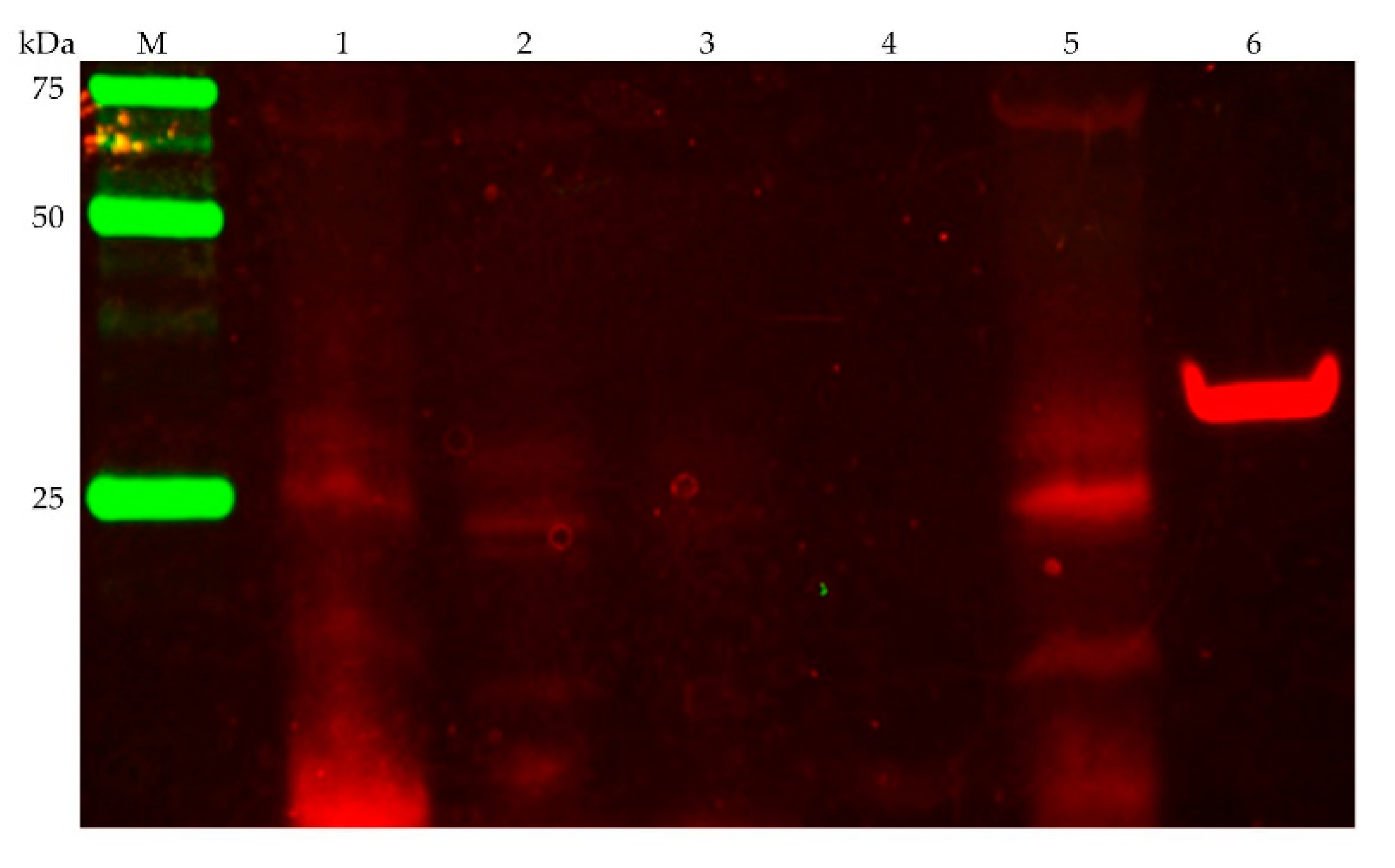
3.2.1. Detection of the Membrane-Bound Subunits NqrB and NqrC
3.2.2. NADH Dehydrogenase Activity Is Inhibited by Micromolar Ag+
3.2.3. NADH Dehydrogenase Activity Is Stimulated by Na+
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jami, E.; Mizrahi, I. Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Morrison, M.; Yu, Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 2011, 76, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Henrissat, B.; Stanton, T.B. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Paslier, D.; Yamada, T.; Mende, D.R. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Rajilic-Stojanovic, M.; de Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef]
- Miyazaki, K.; Martin, J.C.; Marinsek-Logar, R.; Flint, H.J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe 1997, 3, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Ogata, K.; Tajima, K.; Nakamura, M.; Nagamine, T.; Aminov, R.I.; Benno, Y. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr. Microbiol. 2000, 41, 45–49. [Google Scholar] [CrossRef]
- Purushe, J.; Fouts, D.E.; Morrison, M.; White, B.A.; Mackie, R.I.; Coutinho, P.M.; Henrissat, B.; Nelson, K.E. Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: Insights into their environmental niche. Microb. Ecol. 2010, 60, 721–729. [Google Scholar] [CrossRef]
- Avgustin, G.; Wallace, R.J.; Flint, H.J. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int. J. Syst. Bacteriol. 1997, 47, 284–288. [Google Scholar] [CrossRef]
- Griswold, K.E.; White, B.A.; Mackie, R.I. Diversity of extracellular proteolytic activities among Prevotella species from the rumen. Curr. Microbiol. 1999, 39, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Zorec, M.; Vodovnik, M.; Marinšek-Logar, R. Potential of selected rumen bacteria for cellulose and hemicellulose degradation. Food Technol. Biotech. 2014, 52, 210–221. [Google Scholar]
- Takahashi, N.; Yamada, T. Pathways for amino acid metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immun. 2000, 15, 96–102. [Google Scholar] [CrossRef]
- Takahashi, N.; Yamada, T. Glucose metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immun. 2000, 15, 188–195. [Google Scholar] [CrossRef]
- Franke, T.; Deppenmeier, U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol. Microbiol. 2018, 109, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Millen, D.D.; De Beni Arrigoni, M.; Pacheco, R.D.L. Microbiology of the rumen. In Rumenology; Springer International Publishing: Switzerland, 2016; pp. 39–40. [Google Scholar]
- Deusch, S.; Camarinha-Silva, A.; Conrad, J.; Beifuss, U.; Rodehutscord, M.; Seifert, J. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Reyes-Prieto, A.; Barquera, B.; Juárez, O. Origin and evolution of the sodium-pumping NADH:Ubiquinone oxidoreductase. PLoS ONE 2014, 9, e96696. [Google Scholar] [CrossRef]
- Barquera, B. The sodium pumping NADH:quinone oxidoreductase (Na+-NQR), a unique redox-driven ion pump. J. Bioenerg. Biomembr. 2014, 46, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Steuber, J.; Vohl, G.; Casutt, M.S.; Vorburger, T.; Diederichs, K.; Fritz, G. Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase. Nature 2014, 516, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Steuber, J.; Vohl, G.; Muras, V.; Toulouse, C.; Claussen, B.; Vorburger, T.; Fritz, G. The structure of Na+-translocating of NADH:ubiquinone oxidoreductase of Vibrio cholerae: Implications on coupling between electron transfer and Na+ transport. Biol. Chem. 2015, 396, 1015–1030. [Google Scholar] [CrossRef]
- Steuber, J.; Halang, P.; Vorburger, T.; Steffen, W.; Vohl, G.; Fritz, G. Central role of the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) in sodium bioenergetics of Vibrio cholerae. Biol. Chem. 2014, 395, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Zafar, H.; Saier, M.H., Jr. Comparative genomics of transport proteins in seven Bacteroides species. PLoS ONE 2018, 13, e0208151. [Google Scholar] [CrossRef]
- Hackmann, T.J.; Ngugi, D.K.; Firkins, J.L.; Tao, J. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ. Microbiol. 2017, 19, 4670–4683. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Tsirigos, K.D.; Elofsson, A.; Bagos, P.G. PRED-TMBB2: improved topology prediction and detection of beta-barrel outer membrane proteins. Bioinformatics 2016, 32, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Hobson, P.N. Rumen Bacteria. In Methods in Microbiology; Norris, J.R., Ribbons, D.W., Eds.; Academic Press: New York, NY, USA, 1969; Volume 3, pp. 133–149. [Google Scholar]
- Bryant, M.P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 1972, 25, 1324–1328. [Google Scholar] [CrossRef]
- Varel, V.H.; Bryant, M.P. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 1974, 28, 251–257. [Google Scholar]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Tao, M.; Casutt, M.S.; Fritz, G.; Steuber, J. Oxidant-induced formation of a neutral flavosemiquinone in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. Biochim. Biophys. Acta Bioenergetics 2008, 1777, 696–702. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991, 199, 223–231. [Google Scholar] [CrossRef]
- Jehmlich, N.; Schmidt, F.; Hartwich, M.; von Bergen, M.; Richnow, H.H.; Vogt, C. Incorporation of carbon and nitrogen atoms into proteins measured by protein-based stable isotope probing (Protein-SIP). Rapid Commun. Mass Sp. 2008, 22, 2889–2897. [Google Scholar] [CrossRef]
- Vorburger, T.; Nedielkov, R.; Brosig, A.; Bok, E.; Schunke, E.; Steffen, W.; Mayer, S.; Götz, F.; Möller, H.M.; Steuber, J. Role of the Na+-translocating NADH:quinone oxidoreductase in voltage generation and Na+ extrusion in Vibrio cholerae. Biochim. Biophys. Acta Bioenergetics 2016, 1857, 473–482. [Google Scholar] [CrossRef]
- Tanca, A.; Palomba, A.; Pisanu, S.; Deligios, M.; Fraumene, C.; Manghina, V.; Pagnozzi, D.; Addis, M.F.; Uzzau, S. A straightforward and efficient analytical pipeline for metaproteome characterization. Microbiome 2014, 2. [Google Scholar] [CrossRef]
- Biegel, E.; Schmidt, S.; González, J.M.; Müller, V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 2011, 68, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Juárez, O.; Barquera, B. Insights into the mechanism of electron transfer and sodium translocation of the Na+-pumping NADH:quinone oxidoreductase. Biochim. Biophys. Acta Bioenergetics 2012, 1817, 1823–1832. [Google Scholar] [CrossRef]
- Hess, V.; Schuchmann, K.; Muller, V. The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J. Biol. Chem. 2013, 288, 31496–31502. [Google Scholar] [CrossRef] [PubMed]
- Dimroth, P. Mechanisms of sodium transport in bacteria. Philos. Trans. R. Soc. B 1990, 326, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Dimroth, P. Primary sodium ion translocating enzymes. Biochim. Biophys. Acta 1997, 1318, 11–51. [Google Scholar] [CrossRef]
- Leif, H.; Sled, V.D.; Ohnishi, T.; Weiss, H.; Friedrich, T. Isolation and characterization of the proton-translocating NADH: ubiquinone oxidoreductase from Escherichia coli. Eur. J. Biochem. 1995, 230, 538–548. [Google Scholar] [CrossRef]
- Moparthi, V.K.; Hägerhäll, C. The evolution of respiratory chain complex I from a smaller last common ancestor consisting of 11 protein subunits. J. Mol. Evol. 2011, 72, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nakayama, Y.; Yasui, M.; Maeda, M.; Furuishi, K.; Unemoto, T. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na+-translocating NADH:quinone reductase from Vibrio alginolyticus. FEBS Lett. 2001, 488, 5–8. [Google Scholar] [CrossRef]
- Casutt, M.S.; Huber, T.; Brunisholz, R.; Tao, M.; Fritz, G.; Steuber, J. Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 2010, 285, 27088–27099. [Google Scholar] [CrossRef]
- Casutt, M.S.; Wendelspiess, S.; Steuber, J.; Fritz, G. Crystallization of the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Acta Crystallogr. F 2010, 66, 1677–1679. [Google Scholar] [CrossRef] [PubMed]
- Steuber, J.; Krebs, W.; Dimroth, P. The Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio alginolyticus—Redox states of the FAD prosthetic group and mechanism of Ag+ inhibition. Eur. J. Biochem. 1997, 249, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Turk, K.; Hase, C.C.; Fritz, G.; Steuber, J. Quinone reduction by the Na+-translocating NADH dehydrogenase promotes extracellular superoxide production in Vibrio cholerae. J. Bacteriol. 2007, 189, 3902–3908. [Google Scholar] [CrossRef] [PubMed]
- Krebs, W.; Steuber, J.; Gemperli Anja, C.; Dimroth, P. Na+ translocation by the NADH:ubiquinone oxidoreductase (complex I) from Klebsiella pneumoniae. Mol. Microbiol. 2002, 33, 590–598. [Google Scholar] [CrossRef]
- Toulouse, C.; Claussen, B.; Muras, V.; Fritz, G.; Steuber, J. Strong pH dependence of coupling efficiency of the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) of Vibrio cholerae. Biol. Chem. 2017, 398, 251–260. [Google Scholar] [CrossRef]
- Unemoto, T.; Ogura, T.; Hayashi, M. Modifications by Na+ and K+, and the site of Ag+ inhibition in the Na+-translocating NADH:quinone reductase from a marine Vibrio alginolyticus. Biochim. Biophys. Acta Bioenergetics 1993, 1183, 201–205. [Google Scholar] [CrossRef]
- Tokuda, H.; Unemoto, T. Na+ is translocated at NADH:quinone oxidoreductase segment in the respiratory chain of Vibrio alginolyticus. J. Biol. Chem. 1984, 259, 7785–7790. [Google Scholar] [PubMed]
- Marreiros, B.C.; Batista, A.P.; Duarte, A.M.S.; Pereira, M.M. A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. Biochim. Biophys. Acta Bioenergetics 2013, 1827, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Steffen, W.; Steuber, J. Cation transport by the respiratory NADH:quinone oxidoreductase (complex I): Facts and hypotheses. Biochem. Soc. T. 2013, 41, 1280–1287. [Google Scholar] [CrossRef]
- Swartz, T.H.; Ikewada, S.; Ishikawa, O.; Ito, M.; Krulwich, T.A. The Mrp system: A giant among monovalent cation/proton antiporters? Extremophiles 2005, 9, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B.; Strobel, H.J.; Driessen, A.J.; Konings, W.N. Sodium-dependent transport of neutral amino acids by whole cells and membrane vesicles of Streptococcus bovis, a ruminal bacterium. J. Bacteriol. 1988, 170, 3531–3536. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A. Nutrient transport by ruminal bacteria: A review. J. Anim. Sci. 1994, 72, 3019–3031. [Google Scholar] [CrossRef]
- Lu, Z.; Imlay, J.A. The Fumarate Reductase of Bacteroides thetaiotaomicron, unlike that of Escherichia coli, is configured so that it does not generate reactive oxygen species. mBio 2017, 8, e01873-16. [Google Scholar] [CrossRef]
- Stewart, C.S. The rumen bacteria. In The Rumen Microbial Ecosystem; Hobson, P.N., Stewart, C.S., Eds.; Springer: Heidelberg, Germany, 1997; pp. 10–72. [Google Scholar]
- Mitsumori, M.; Shinkai, T.; Takenaka, A.; Enishi, O.; Higuchi, K.; Kobayashi, Y.; Nonaka, I.; Asanuma, N.; Denman, S.E.; McSweeney, C.S. Responses in digestion, rumen fermentation and microbial populations to inhibition of methane formation by a halogenated methane analogue. Br. J. Nutr. 2012, 108, 482–491. [Google Scholar] [CrossRef]
| Uniprot Accession no. | Identified Proteins Species | Score (Max Quant) | Sequence Coverage (%) | Unique Peptides | Total Peptides |
|---|---|---|---|---|---|
| D5ESF6 | NqrF | 40.28 | 32.00 | 4 | 9 |
| Prevotella ruminicola | |||||
| D5ESG1 | NqrA | 11.67 | 19.60 | 5 | 8 |
| Prevotella ruminicola | |||||
| D5ESF9 | NqrC | 5.05 | 12.20 | 3 | 3 |
| Prevotella ruminicola | |||||
| A0A0D0J042 | NqrF | 3.99 | 11.30 | 2 | 3 |
| Prevotella sp. P5-119 | |||||
| R5P524 | NqrA | 2.00 | 8.30 | 1 | 3 |
| Prevotella sp. CAG:1092 | |||||
| D5ESG0 | NqrB | 1.67 | 2.60 | 1 | 1 |
| Prevotella ruminicola | |||||
| D1VWD5 | NqrA | -2.00 | 7.00 | 2 | 3 |
| Prevotella timonensis | |||||
| A0A099BUQ4 | NqrA | -2.00 | 6.00 | 1 | 3 |
| Prevotella sp. S7 MS 2 | |||||
| D5EZ70 | El. transp. complex RnfG | 3.98 | 16.60 | 2 | 2 |
| Prevotella ruminicola | |||||
| D5EWF4 | Succ. dehyd./fum. reduc., flavopr. | 251.44 | 41.3 | 9 | 25 |
| Prevotella ruminicola | |||||
| D5EWF3 | Succ. dehyd./fum. reduc., Fe-S pr. | 133.55 | 51.6 | 1 | 13 |
| Prevotella ruminicola | |||||
| D3HYQ8 | Succ. dehyd./fum. reduc. Fe-S sub. | 9.28 | 53.6 | 0 | 13 |
| Prevotella buccae | |||||
| R6CJ62 | Fumarate reductase, Fe-S pr. | 9.13 | 47.2 | 0 | 11 |
| Prevotella copri | |||||
| A0A096C9D9 | Succinate dehydrogenase | 7.36 | 22.7 | 1 | 15 |
| Prevotella amnii | |||||
| D8DXM5 | Fumarate reductase, Fe-S pr. | 5.58 | 45.2 | 0 | 11 |
| Prevotella bryantii B14 | |||||
| L0JDX3 | Menaqui.-Succ. dehyd./fum. reduc. | 3.93 | 17.6 | 1 | 11 |
| Prevotella dentalis | |||||
| L1MZ30 | Succ. dehyd./fum. reduc., flavopr. | 3.78 | 26.4 | 1 | 16 |
| Prevotella saccharolytica | |||||
| R7F4R5 | Succ. dehyd./fum. reduc., flavopr. | 2.67 | 19 | 0 | 12 |
| Prevotella sp. CAG:485 | |||||
| D7NDJ3 | Fumarate reductase, Fe-S pr. | 2.50 | 40.5 | 2 | 10 |
| Prevotella oris | |||||
| U2QH41 | Succ. dehyd./fum. reduc., flavopr. | 1.45 | 17.9 | 1 | 12 |
| Prevotella baroniae | |||||
| A0A096BN50 | Succinate dehydrogenase | 1.18 | 46.4 | 0 | 12 |
| Prevotella buccalis | |||||
| F8NCB8 | Succinate dehydrogenase sub. B | -2.00 | 17.5 | 0 | 4 |
| Prevotella multisaccharivorax | |||||
| R5GTK6 | Succ. dehyd./fum. reduc. Fe-S sub. | -2.00 | 44 | 0 | 10 |
| Prevotella sp. CAG:755 |
| SampleFraction | Uniprot Accession no. | Identified Proteins of Prevotella bryantii B14 | Score (Mascot/PD) | Sequence Coverage (%) | Unique Peptides | Total Peptides |
|---|---|---|---|---|---|---|
| CD | D8DWC1 | NqrA | 275.00 | 17.37 | 7 | 7 |
| D8DWB9 | NqrC | 151.17 | 30.95 | 6 | 6 | |
| D8DYE1 | El. transp. complex RnfG (FMN) | 95.90 | 23.66 | 3 | 3 | |
| D8DWN9 | NuoCD | 117.97 | 7.82 | 3 | 3 | |
| D8DWN7 | NuoI | 63.74 | 7.25 | 2 | 2 | |
| D8DWP0 | NuoB | 57.69 | 9.45 | 2 | 2 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 619.21 | 51.19 | 13 | 13 | |
| D8DXM6 | Succinate dehydrogenase | 475.15 | 22 | 15 | 15 | |
| MP | D8DWB9 | NqrC | 137.82 | 17.14 | 3 | 3 |
| D8DWC1 | NqrA | 113.57 | 11.58 | 5 | 5 | |
| D8DYE1 | El. transp. complex RnfG (FMN) | 196.68 | 32.82 | 5 | 5 | |
| D8DXV4 | El. transp. complex RnfG | 110.26 | 15.26 | 2 | 2 | |
| D8DWN9 | NuoCD | 36.65 | 3.82 | 2 | 2 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 477.08 | 51.19 | 14 | 14 | |
| D8DXM6 | Succinate dehydrogenase | 243.90 | 15.48 | 11 | 11 |
| NQR Subunit | Uniprot Accession No. | RNF Subunit | Uniprot Accession No. | Identity (%) | NDH-I Subunit | Uniprot Accession No. | Identity (%) |
|---|---|---|---|---|---|---|---|
| NqrA | D8DWC1 | RnfC | D8DXV6 | 25.22 | NuoH | D8DWN8 | 18.44 |
| NqrB | D8DWC0 | RnfD | D8DXV5 | 30.28 | NuoK | D8DWN5 | 26.88 |
| NqrC | D8DWB9 | RnfG | D8DYE1 | 27.05 | NuoI | D8DWN7 | 26.83 |
| NqrD | D8DWB8 | RnfE | D8DXV3 | 37.36 | NuoN | D8DX02 | 19.08 |
| NqrE | D8DWB7 | RnfA | D8DXV2 | 44.50 | NuoL | A0A1H9A8K0 | 16.67 |
| NqrF | D8DWB6 | RnfB | D8DXV7 | 17.34 | NuoCD | D8DWN9 | 17.48 |
| Protein Band | Uniprot Accession No. | Identified Proteins of Prevotella bryantii B14 | Score (Mascot/PD) | Sequence Coverage (%) | Unique Peptides | Total Peptides |
|---|---|---|---|---|---|---|
| Triton 1%—A | D8DWC1 | NqrA | 259.37 | 24.50 | 10 | 10 |
| D8DXV4 | El. transp. complex RnfG | 87.48 | 16.84 | 2 | 2 | |
| D8DWN8 | NuoH | 98.71 | 5.77 | 2 | 2 | |
| D8DXM6 | Succinate dehydrogenase | 1843.34 | 41.88 | 32 | 32 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 1272.00 | 63.89 | 16 | 16 | |
| Triton 1%—B | D8DWC1 | NqrA | 1126.93 | 60.58 | 26 | 27 |
| D8DWB6 | NqrF | 386.31 | 18.01 | 7 | 7 | |
| D8DWB9 | NqrC | 272.89 | 40.48 | 7 | 8 | |
| D8DWC0 | NqrB | 55.66 | 11.43 | 3 | 3 | |
| D8DWB8 | NqrD | 41.65 | 4.78 | 2 | 2 | |
| D8DWN8 | NuoH | 115.46 | 9.62 | 4 | 4 | |
| D8DWN7 | NuoI | 90.35 | 16.58 | 3 | 3 | |
| D8DX02 | NuoN | 81.47 | 4.50 | 2 | 2 | |
| D8DWP0 | NuoB | 70.53 | 6.30 | 2 | 2 | |
| D8DXM6 | Succinate dehydrogenase | 814.30 | 29.59 | 22 | 22 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 605.93 | 48.02 | 12 | 12 | |
| Triton 1%—C | D8DWC1 | NqrA | 1233.90 | 69.04 | 33 | 33 |
| D8DWB9 | NqrC | 348.79 | 41.43 | 8 | 9 | |
| D8DWB6 | NqrF | 160.70 | 10.19 | 4 | 4 | |
| D8DWC0 | NqrB | 44.90 | 6.49 | 2 | 2 | |
| D8DXV4 | El. transp. complex RnfG | 88.64 | 15.26 | 2 | 2 | |
| D8DWN8 | NuoH | 133.20 | 9.89 | 5 | 5 | |
| A0A1H9A8K0 | NuoL | 110.73 | 5.13 | 3 | 3 | |
| D8DWP0 | NuoB | 66.30 | 6.3 | 2 | 2 | |
| D8DWN9 | NuoCD | 61.14 | 4.2 | 2 | 2 | |
| D8DWN7 | NuoI | 39.70 | 11.4 | 2 | 2 | |
| D8DXM6 | Succinate dehydrogenase | 120.15 | 6.22 | 5 | 5 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 99.83 | 9.92 | 2 | 2 | |
| Triton 2%—A | D8DWC1 | NqrA | 103.85 | 11.58 | 4 | 4 |
| D8DWN8 | NuoH | 73.76 | 5.77 | 2 | 2 | |
| D8DXM6 | Succinate dehydrogenase | 2189.86 | 41.88 | 32 | 32 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 1159.13 | 60.71 | 15 | 15 | |
| Triton 2%—B | D8DWC1 | NqrA | 791.13 | 45.66 | 19 | 20 |
| D8DWB6 | NqrF | 255.86 | 14.45 | 6 | 6 | |
| D8DWB9 | NqrC | 176.80 | 18.57 | 4 | 4 | |
| D8DWN8 | NuoH | 122.62 | 9.62 | 4 | 4 | |
| D8DWP0 | NuoB | 67.36 | 6.30 | 2 | 2 | |
| D8DWN9 | NuoCD | 63.92 | 4.20 | 2 | 2 | |
| D8DXM6 | Succinate dehydrogenase | 822.69 | 26.25 | 18 | 18 | |
| D8DXM5 | Fumarate reductase, Fe-S pr. | 537.00 | 54.37 | 13 | 13 | |
| Triton 2%—C | D8DWC1 | NqrA | 1218.80 | 63.25 | 30 | 30 |
| D8DWB6 | NqrF | 464.91 | 24.64 | 8 | 9 | |
| D8DWB9 | NqrC | 242.60 | 34.76 | 6 | 7 | |
| D8DWC0 | NqrB | 66.45 | 11.17 | 3 | 3 | |
| D8DWB8 | NqrD | 41.62 | 4.78 | 2 | 2 | |
| D8DXV4 | El. transp. complex RnfG | 25.98 | 11.58 | 2 | 2 | |
| D8DWN8 | NuoH | 118.11 | 7.69 | 4 | 4 | |
| D8DWN7 | NuoI | 104.26 | 16.58 | 3 | 3 | |
| D8DWP0 | NuoB | 78.14 | 8.66 | 3 | 3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deusch, S.; Bok, E.; Schleicher, L.; Seifert, J.; Steuber, J. Occurrence and Function of the Na+-Translocating NADH:Quinone Oxidoreductase in Prevotella spp. Microorganisms 2019, 7, 117. https://doi.org/10.3390/microorganisms7050117
Deusch S, Bok E, Schleicher L, Seifert J, Steuber J. Occurrence and Function of the Na+-Translocating NADH:Quinone Oxidoreductase in Prevotella spp. Microorganisms. 2019; 7(5):117. https://doi.org/10.3390/microorganisms7050117
Chicago/Turabian StyleDeusch, Simon, Eva Bok, Lena Schleicher, Jana Seifert, and Julia Steuber. 2019. "Occurrence and Function of the Na+-Translocating NADH:Quinone Oxidoreductase in Prevotella spp." Microorganisms 7, no. 5: 117. https://doi.org/10.3390/microorganisms7050117