3. Results and Discussion
Through anaerobic isolation and cultivation, a rod-shaped (1.0 μm long and 1.0–4.0 μm wide), non-motile, non-spore-forming and Gram-positive obligate anaerobic microorganism was obtained. Gas chromatograph showed that 4.37 mmol/L butyric acid, 19.88 mmol/L acetic acid, 38.67 mmol/L lactic acid and 2.19 mmol/L benzoic acid were produced after 72 h of fermentation in PYG medium. TF06-26 could grow at 25 °C to 45 °C (optimum 37 °C) with pH 5.0–8.0 (optimum pH 7.0) as well as tolerate 0.3% (
w/
v) cholate and 2.0% (
w/
v) salt. The antibiotic susceptibility test demonstrated that this strain was resistant to kanamycin, amikacin and framycetin, whilst being sensitive towards ampicillin, carbenicillin and cefazolin. The general features of TF06-26 are showed in
Table 1; more details of the growth conditions and antibiotic susceptibility test are shown in
Tables S1 and S2.
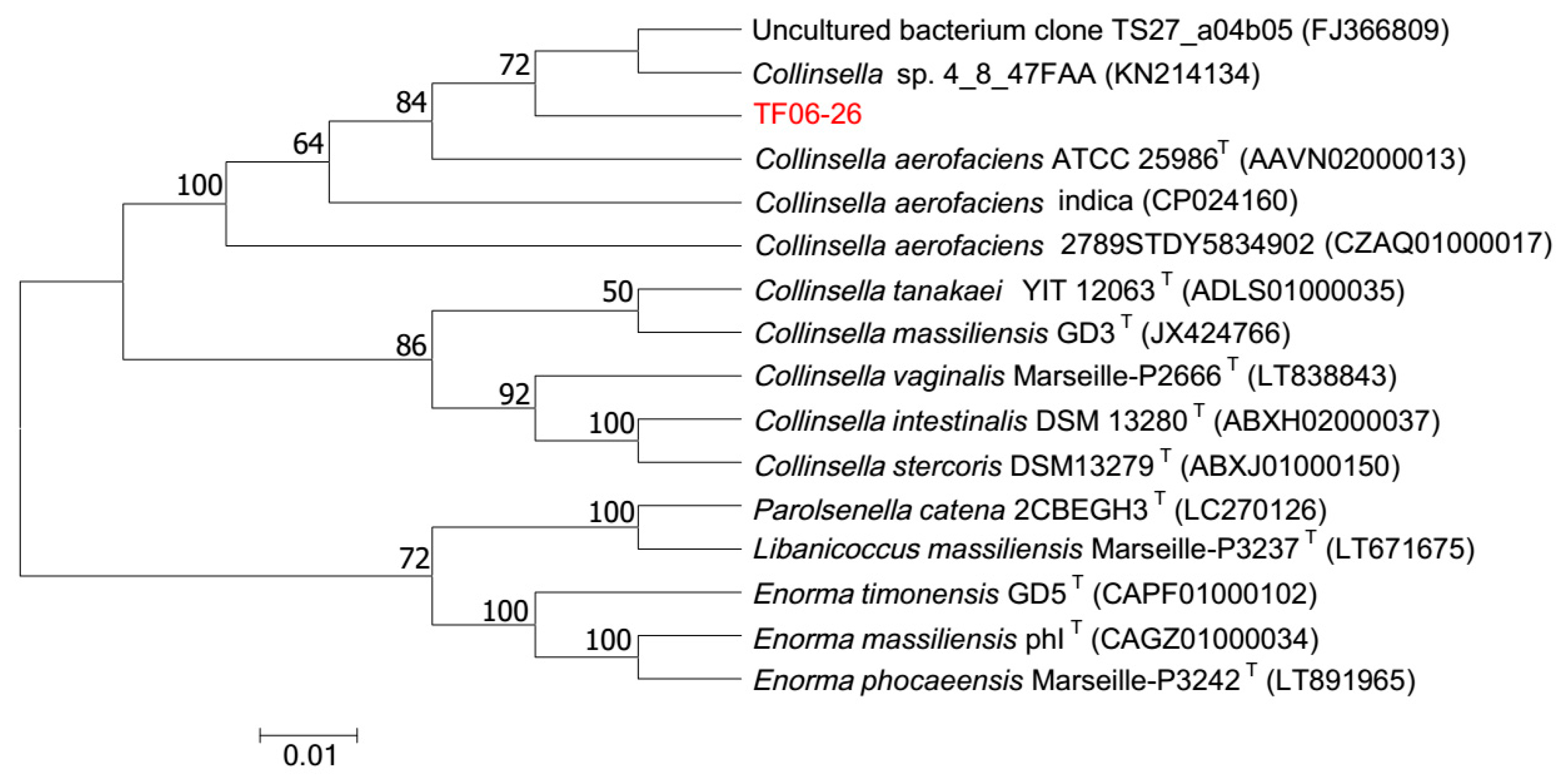
To determine the phylogenetic position of TF06-26, the 1494 bp 16S rRNA gene and 2,260,546 bp genome were obtained. The 16S rRNA gene sequence was compared against the EzBioCloud database, and the sequence alignments showed that TF06-26 was phylogenetically close to the genus
Collinsella, especially the species
C.
aerofaciens. TF06-26 exhibited 99.93%, 99.65% and 99.51% identity similarity with
C.
aerofaciens ATCC 25986
T,
C.
aerofaciens indica and
C.
aerofaciens 2789STDY5834902, respectively, and all values were higher than the threshold (98.65%) for differentiating two species [
27]. A dendrogram depicting the phylogenetic relationships of TF06-26 is shown in
Figure 1. TF06-26 formed a distinct subline associated with
C. sp. 4_8_47FAA (100.00% similarity), the uncultured bacterium clone TS27_a04b05 (96.25% similarity),
C.
aerofaciens ATCC 25986
T,
C.
aerofaciens indica and
C.
aerofaciens 2789STDY5834902. These results suggest that it is likely that TF06-26 can be ascribed to the species
C.
aerofaciens.
Digital DDH relatedness and ANI were performed between TF06-26 and nine type strains of the genus
Collinsella to verify the phylogenetic position of TF06-26. More details of these strains are shown in
Table S3. Consistent with the results of 16s rRNA gene similarity, both digital DDH relatedness and ANI values showed that TF06-26 is most closely related to the type strains of the species
C.
aerofaciens (
Table 2). However, 65.7% DDH relatedness and 93.08% ANI values were slightly below the common threshold for the same species (70% DDH and 95% ANI). Considering the extremely high 16S rRNA similarity, four
C.
aerofaciens strains were added for comparison. These four strains were simultaneously assigned to
C.
aerofaciens by the NCBI genome database and the GTDB [
28], which confirmed their phylogenetic positions. TF06-26 exhibited 71.19–75.9% digital DDH relatedness with these four
C.
aerofaciens strains, which were higher than the 70% threshold (
Table 3). However, 93.25–95.05% OrthoANI values and 93.07–94.93% ANIb values within
C.
aerofaciens strains suggested that a 95% common threshold is not suitable for identifying this species, and 94.22% (TF06-26 with
C.
aerofaciens 2789STDY5834902), 94.13% (TF06-26 with
aerofaciens indica) and 93.43% (TF06-26 with
C.
aerofaciens 2789STDY5608842) OrthoANI values were in the reasonable range among
C.
aerofaciens strains (
Table 4 and
Table S4). Based on the above information, TF06-26 was assigned to the species
C.
aerofaciens.
The ad hoc committee for the reconciliation of approaches to bacterial systematics advised that bacterial isolates within a given species be considered as distinct subspecies if they differ phenotypically [
29]. Carbon source metabolism and growth condition tests were undertaken to compare the phenotype characteristics between TF06-26 and the type strains of the species
C.
aerofaciens. Fermenting D-lactose, sucrose, salicin, D-galactose, D-fructose, D-mannose, arbutin, esculine, cellobiose, D-maltose and gluconate distinguished TF06-26 and
C.
aerofaciens ATCC 25986
T. Additionally, TF06-26 exhibited a stronger ability to adapt to acidic and high-cholate environments with positive growth at pH 5.0 and 2% (
w/
v) cholate, in contrast to
C.
aerofaciens ATCC 25986
T (
Table 5). Furthermore, TF06-26 was able to produce butyric acid and benzoic acid, and these distinct characteristics indicate that TF06-26 is a new subspecies of species
C.
aerofaciens.
As well as comparing strain types, this study also attempted to find different characteristics between TF06-26 and other strains of the species
C. aerofaciens. Since limited phenotypic information of other
C. aerofaciens strains has been reported, a comparative genome analysis was performed to further investigate genomic differences. Four non-type strains and eight additional type strains of the genus
Collinsella were included. The pan-genome of these 14 genomes consisted of 535 core genes, 2622 accessory genes and 4160 unique genes (
Figure S1,
Table S5). When compared with additional type strains, the
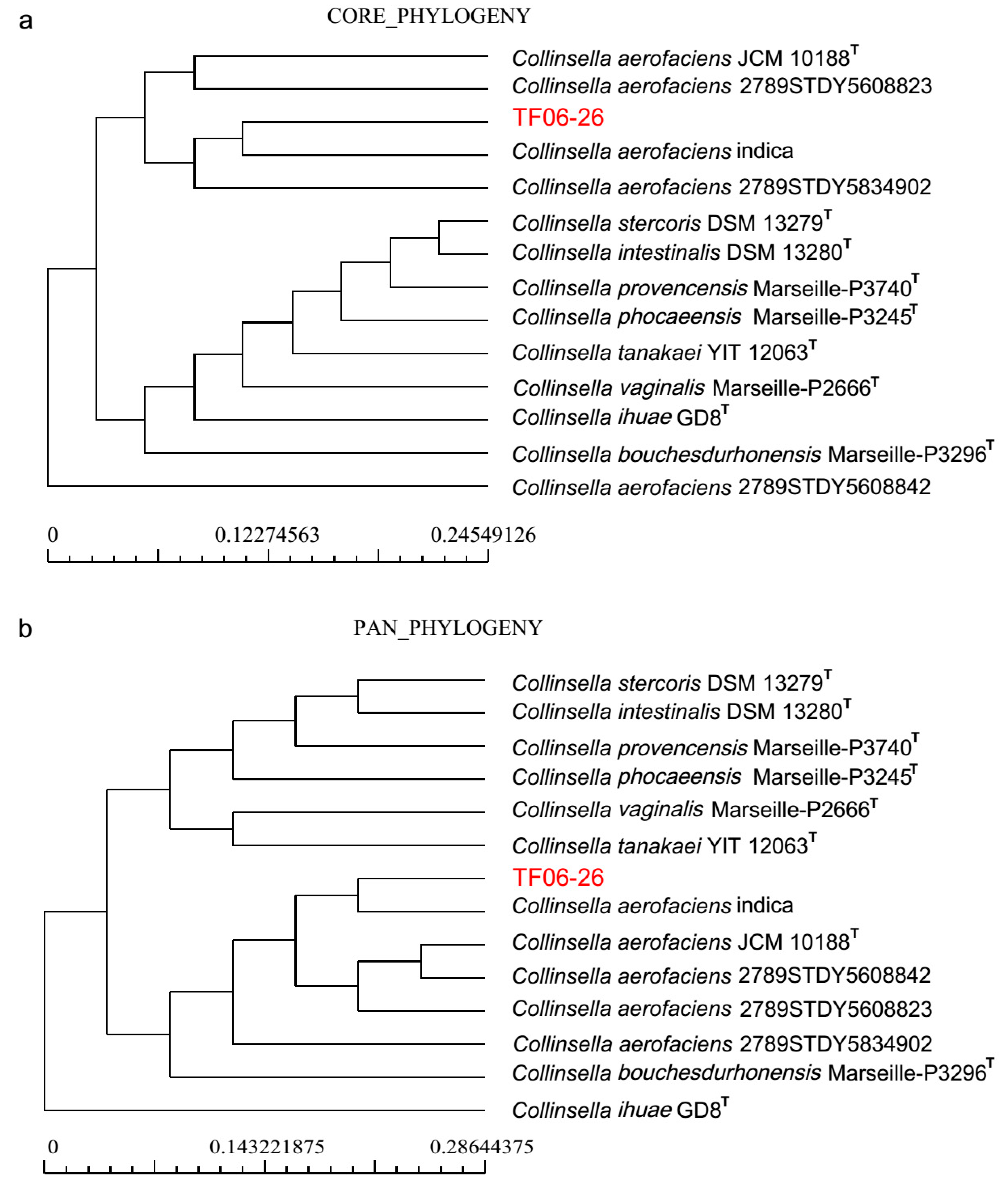
C. aerofaciens strains and TF06-26 had more accessory genes and fewer unique genes. The dendrogram of both core genes and pan genes show that
C.
aerofaciens indica has the closest phylogenetic relationship with TF06-26 (
Figure 2). The KEGG functional annotation of all 14 genomes indicates that there is a higher level of unique genes in carbohydrate metabolism function, and the same trend can be observed for core genes in translation function (
Figure S2a). Identifying COG functional categories showed that core genomes were enriched in the (J) category (translation, ribosomal structure and biogenesis), while accessory genes and unique genes could mostly be ascribed to the (R) category (general function prediction only), the (G) category (carbohydrate transport and metabolism) and the (K) category (transcription) (
Figure S2b). In our new strain TF06-26, 130 genes were specific. Functional annotation of these unique genes showed that butyric acid kinase (EC:2.7.2.7), phosphate butyryltransferase (EC:2.3.1.19), mannose-specific IIB component (EC:2.7.1.191) and mannose-specific IIC component were only coded by TF06-26 (
Table 6). The two former enzymes are related to butyric acid biosynthesis and the two latter enzymes are part of the mannose transportation process. This suggests that only TF06-26 has the ability to produce butyric acid and fermentation mannose within the genus
Collinsella.