EpiExploreR: A Shiny Web Application for the Analysis of Animal Disease Data
Abstract
1. Introduction
2. Materials and Methods
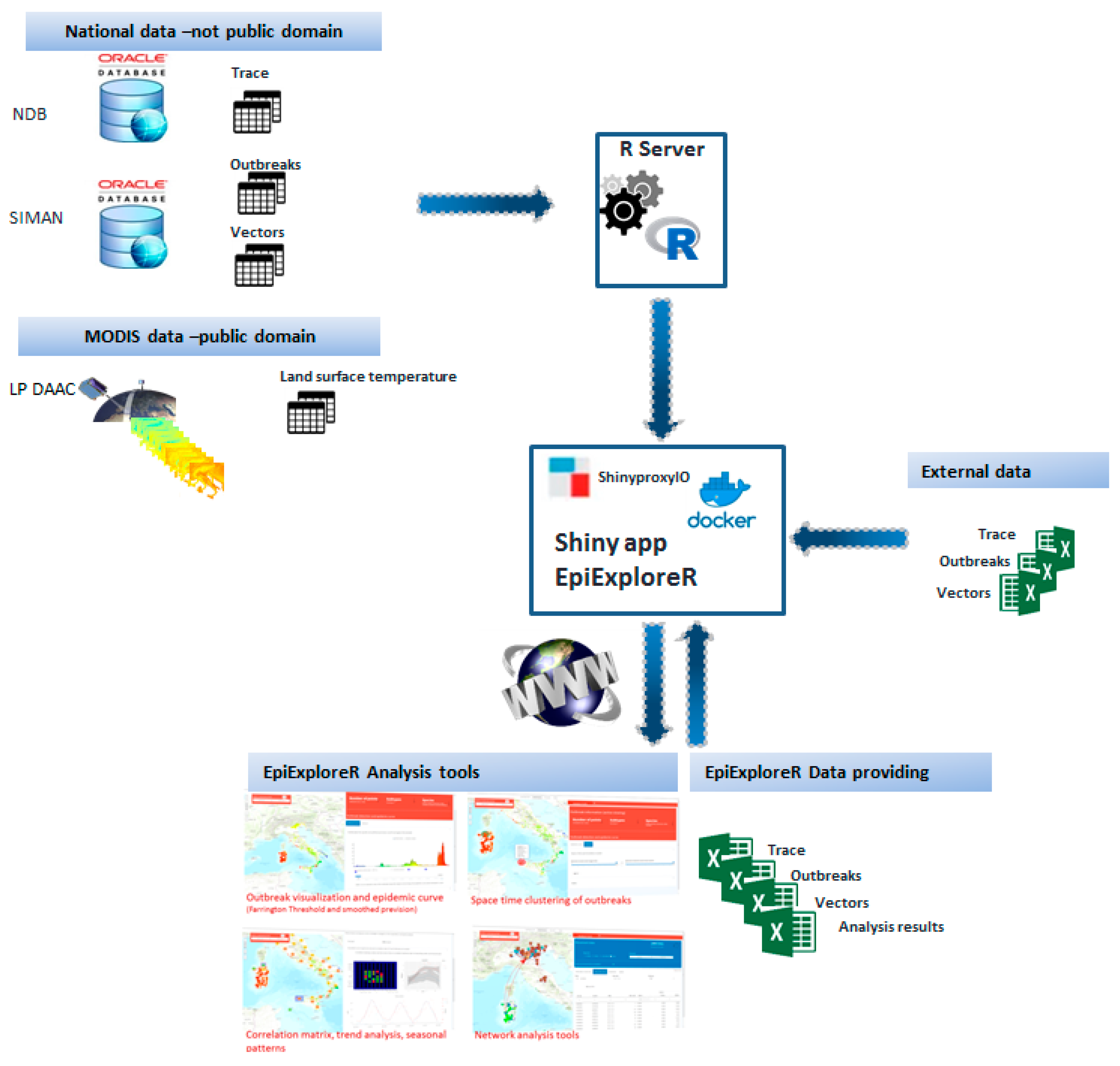
2.1. EpiExploreR Implementation and Development
- Accessing and exploring different sources of geo-referenced, nearly real-time data, including notified outbreaks, surveillance of vectors, animal movements and remotely sensed data;
- Applying base methods for early outbreak detection (e.g., Farrington algorithm, spatiotemporal cluster analysis and data correlation tools);
- Running and calibrating temperature-driven mosquito models;
- Performing network analysis useful in the identification of disease transmission patterns.
2.2. Data Collection and Data Flow
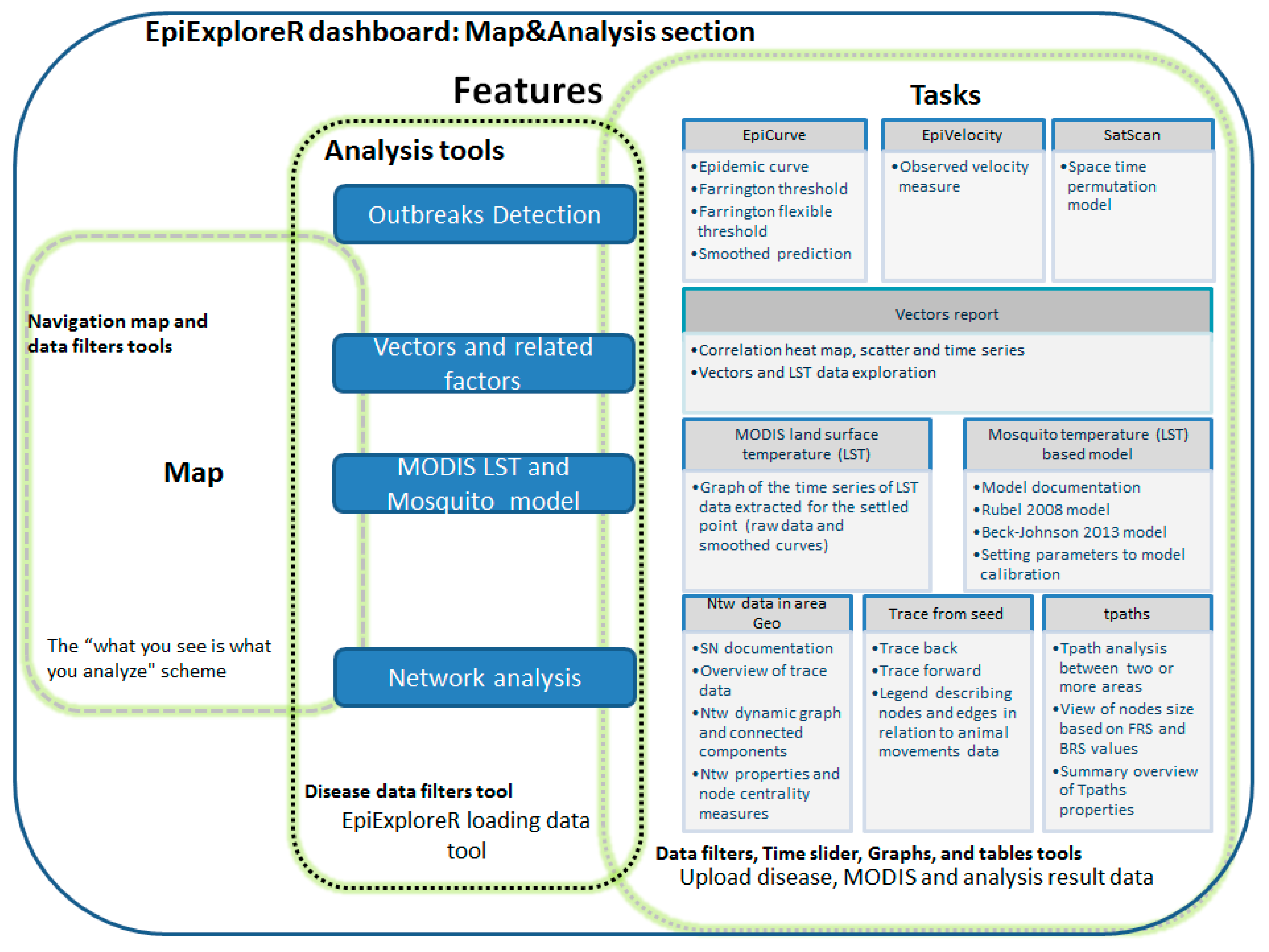
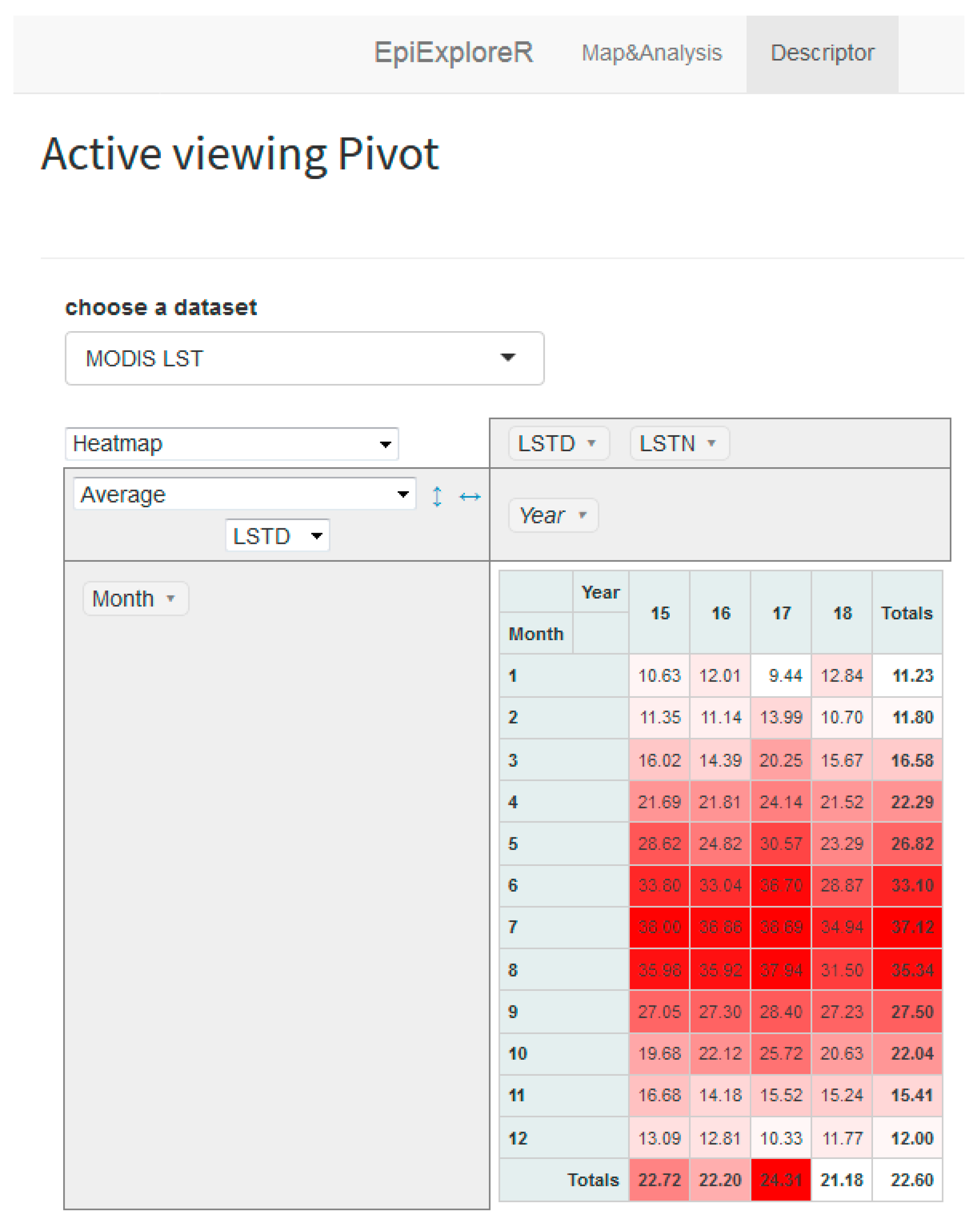
2.3. Spatiotemporal Methods and the Epiexplorer Dashboard Features
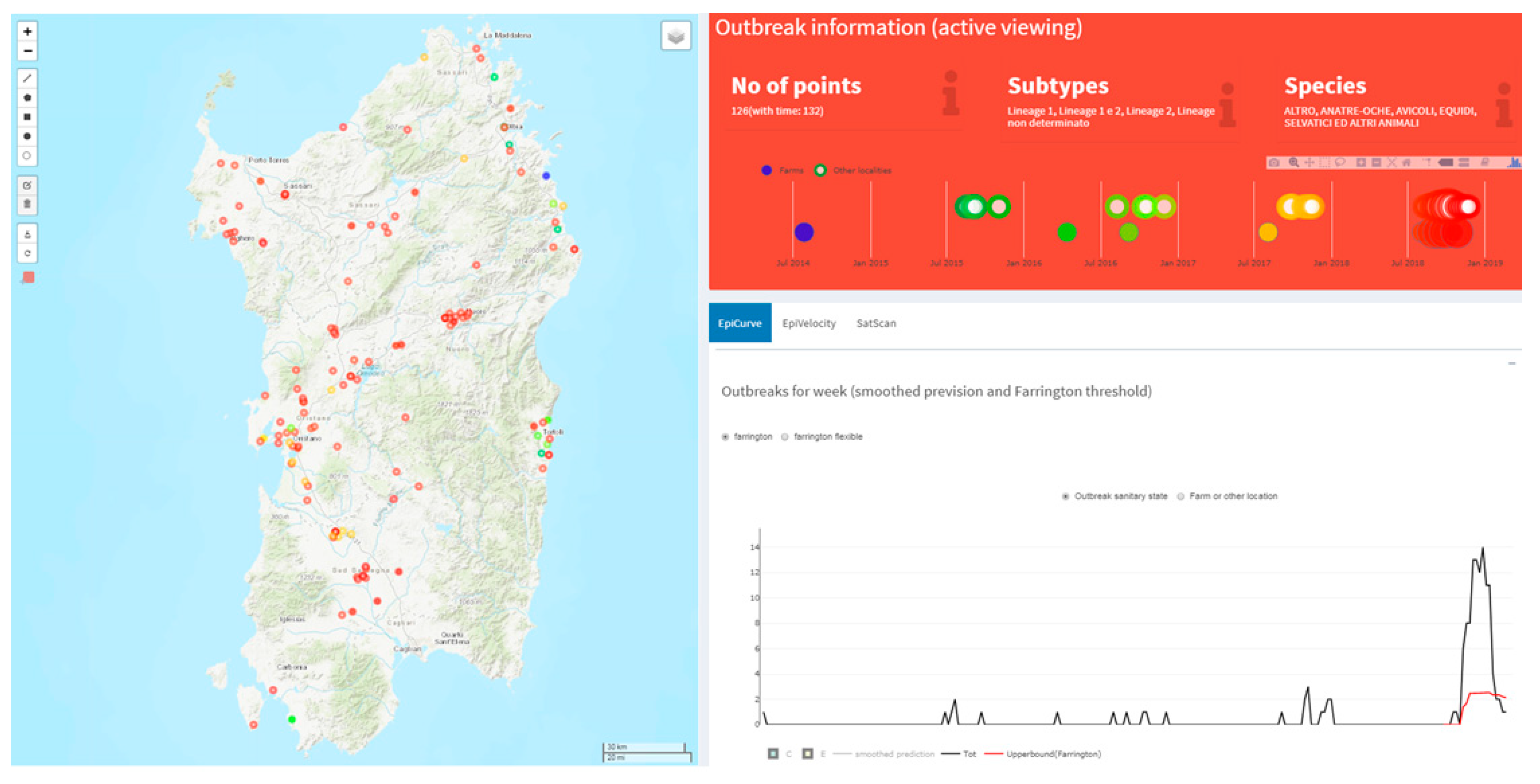
2.3.1. Disease Mapping
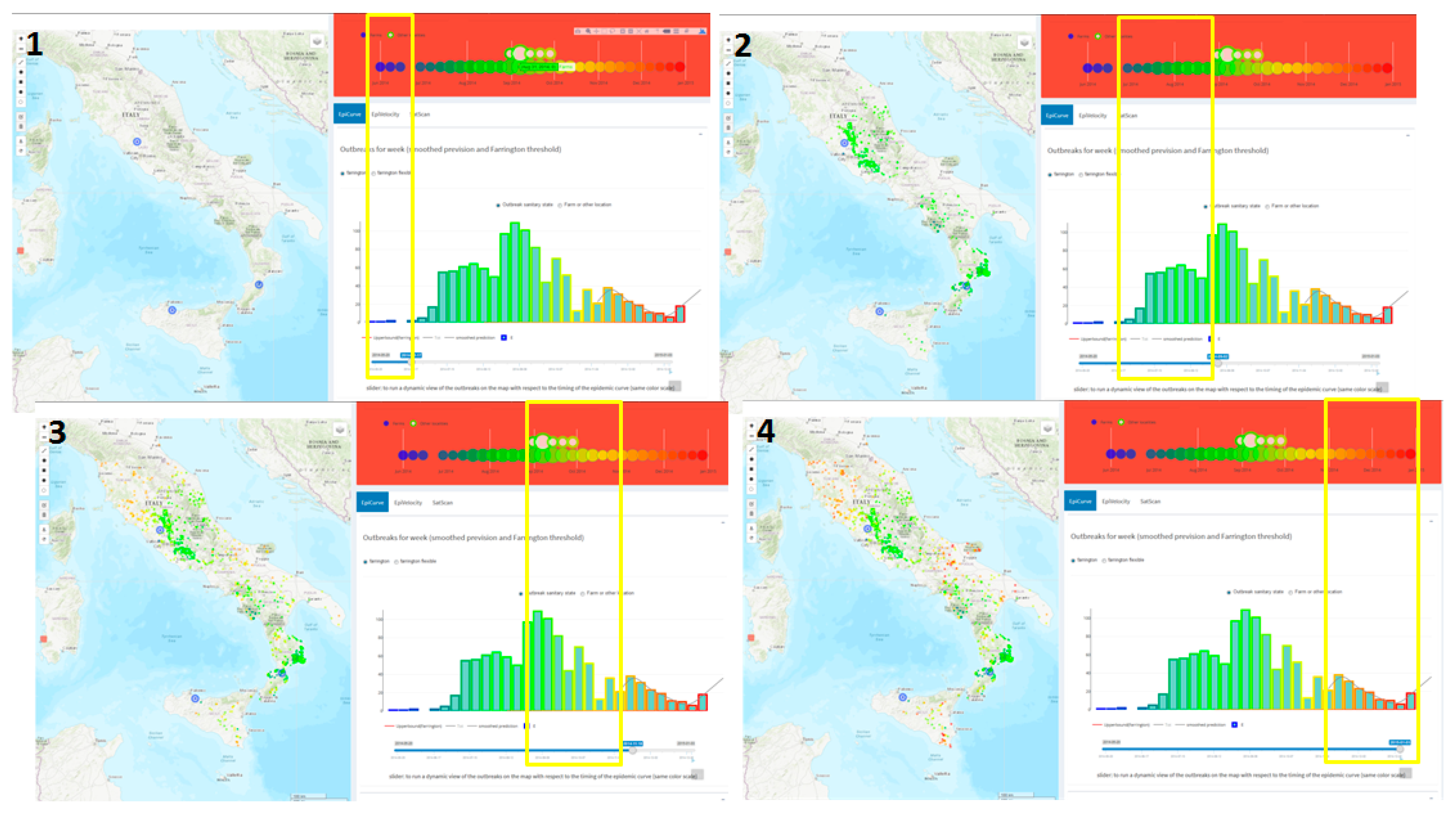
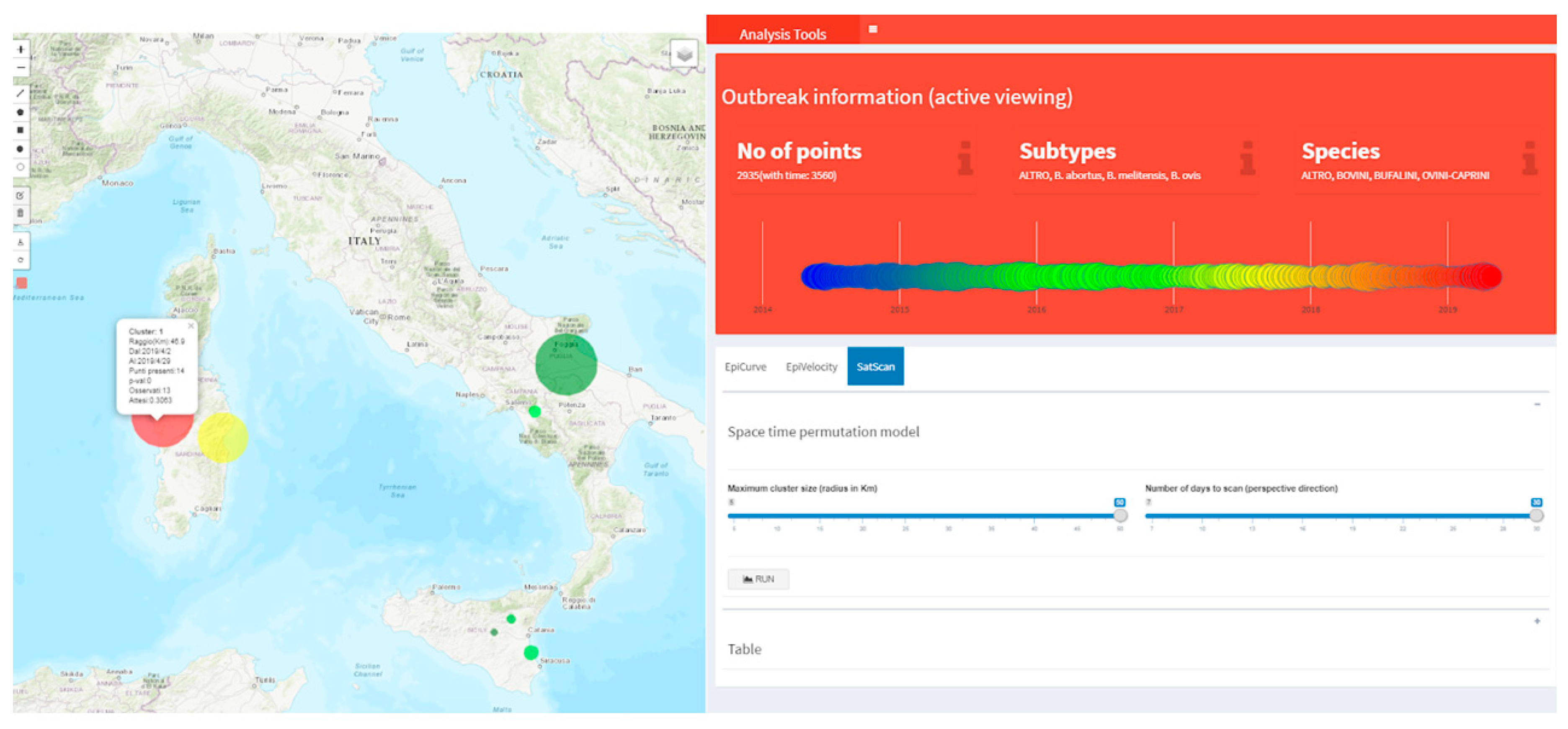
2.3.2. Early Outbreak Detection (EOD) Methods
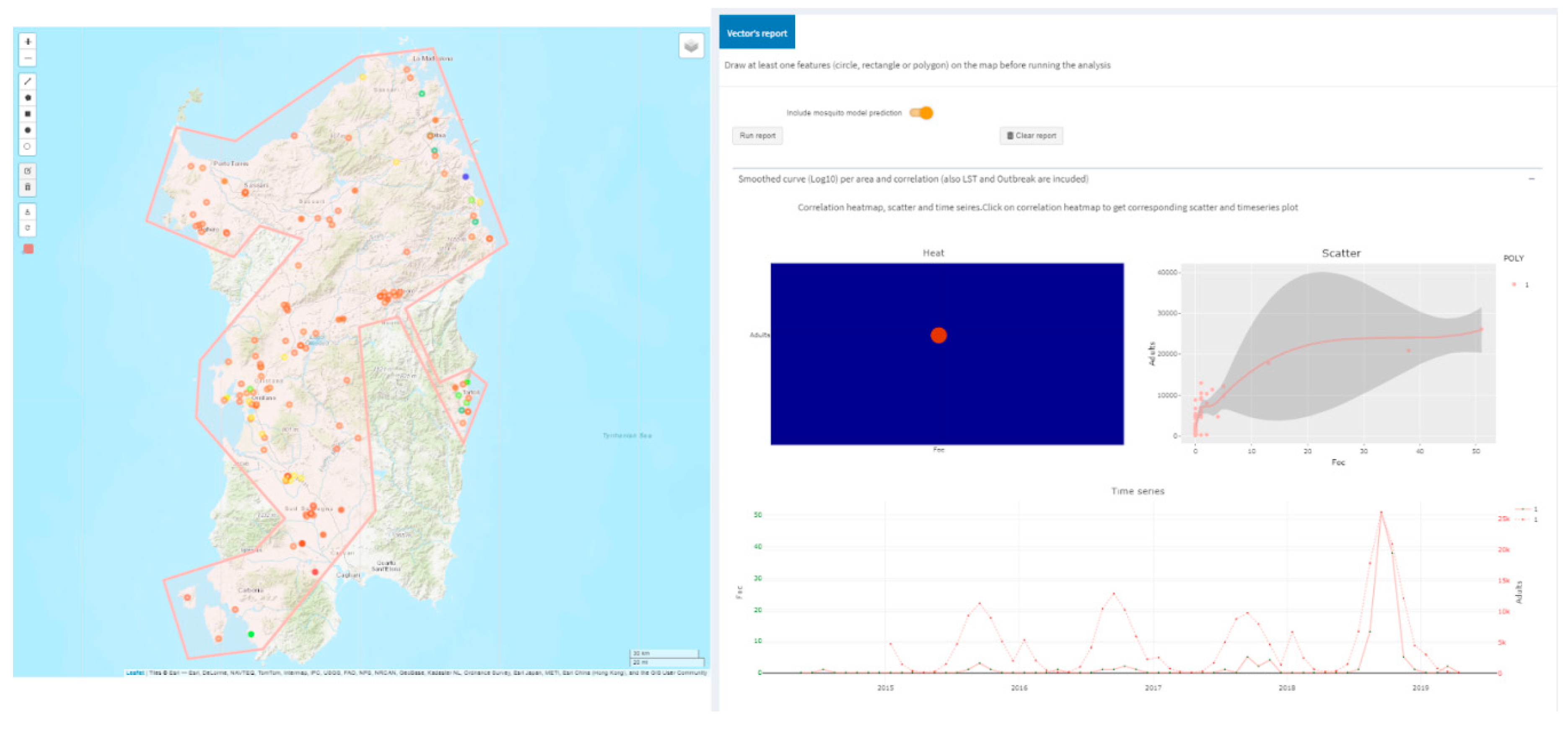
2.3.3. Temperature Driven Mosquito Modeling
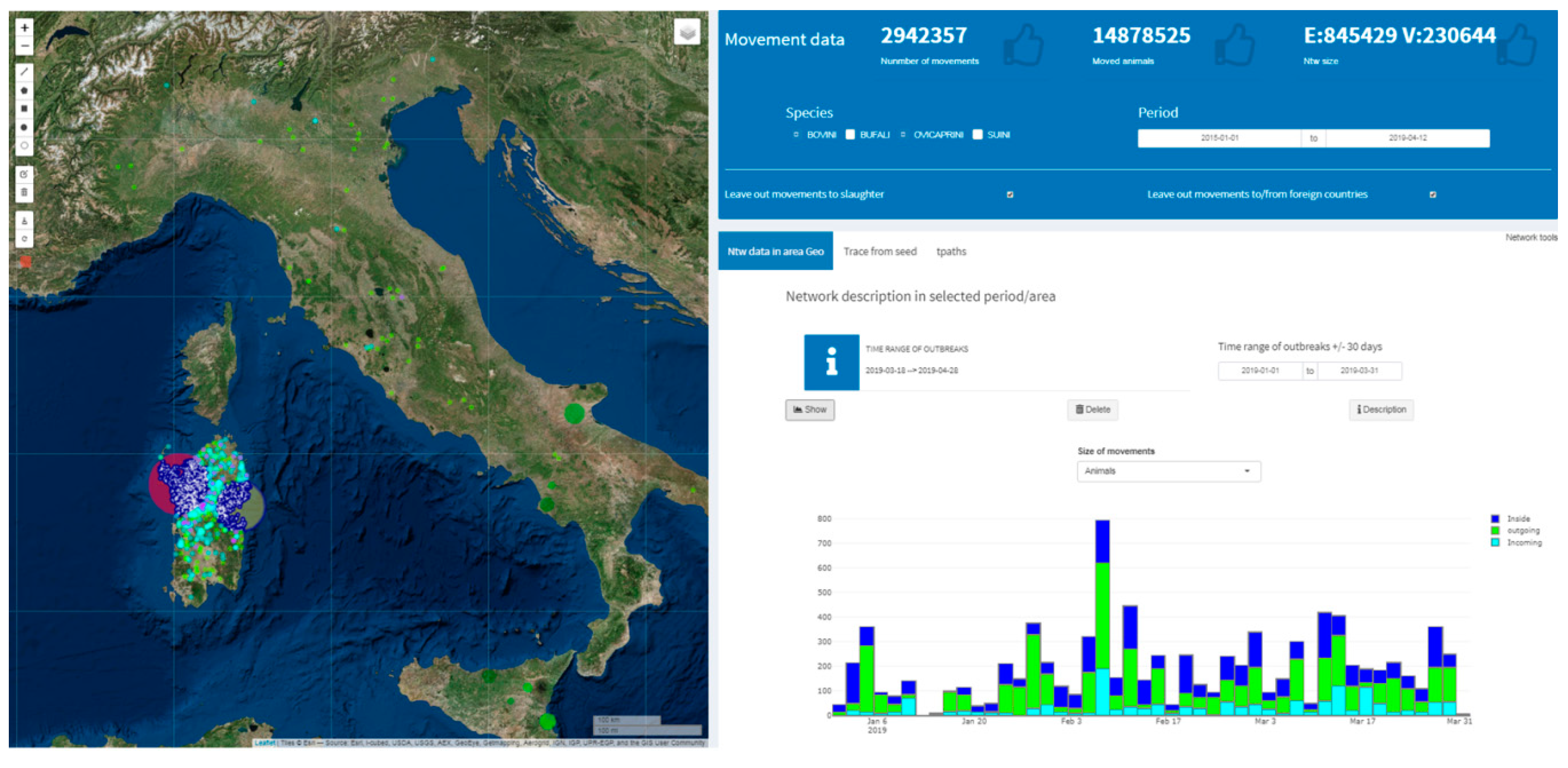
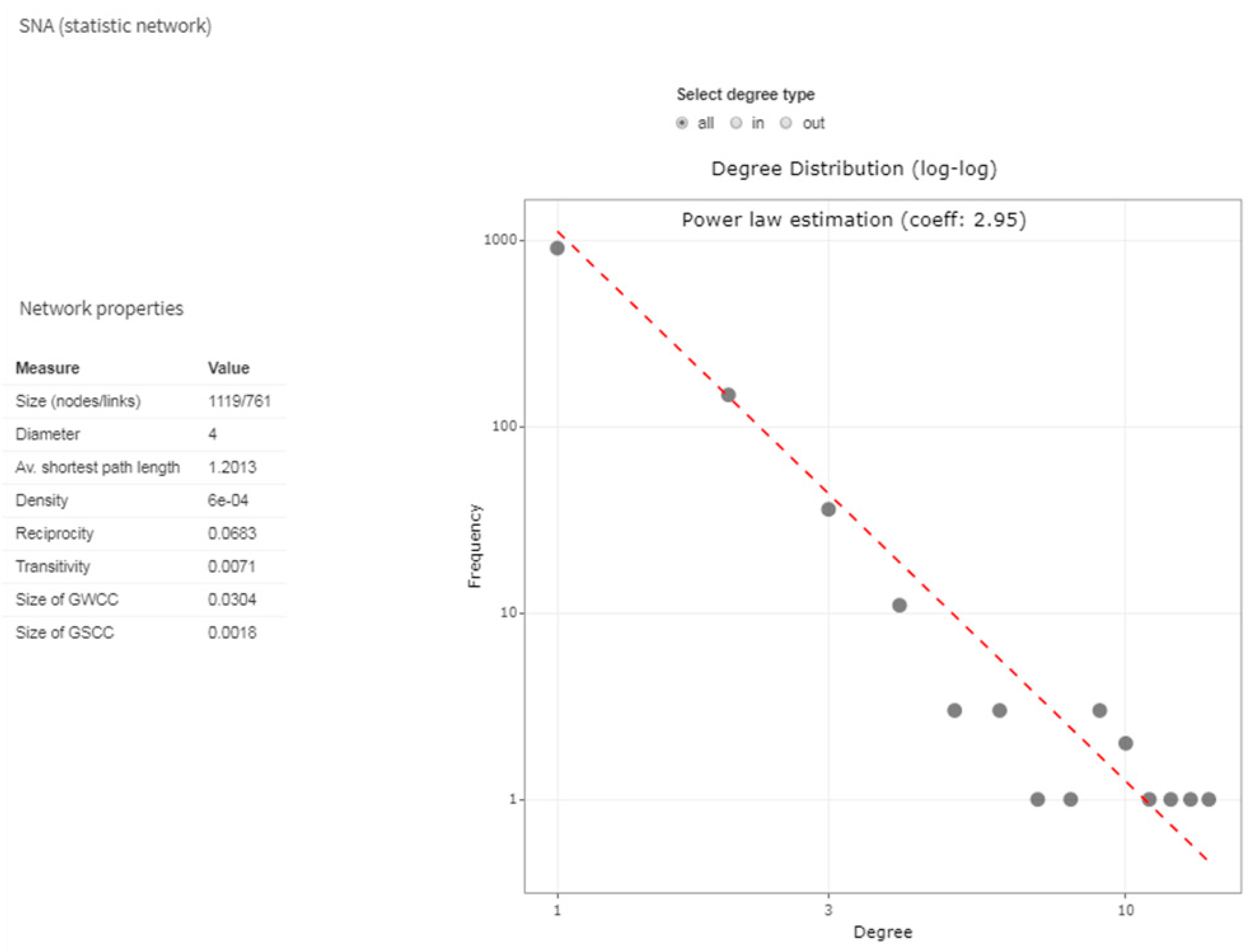
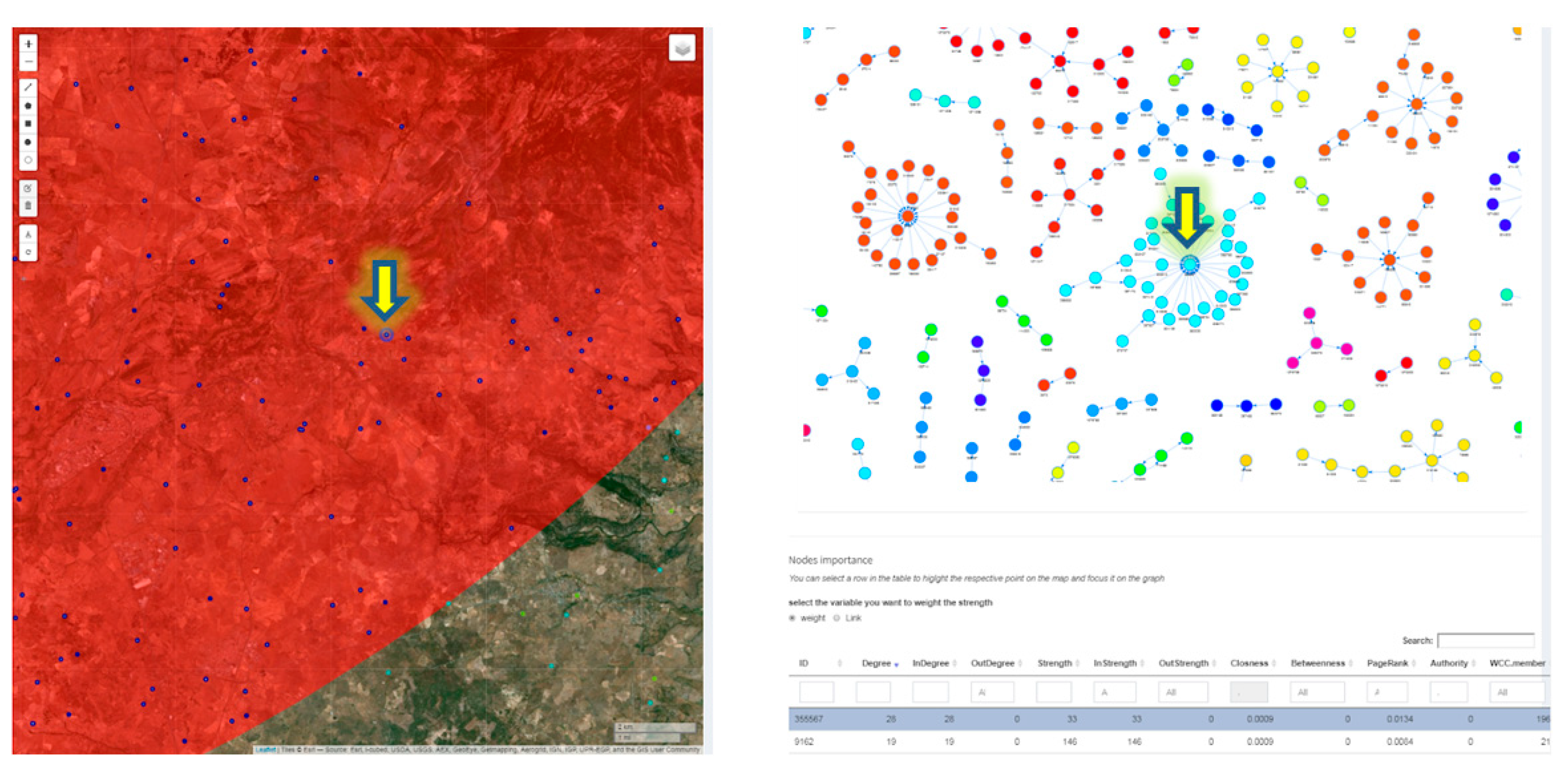
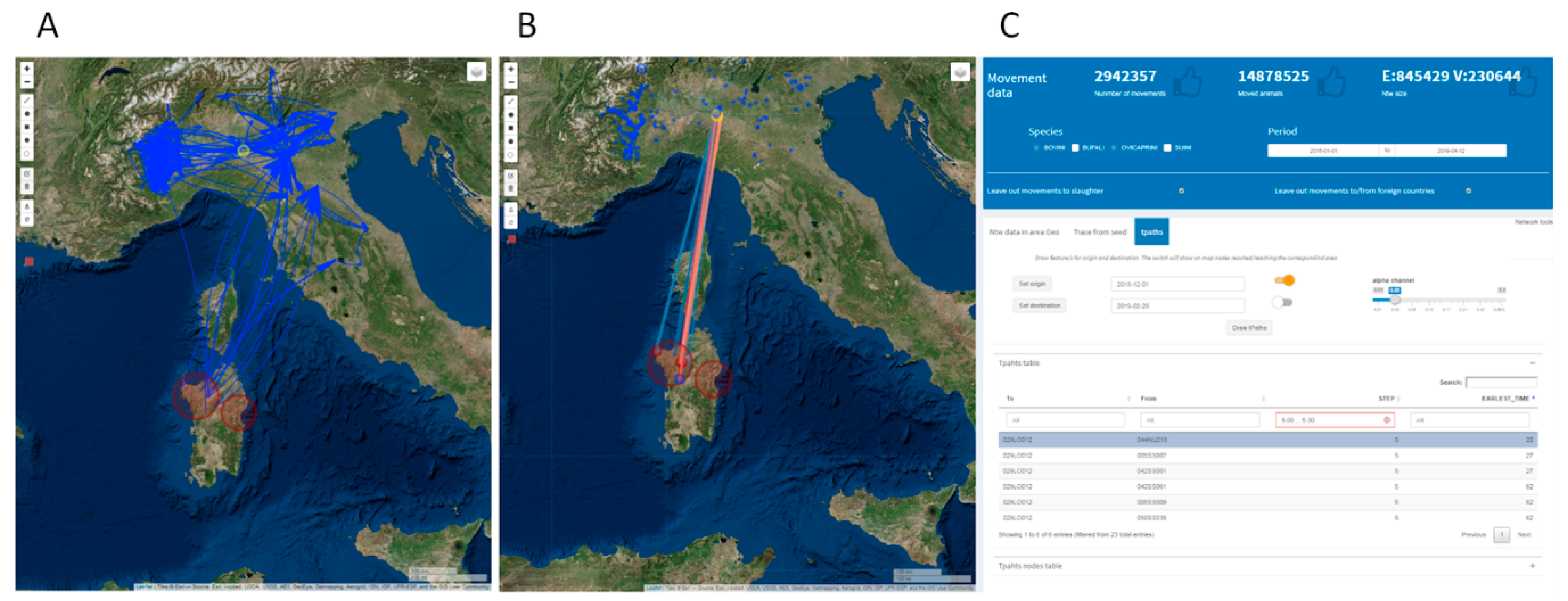
2.3.4. Network Analysis in Livestock Mobility
3. Results
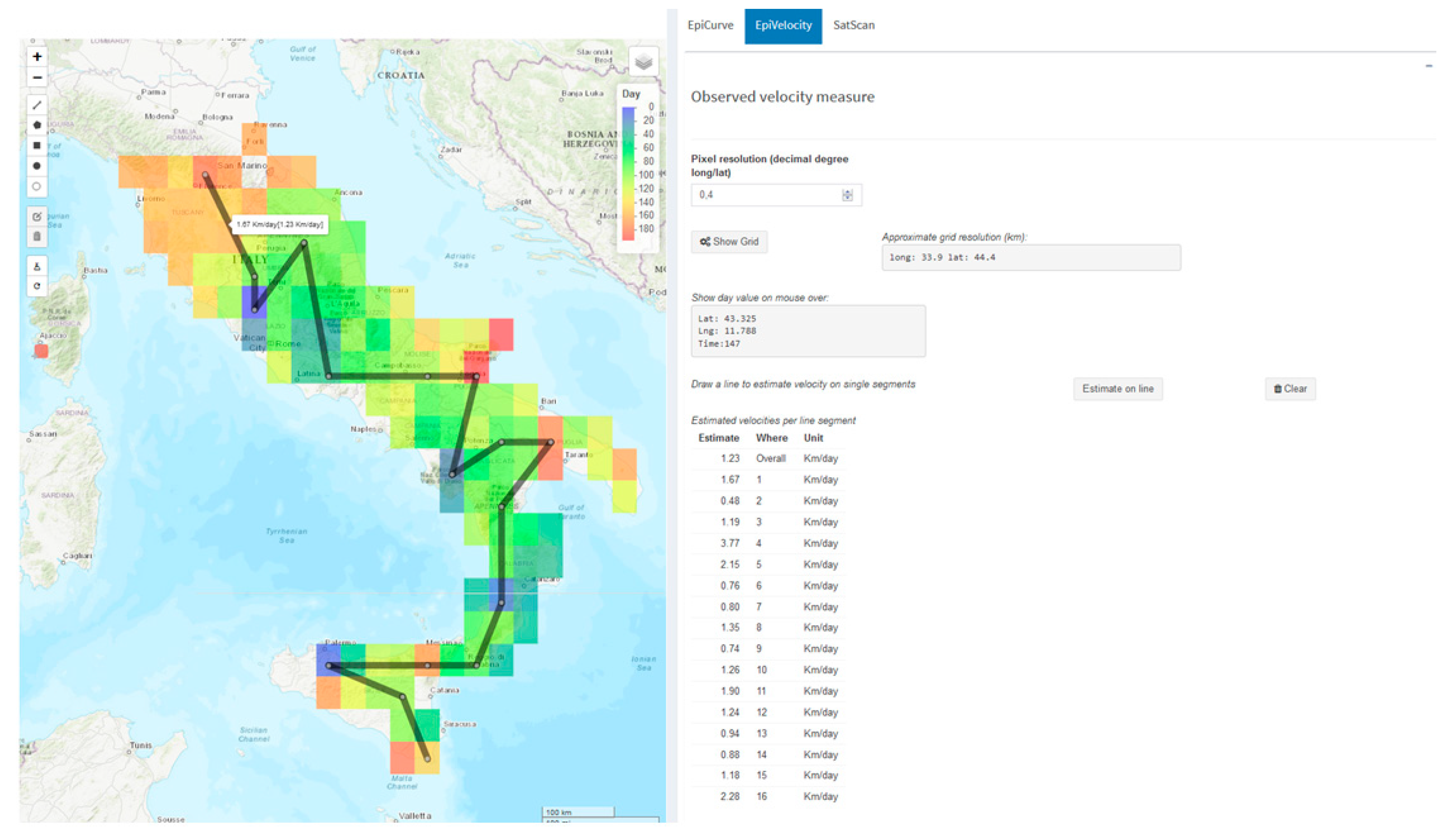
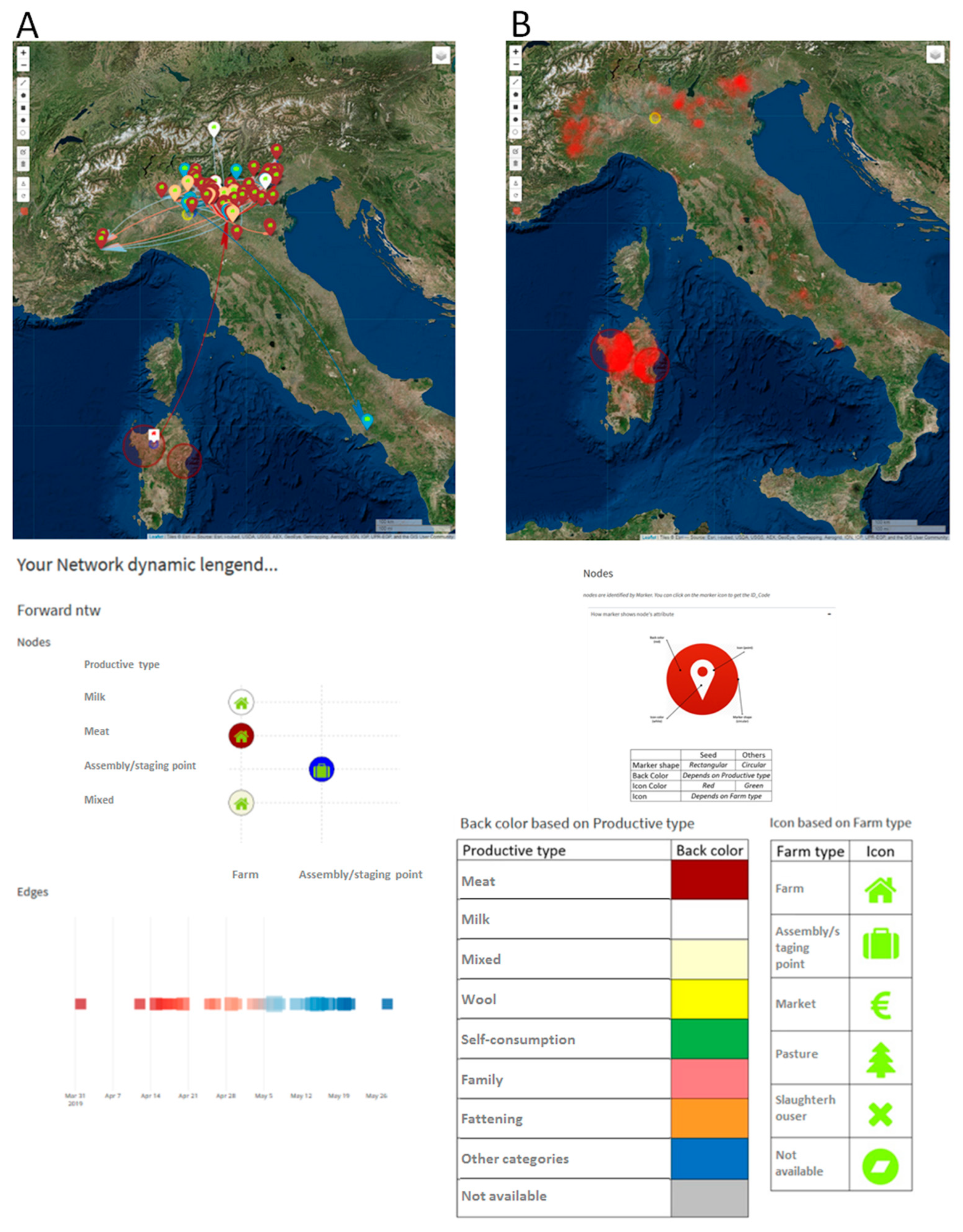
3.1. The Estimated Velocity of the BTV-1 Spreading in Central Italy During 2014
3.2. West Nile Disease (WND) in Sardinia Region
3.3. Alive Cluster Detection for Brucellosis Disease and Evaluation of Its Introduction or Spread in Italy through Animal Movements Network Analysis
4. Discussion
- Load example data to explore the complete set of features for training purposes;
- Load external data to perform the analysis using self-owned data (in both the public and private versions);
- Load data provided by national data sources: NDB and SIMAN (only for private version).
- Upload an additional dataset (e.g., genomic sequences and animal density data) and use the appropriated spatiotemporal and mathematical models.
- Create custom epidemiological reports in HTML-format. Include tools to import and export spatial data (e.g., shapefiles) reproducibility of the performed analyses.
Author Contributions
Funding
Conflicts of Interest
References
- Buliva, E.; Elhakim, M.; Minh, T.; Nguyen, N.; Elkholy, A.; Mala, P.; Abubakar, A.; Malik, S.M.M.R. Emerging and Reemerging Diseases in the World Health Organization (WHO) Eastern Mediterranean Region—Progress, Challenges, and WHO Initiatives. Front. Public Health 2017, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Rabozzi, G.; Bonizzi, L.; Crespi, E.; Somaruga, C.; Sokooti, M.; Tabibi, R.; Vellere, F.; Brambilla, G.; Colosio, C. Emerging Zoonoses: The “One Health Approach.”. Saf. Health Work 2012, 3, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Belay, E.D.; Kile, J.C.; Hall, A.J.; Barton-Behravesh, C.; Parsons, M.B.; Salyer, S.; Walke, H. Zoonotic Disease Programs for Enhancing Global Health Security. Emerg Infect Dis. 2017, 23, S65. [Google Scholar] [CrossRef] [PubMed]
- OIE World Animal Health Information System (WAHIS). Available online: https://www.oie.int/en/animal-health-in-the-world/wahis-portal-animal-health-data/ (accessed on 10 December 2019).
- Animal Disease Notification System (ADNS)—European Commission. Available online: https://ec.europa.eu/food/animals/animal-diseases/not-system_en (accessed on 24 August 2017).
- TRACES: TRAde Control and Expert System—Food Safety—European Commission. Available online: https://ec.europa.eu/food/animals/traces_en (accessed on 5 April 2018).
- LP DAAC. NASA Land Data Products and Services. Available online: https://lpdaac.usgs.gov/ (accessed on 1 June 2017).
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2015, 43, D30–D35. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Surveillance Atlas of Infectious Diseases. 2018. Available online: https://atlas.ecdc.europa.eu/public/index.aspx (accessed on 31 December 2018).
- Pollett, S.; Althouse, B.M.; Forshey, B.; Rutherford, G.W.; Jarman, R.G. Internet-based biosurveillance methods for vector-borne diseases: Are they novel public health tools or just novelties? PLoS Negl. Trop. Dis. 2017, 11, e0005871. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.N.; Etter, E. Epidemiological surveillance methods for vector-borne diseases. Rev. Sci. Tech. Off. Int. Epizoot. 2015, 34, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, D.U.; Stevens, K.B. Spatial and temporal epidemiological analysis in the Big Data era. Prev. Vet. Med. 2015, 122, 213–220. [Google Scholar] [CrossRef]
- Christaki, E. New technologies in predicting, preventing and controlling emerging infectious diseases. Virulence 2015, 6, 558–565. [Google Scholar] [CrossRef]
- Steele, L.; Orefuwa, E.; Dickmann, P. Drivers of earlier infectious disease outbreak detection: A systematic literature review. Int. J. Infect. Dis. 2016, 53, 15–20. [Google Scholar] [CrossRef]
- Choi, J.; Cho, Y.; Shim, E.; Woo, H. Web-based infectious disease surveillance systems and public health perspectives: A systematic review. BMC Public Health 2016, 16, 1238. [Google Scholar] [CrossRef]
- Carroll, L.N.; Au, A.P.; Detwiler, L.T.; Fu, T.; Painter, I.S.; Abernethy, N.F. Visualization and analytics tools for infectious disease epidemiology: A systematic review. J. Biomed. Inf. 2014, 51, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Le Comber, S.C.; Fry, H.; Bull, M.; Leach, S.; Hayward, A.C. Spatial methods for infectious disease outbreak investigations: Systematic literature review. Eurosurveillance 2015, 20. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. The R Project for Statistical Computing 2013. Available online: http://www.R-project.org/ (accessed on 5 April 2018).
- Bender-deMoll, S.; Morris, M. tsna: Tools for Temporal Social Network Analysis. 2016. Available online: https://CRAN.R-project.org/package=tsna (accessed on 5 April 2018).
- Höhle, M. R Package “Surveillance”. 2014. Available online: http://surveillance.r-forge.r-project.org/ (accessed on 5 April 2018).
- Pebesma, E.; Bivand, R.; Rowlingson, B.; Gomez-Rubio, V.; Hijmans, R.; Sumner, M.; MacQueen, D.; Lemon, J.; O’Brien, J.; O’Rourke, J. sp: Classes and Methods for Spatial Data. 2018. Available online: https://CRAN.R-project.org/package=sp (accessed on 5 April 2018).
- Kleinman, K. rsatscan: Tools, Classes, and Methods for Interfacing with SaTScan Stand-Alone Software. 2015. Available online: https://CRAN.R-project.org/package=rsatscan (accessed on 5 April 2018).
- Butts, C.T.; Hunter, D.; Handcock, M.; Bender-deMoll, S.; Horner, J. network: Classes for Relational Data. 2018. Available online: https://CRAN.R-project.org/package=network (accessed on 5 April 2018).
- Csardi, G.; Nepusz, T. R Package ‘Igraph’. 2006. Available online: http://igraph.org (accessed on 5 April 2018).
- Muellner, U.; Fournié, G.; Muellner, P.; Ahlstrom, C.; Pfeiffer, D.U. epidemix—An interactive multi-model application for teaching and visualizing infectious disease transmission. Epidemics 2018, 23, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Moraga, P. SpatialEpiApp : A Shiny web application for the analysis of spatial and spatio-temporal disease data. Spat. Spatio-Temporal Epidemiol. 2017, 23, 47–57. [Google Scholar] [CrossRef]
- Nöremark, M.; Widgren, S. EpiContactTrace: An R-package for contact tracing during livestock disease outbreaks and for risk-based surveillance. BMC Vet. Res. 2014, 10, 71. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). EpiSignalDetection Tool. 17 December 2018. Available online: http://ecdc.europa.eu/en/publications-data/episignaldetection-tool (accessed on 31 December 2018).
- Jombart, T.; Aanensen, D.M.; Baguelin, M.; Birrell, P.; Cauchemez, S.; Camacho, A.; Colijn, C.; Collins, C.; Cori, A.; Didelot, X.; et al. OutbreakTools: A new platform for disease outbreak analysis using the R software. Epidemics 2014, 7, 28–34. [Google Scholar] [CrossRef]
- Groendyke, C.; Welch, D. epinet: An R Package to Analyze Epidemics Spread across Contact Networks. J. Stat. Softw. 2018, 83, 1–22. [Google Scholar] [CrossRef]
- Kulldorff, M. SaTScan—Software for the Spatial, Temporal, and Space-Time Scan Statistics. 2018. Available online: https://www.satscan.org/ (accessed on 5 April 2018).
- Chang, W.; Cheng, J.; Allaire, J.; Xie, Y.; McPhearson, J. R Package “Shiny”. 2016. Available online: https://shiny.rstudio.com/ (accessed on 5 April 2018).
- Attali, D. shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds. 2018. Available online: https://CRAN.R-project.org/package=shinyjs (accessed on 5 April 2018).
- Chang, W.; Borges Ribeiro, B. shinydashboard: Create Dashboards with “Shiny”. 2018. Available online: https://CRAN.R-project.org/package=shinydashboard (accessed on 5 April 2018).
- Chang, W. shinythemes: Themes for Shiny. 2018. Available online: https://CRAN.R-project.org/package=shinythemes (accessed on 5 April 2018).
- Perrier, V.; Meyer, F.; Granjon, D. shinyWidgets: Custom Inputs Widgets for Shiny. 2019. Available online: https://CRAN.R-project.org/package=shinyWidgets (accessed on 5 April 2018).
- Sail, A.; Hass, L. shinycssloaders: Add CSS Loading Animations to “Shiny” Outputs. 2017. Available online: https://CRAN.R-project.org/package=shinycssloaders (accessed on 5 April 2018).
- Almende, B.V.; Thieurmel, B.; Robert, T. visNetwork: Network Visualization Using “Vis.js” Library. 2018. Available online: https://CRAN.R-project.org/package=visNetwork (accessed on 5 April 2018).
- Cheng, J.; Xie, Y.; Wickham, H.; Agafonkin, V. leaflet: Create Interactive Web Maps with the JavaScript “Leaflet” Library. 2018. Available online: https://CRAN.R-project.org/package=leaflet (accessed on 5 April 2018).
- Hijmans, R.J.; Etten, J.V.; Sumner, M.; Cheng, J.; Bevan, A.; Bivand, R.; Busetto, L.; Canty, M.; Forrest, D.; Ghosh, A.; et al. raster: Geographic Data Analysis and Modeling. 2019. Available online: https://CRAN.R-project.org/package=raster (accessed on 5 April 2018).
- Sievert, C.; Parmer, C.; Hocking, T.; Chamberlain, S.; Ram, K.; Corvellec, M.; Despouy, P. plotly: Create Interactive Web Graphics via “plotly.js”. 2018. Available online: https://CRAN.R-project.org/package=plotly (accessed on 5 April 2018).
- Wickham, H.; Chang, W. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2018. Available online: https://CRAN.R-project.org/package=ggplot2 (accessed on 5 April 2018).
- Martoglio, E.; Kruchten, N.; Chinnasamy, N.; Russell, K. rpivotTable: Build Powerful Pivot Tables and Dynamically Slice & Dice your Data. 2018. Available online: https://CRAN.R-project.org/package=rpivotTable (accessed on 5 April 2018).
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. 2019. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 5 April 2018).
- Yu, G.; Ekstrøm, C.T. emojifont: Emoji and Font Awesome in Graphics. 2018. Available online: https://CRAN.R-project.org/package=emojifont (accessed on 5 April 2018).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes. 2014. Available online: https://CRAN.R-project.org/package=RColorBrewer (accessed on 5 April 2018).
- Xie, Y.; Cheng, J.; Tan, X.; Allaire, J.J.; Girlich, M.; Ellis, G.F.; Rauh, J.; Reavis, B.; Gersen, L.; Szopka, B.; et al. DT: A Wrapper of the JavaScript Library “DataTables”. 2018. Available online: https://CRAN.R-project.org/package=DT (accessed on 5 April 2018).
- Bivand, R.; Keitt, T.; Rowlingson, B.; Pebesma, E.; Sumner, M.; Hijmans, R.; Rouault, E.; Warmerdam, F.; Ooms, J.; Rundel, C. rgdal: Bindings for the “Geospatial” Data Abstraction Library. 2019. Available online: https://CRAN.R-project.org/package=rgdal (accessed on 5 April 2018).
- Olsen, A. bezier: Toolkit for Bezier Curves and Splines. 2018. Available online: https://CRAN.R-project.org/package=bezier (accessed on 5 April 2018).
- Karambelkar, B.; Schloerke, B.; Bangyou, Z.; Robin, C.; Markus, V.; Markus, D.; Thasler, H.; Wilhelm, D.; Risk, K.; Wisniewski, T.; et al. leaflet.extras: Extra Functionality for “Leaflet” Package. 2018. Available online: https://CRAN.R-project.org/package=leaflet.extras (accessed on 5 April 2018).
- Bivand, R.; Rundel, C.; Pebesma, E.; Stuetz, R.; Hufthammer, K.O.; Giraudoux, P.; Davis, M.; Santilli, S. rgeos: Interface to Geometry Engine—Open Source (‘GEOS’). 2018. Available online: https://CRAN.R-project.org/package=rgeos (accessed on 5 April 2018).
- Wood, S. mgcv: Mixed GAM Computation Vehicle with Automatic Smoothness Estimation. 2019. Available online: https://CRAN.R-project.org/package=mgcv (accessed on 5 April 2018).
- Ooms, J. V8: Embedded JavaScript Engine for R. 2019. Available online: https://CRAN.R-project.org/package=V8 (accessed on 5 April 2018).
- Dragulescu, A.A.; Arendt, C. xlsx: Read, Write, Format Excel 2007 and Excel 97/2000/XP/2003 Files. 2018. Available online: https://CRAN.R-project.org/package=xlsx (accessed on 5 April 2018).
- Lang, D.T. RCurl: General Network (HTTP/FTP/...) Client Interface for R. 2018. Available online: https://CRAN.R-project.org/package=RCurl (accessed on 5 April 2018).
- Vaidyanathan, R.; Xie, Y.; Allaire, J.J.; Cheng, J.; Russell, K. htmlwidgets: HTML Widgets for R. 2018. Available online: https://CRAN.R-project.org/package=htmlwidgets (accessed on 5 April 2018).
- Kahle, D.; Wickham, H.; Jackson, S.; Korpela, M. ggmap: Spatial Visualization with ggplot2. 2019. Available online: https://CRAN.R-project.org/package=ggmap (accessed on 5 April 2018).
- SIMAN. Available online: https://www.vetinfo.sanita.it/j6_siman/common/welcome.do%3bjsessionid=6F6B23878054B503BAF770EB29E3BE42-n1.tomcatprod2 (accessed on 5 April 2018).
- Sistema Informativo Veterinario. Available online: https://www.vetinfo.sanita.it/sso_portale/accesso.pl (accessed on 5 April 2018).
- Farrington, C.P.; Andrews, N.J.; Beale, A.D.; Catchpole, M.A. A Statistical Algorithm for the Early Detection of Outbreaks of Infectious Disease. J. R. Stat. Soc. Ser. A Stat. Soc. 1996, 159, 547–563. [Google Scholar] [CrossRef]
- Noufaily, A.; Enki, D.G.; Farrington, P.; Garthwaite, P.; Andrews, N.; Charlett, A. An improved algorithm for outbreak detection in multiple surveillance systems. Stat. Med. 2013, 32, 1206–1222. [Google Scholar] [CrossRef]
- Mallows, C.L. Non-Null Ranking Models. I. Biometrika 1957, 44, 114–130. [Google Scholar] [CrossRef]
- Jain, A.K. Data clustering: 50 years beyond K-means. Pattern Recognit. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Kulldorff, M.; Heffernan, R.; Hartman, J.; Assunção, R.; Mostashari, F. A Space–Time Permutation Scan Statistic for Disease Outbreak Detection. PLoS Med. 2005, 2, e59. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.; Kulldorff, M.; Klassen, A.C. A spatial scan statistic for ordinal data. Stat. Med. 2007, 26, 1594–1607. [Google Scholar] [CrossRef] [PubMed]
- Kulldorff, M.; Huang, L.; Konty, K. A scan statistic for continuous data based on the normal probability model. Int. J. Health Geogr. 2009, 8, 58. [Google Scholar] [CrossRef]
- Kulldorff, M. A spatial scan statistic. Commun. Stat. Theory Methods 1997, 26, 1481–1496. [Google Scholar] [CrossRef]
- Kulldorff, M.; Nagarwalla, N. Spatial disease clusters: Detection and inference. Stat. Med. 1995, 14, 799–810. [Google Scholar] [CrossRef]
- Rubel, F.; Brugger, K.; Hantel, M.; Chvala-Mannsberger, S.; Bakonyi, T.; Weissenböck, H.; Nowotny, N. Explaining Usutu virus dynamics in Austria: Model development and calibration. Prev. Vet. Med. 2008, 85, 166–186. [Google Scholar] [CrossRef]
- Beck-Johnson, L.M.; Nelson, W.A.; Paaijmans, K.P.; Read, A.F.; Thomas, M.B.; Bjørnstad, O.N. The Effect of Temperature on Anopheles Mosquito Population Dynamics and the Potential for Malaria Transmission. PLoS ONE 2013, 8, e79276. [Google Scholar] [CrossRef]
- Natale, F.; Giovannini, A.; Savini, L.; Palma, D.; Possenti, L.; Fiore, G.; Calistri, P. Network analysis of Italian cattle trade patterns and evaluation of risks for potential disease spread. Prev. Vet. Med. 2009, 92, 341–350. [Google Scholar] [CrossRef]
- Dubé, C.; Ribble, C.; Kelton, D.; McNab, B. Introduction to network analysis and its implications for animal disease modelling. Rev. Sci. Tech. Int. Epiz. 2011, 30, 425–436. [Google Scholar] [CrossRef]
- Craft, M.E.; Caillaud, D. Network Models: An Underutilized Tool in Wildlife Epidemiology? Interdiscip. Perspect. Infect. Dis. 2011, 2011, 676949. [Google Scholar] [CrossRef] [PubMed]
- Lentz, H.H.; Koher, A.; Hövel, P.; Gethmann, J.; Sauter-Louis, C.; Selhorst, T.; Conraths, F.J. Disease Spread through Animal Movements: A Static and Temporal Network Analysis of Pig Trade in Germany. PLoS ONE 2016, 11, e0155196. [Google Scholar] [CrossRef] [PubMed]
- Network Science by Albert-László Barabási. Available online: http://networksciencebook.com/ (accessed on 5 April 2018).
- Pastor-Satorras, R.; Vespignani, A. Epidemic Spreading in Scale-Free Networks. Phys. Rev. Lett. 2001, 86, 3200. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, B.; Wang, L.; Morris, M. Forward Reachable Sets: Analytically Derived Properties of Connected Components for Dynamic Networks. ArXiv160503241 Q-Bio. 2016. Available online: http://arxiv.org/abs/1605.03241 (accessed on 10 December 2019).
- Pioz, M.; Guis, H.; Calavas, D.; Durand, B.; Abrial, D.; Ducrot, C. Estimating front-wave velocity of infectious diseases: A simple, efficient method applied to bluetongue. Vet. Res. 2011, 42, 60. [Google Scholar] [CrossRef]
- Nicolas, G.; Tisseuil, C.; Conte, A.; Allepuz, A.; Pioz, M.; Lancelot, R.; Gilbert, M. Environmental heterogeneity and variations in the velocity of bluetongue virus spread in six European epidemics. Prev. Vet. Med. 2018, 149, 1–9. [Google Scholar] [CrossRef]
- Stefano, C.; Sandro, R.; Maria, C.A.; Federica, L.; Giorgio, M. Reoccurrence of West Nile Virus Disease in Humans and Successive Entomological Investigation in Sardinia, Italy, 2017. J. Anim. Sci. Res. 2017, 2. [Google Scholar] [CrossRef]
- Savini, L.; Candeloro, L.; Conte, A.; De Massis, F.; Giovannini, A. Development of a forecasting model for brucellosis spreading in the Italian cattle trade network aimed to prioritise the field interventions. Sendiña-Nadal I, editor. PLoS ONE 2017, 12, e0177313. [Google Scholar] [CrossRef]
- Darbon, A.; Valdano, E.; Poletto, C.; Giovannini, A.; Savini, L.; Candeloro, L.; Colizza, V. Network-based assessment of the vulnerability of Italian regions to bovine brucellosis. Prev. Vet. Med. 2018, 158, 25–34. [Google Scholar] [CrossRef]


| Ref. | Description | Tool or Task | |
|---|---|---|---|
| Software | |||
| R | [18] | Language and environment for statistical computing and graphics. | User interface |
| SaTScan | [31] | Software that analyzes spatial, temporal and space-time data using scan statistics. | SatScan |
| R-package | |||
| surveillance | [20] | Temporal and Spatiotemporal Modeling and Monitoring of Epidemic Phenomena. | EpiCurve |
| shiny | [32] | Web Application Framework for R. | User interface |
| shinyjs | [33] | Perform common useful JavaScript operations in Shiny apps that will greatly improve the apps without having to know any JavaScript. | User interface |
| shinydashboard | [34] | Create dashboard with Shiny. | User interface |
| shinythemes | [35] | Themes for Shiny. | User interface |
| shinyWidgets | [36] | Custom Inputs Widgets for Shiny. | User interface |
| shinycssloaders | [37] | Add CSS Loading Animations to ‘Shiny’ Outputs. | User interface |
| sp | [21] | Classes and Methods for Spatial Data. | Multiple |
| rsatscan | [22] | Tools, Classes and Methods for Interfacing with SaTScan Stand-Alone Software. | SatScan |
| network | [23] | Tools to create and modify network objects. | Ntw data in area Geo |
| tsna | [19] | Temporal SNA tools for continuous- and discrete-time longitudinal networks. | Trace from seed and TPath |
| visNetwork | [38] | It allows an interactive visualization of networks. | Ntw data in area Geo |
| igraph | [24] | Routines for simple graphs and network analysis. | Ntw data in area Geo |
| leaflet | [39] | Create and customize interactive maps using the ‘Leaflet’ JavaScript library and the ‘htmlwidgets’ package. | User interface |
| raster | [40] | Reading, writing, manipulating, analyzing and modeling of gridded spatial data. | EpiVelocity |
| plotly | [41] | Create Interactive Web Graphics via ‘plotly.js’. | Graphs |
| ggplot2 | [42] | Create Elegant Data Visualizations Using the Grammar of Graphics. | Graphs |
| rpivotTable | [43] | Build Powerful Pivot Tables and Dynamically Slice and Dice your Data. | Descriptor section |
| dplyr | [44] | A fast, consistent tool for working with data frame-like objects, both in memory and out of memory. | Multiple |
| emojifont | [45] | An implementation of using emoji and fontawesome for using in both base and ‘ggplot2’ graphics. | Ntw data in area Geo |
| RColorBrewer | [46] | Provides color schemes for maps. | Map |
| DT | [47] | Data objects in R can be rendered as HTML tables using the JavaScript library ‘DataTables’ (typically via R Markdown or Shiny). | Tables |
| rgdal | [48] | Bindings for the ‘Geospatial’ Data Abstraction Library. | Multiple |
| bezier | [49] | Toolkit for Bezier Curves and Splines. | Ntw data in area Geo |
| leaflet.extras | [50] | Extra Functionality for ‘leaflet’ Package. | User interface |
| rgeos | [51] | Interface to Geometry Engine—Open Source (‘GEOS’). | Multiple |
| mgcv | [52] | Mixed GAM Computation Vehicle with Automatic Smoothness Estimation. | EpiCurve |
| v8 | [53] | An R interface to Google’s open source JavaScript engine. | Multiple |
| xlsx | [54] | Read, Write, Format Excel 2007 and Excel 97/2000/XP/2003 Files. | Data download/upload |
| RCurl | [55] | General Network (HTTP/FTP/...) Client Interface for R. | Data download/upload |
| htmlwidgets | [56] | A framework for creating HTML widgets. | User interface |
| stats4 | [18] | Statistical Functions using S4 classes. | Multiple |
| ggmap | [57] | A collection of functions to visualize spatial data and models on top of static maps from various online sources (e.g., Google Maps and Stamen Maps). | Map |
| App Analysis Tool/Task | File Name | Worksheet Name | Data Description * |
|---|---|---|---|
| Outbreak detection/EpiCurve | outbreak.data.xlsx | Outbreaks | Outbreak disease data |
| Vectors and related factors /Vectors report | ento.data.xlsx | Ento | Entomological data |
| Vectors and related factors /Vectors report | ento.data.xlsx | Outbreaks | Outbreak disease data |
| Vectors and related factors /Vectors report | ento.data.xlsx | LST_RAW | LST data (at 8 day temporal resolution) |
| Vectors and related factors /Vectors report | ento.data.xlsx | LST_Month | LST data (monthly temperature average) |
| MODIS LST and Mosquito model/MODIS Land surface temperature (LST) | LSTpoint.data.xlsx | PointCoordinates | Coordinates of the user-defined point |
| MODIS LST and Mosquito model/MODIS Land surface temperature (LST) | LSTpoint.data.xlsx | LST.Observed | TLS data for the set point (8 days temporal resolution values) |
| MODIS LST and Mosquito model/MODIS Land surface temperature (LST) | LSTpoint.data.xlsx | LST.interpolation | Interpolated daily TLS values |
| MODIS LST and Mosquito model/MODIS Land surface temperature (LST) | LSTpoint.data.xlsx | LST.NA | TLS data missing |
| MODIS LST and Mosquito model/MODIS Land surface temperature (LST) based model | MosquitoModel.xlsx | MosquitoModel | Mosquito model results: Larvae/Adults daily data and related mean temperature values |
| Network Analysis/Ntw data in area Geo | NodeCentralities.xlsx | Nodes Centralities | Nodes data and related centrality measures values |
| Network Analysis/Ntw data in area Geo | NodeCentralities.xlsx | Static edges | Contacts data of the static network |
| Network Analysis/Ntw data in area Geo | NodeCentralities.xlsx | Nodes | Nodes data of the static network |
| Network Analysis/Trace from seed | Subntw.xlsx | TraceFromSeed | Movements data related to the specified seed in back and forward in the established timeframe |
| Network Analysis/Tpaths | tpaths.Tables.xlsx | Selection | Data related to the Tpath analysis: start/end date, species, (from/to) slaughter/foreign state movements (included/excluded) |
| Network Analysis/Tpaths | tpaths.Tables.xlsx | Tpath edges table | Tpath analysis results in terms of edges involved |
| Network Analysis/tpaths | tpaths.Tables.xlsx | Tpath nodes table | Classification of nodes included in the Tpath analysis in terms of their FRS values (origin area), BRS values (destination area) and DEG values for bridge nodes (external to the origin and destination areas) |
| Name | Description * |
|---|---|
| Network Properties at the Global Level | |
| Size | The number of nodes and edges. |
| Diameter | The length of the longest path (in number of edges) between two nodes. |
| Average shortest path length | Refers to the average of all the shortest distance (number of edges) between each pair of reachable nodes in the network [75]. |
| Density | The number of edges in the network over all the possible edges that could exist in the network. |
| Reciprocity | Measures the mutual edge relation: the probability that if node i is connected to node j, node j is also connected to node i. |
| Transitivity | Measures that probability that adjacent nodes of a network are connected (also known as clustering coefficient). |
| Network communities | The networks often have different clusters or communities of nodes that are more densely connected to each other than to the rest of the network. |
| Network Properties at Local Level (the Weighted Measures are Calculated Considering as Edge Weight Alternatively the Number of Animals Moved or Number of Movements) | |
| Degree | The number of adjacent edges to each node. It is considered as InDegree and OutDegree: InDegree is a count of the number of incoming edges to the node and OutDegree is the number of outgoing edges from the node. |
| Strength | A weighted measure of degree that takes into account the number of edges going from one node to another or the number of animals moved. |
| Closeness | Measures how many steps are required to access every other node from a given node. |
| Betweenness | The number of shortest paths between nodes, passing through a particular node. |
| Page rank | Approximates the probability that any message will arrive to a particular node. |
| Authority score | A node has high authority when it is linked to many other nodes, in turn linked to many other nodes. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savini, L.; Candeloro, L.; Perticara, S.; Conte, A. EpiExploreR: A Shiny Web Application for the Analysis of Animal Disease Data. Microorganisms 2019, 7, 680. https://doi.org/10.3390/microorganisms7120680
Savini L, Candeloro L, Perticara S, Conte A. EpiExploreR: A Shiny Web Application for the Analysis of Animal Disease Data. Microorganisms. 2019; 7(12):680. https://doi.org/10.3390/microorganisms7120680
Chicago/Turabian StyleSavini, Lara, Luca Candeloro, Samuel Perticara, and Annamaria Conte. 2019. "EpiExploreR: A Shiny Web Application for the Analysis of Animal Disease Data" Microorganisms 7, no. 12: 680. https://doi.org/10.3390/microorganisms7120680
APA StyleSavini, L., Candeloro, L., Perticara, S., & Conte, A. (2019). EpiExploreR: A Shiny Web Application for the Analysis of Animal Disease Data. Microorganisms, 7(12), 680. https://doi.org/10.3390/microorganisms7120680





