Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Induction of VBNC E. coli O157:H7 by Low Temperature
2.2. Preparation of RNA-Seq Samples
2.3. RNA Extraction, Library Construction, and Sequencing
2.4. RNA-Seq Data Analysis
2.5. Accession Number
3. Results
3.1. Comparison with the Ribosomal RNA Database
3.2. Gene Expression Level in the Samples
3.3. Statistics of the Differentially Expressed Gene between the Samples
3.4. GO Functional Analysis
3.5. KEGG Pathway Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wei, C.; Zhong, J.; Hu, T.; Zhao, X. Simultaneous detection of Escherichia coli O157: H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ye, C.; Lin, H.; Lv, L.; Yu, X. UV disinfection induces a VBNC state in Escherichia coli and Pseudomonas aeruginosa. Environ. Sci. Technol. 2015, 49, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, R.; Li, L.; Peters, B.M.; Li, B.; Lin, C.W.; Chuang, T.-L.; Chen, D.; Zhao, X.; Xiong, Z. Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli O157 under cryopreservation. Res. Microbial. 2017, 168, 188–193. [Google Scholar] [CrossRef]
- Dinu, L.D.; Bach, S. Detection of viable but non-culturable Escherichia coli O157: H7 from vegetable samples using quantitative PCR with propidium monoazide and immunological assays. Food Control 2013, 31, 268–273. [Google Scholar] [CrossRef]
- Chaisowwong, W.; Kusumoto, A.; Hashimoto, M.; Harada, T.; Maklon, K.; Kawamoto, K. Physiological characterization of Campylobacter jejuni under cold stresses conditions: Its potential for public threat. J. Vet. Med. Sci. 2012, 74, 43–50. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Detection of viable but non-culturable Escherichia coli O157: H7 by PCR in combination with propidium monoazide. 3 Biotech 2018, 8, 28. [Google Scholar] [CrossRef]
- Kong, I.-S.; Bates, T.C.; Hülsmann, A.; Hassan, H.; Smith, B.E.; Oliver, J.D. Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS Microbial. Ecol. 2004, 50, 133–142. [Google Scholar] [CrossRef]
- Masmoudi, S.; Denis, M.; Maalej, S. Inactivation of the gene katA or sodA affects the transient entry into the viable but non-culturable response of Staphylococcus aureus in natural seawater at low temperature. Mar. Pollut. Bull. 2010, 60, 2209–2214. [Google Scholar] [CrossRef]
- Asakura, H.; Ishiwa, A.; Arakawa, E.; Makino, S.I.; Okada, Y.; Yamamoto, S.; Igimi, S. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ. Microbial. 2007, 9, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wang, Y.; An, H.; Hao, Y.; Hu, X.; Liao, X. New insights into the formation of viable but nonculturable Escherichia coli O157: H7 induced by high-pressure CO2. Mbio 2016, 7, e00961-16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Deng, Y.; Peters, B.M.; Li, L.; Li, B.; Chen, L.; Xu, Z.; Shirtliff, M.E. Transcriptomic analysis on the formation of the viable putative non-culturable state of beer-spoilage Lactobacillus acetotolerans. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Sun, F.; Wang, Y.; Hashmi, M.Z.; Guo, L.; Ding, L.; Shen, C. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci. Rep. 2015, 5, 18590. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Di Toro, M.R.; Grieco, F.; Michelotti, V.; Salma, M.; Lamontanara, A.; Russo, P.; Orrù, L.; Alexandre, H.; Spano, G. Viable But Not Culturable (VBNC) state of Brettanomyces bruxellensis in wine: New insights on molecular basis of VBNC behaviour using a transcriptomic approach. Food Microbial. 2016, 59, 196–204. [Google Scholar] [CrossRef]
- Kogure, K.; Simidu, U.; Taga, N. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 1979, 25, 415–420. [Google Scholar] [CrossRef]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009, 38, 1767–1771. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Boil. 2013, 14, R36. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, A.; Marzi, S.; Myasnikov, A.G.; Fabbretti, A.; Yusupov, M.; Gualerzi, C.O.; Klaholz, B.P. Structure of the 30S translation initiation complex. Nature 2008, 455, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Studer, S.M.; Joseph, S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell 2006, 22, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Sijbrandi, R.; Urbanus, M.L.; Corinne, M.; Bernstein, H.D.; Oudega, B.; Otto, B.R.; Luirink, J. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J. Biol. Chem. 2003, 278, 4654–4659. [Google Scholar] [CrossRef]
- Grentzmann, G.; Brechemier-Baey, D.; Heurgué-Hamard, V.; Buckingham, R.H. Function of polypeptide chain release factor RF-3 in Escherichia coli. RF-3 action in termination is predominantly at UGA-containing stop signals. J. Biol. Chem. 1995, 270, 10595. [Google Scholar] [CrossRef]
- del Mar Lleò, M.; Pierobon, S.; Tafi, M.C.; Signoretto, C.; Canepari, P. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl. Environ. Microbiol. 2000, 66, 4564–4567. [Google Scholar] [CrossRef]
- Makino, S.I.; Kii, T.; Asakura, H.; Shirahata, T.; Ikeda, T.; Takeshi, K.; Itoh, K. Does Enterohemorrhagic Escherichia coli O157:H7 Enter the Viable but Nonculturable State in Salted Salmon Roe? Appl. Environ. Microbiol. 2000, 66, 5536–5539. [Google Scholar] [CrossRef]
- Asakura, H.; Makino, S.I.; Takagi, T.; Kuri, A.; Kurazono, T.; Watarai, M.; Shirahata, T. Passage in mice causes a change in the ability of Salmonella enterica serovar Oranienburg to survive NaCl osmotic stress: Resuscitation from the viable but non-culturable state. Fems Microbiol. Lett. 2002, 212, 87–93. [Google Scholar] [CrossRef]
- Lamas, A.; Regal, P.; Vázquez, B.; Miranda, J.M.; Franco, C.M.; Cepeda, A. Transcriptomics: A powerful tool to evaluate the behavior of foodborne pathogens in the food production chain. Food Res. Int. 2019, 125, 108543. [Google Scholar] [CrossRef] [PubMed]



| Sample | Ranking | Gene | FPKM | Reads | NR Annotation |
|---|---|---|---|---|---|
| LP | 1 | Z_RS10405 | 29,197.3 | 64,534.96 | Glutamate decarboxylase beta (E. coli O25b:H4-ST131) (E. coli) |
| 2 | raiA | 14,869.5 | 8023 | Translation inhibitor protein RaiA (E. coli O157:H7 str. Sakai) (E. coli) | |
| 3 | phoE | 14,297.2 | 24,902 | Porin protein PhoE (E. coli) | |
| 4 | ompA | 13,175.65 | 21,639 | Outer membrane protein A (E. coli MS 107-1) (E. coli) | |
| 5 | fliC | 12,644.53 | 35,070 | flagellin FliC (E. coli) | |
| VBNC | 1 | ompA | 12,956.99 | 26,842 | Outer membrane protein A (E. coli MS 107-1) (E. coli) |
| 2 | rpoA | 9133.91 | 17,995 | DNA-directed RNA polymerase subunit alpha (E. coli O157:H7 str. Sakai) (E. coli) | |
| 3 | tuf | 9112.96 | 21,490.06 | Elongation factor Tu (E. coli CFT073) (E. coli) | |
| 4 | secY | 8488.26 | 22,500 | Preprotein translocase subunit SecY (Shigella boydii) | |
| 5 | Z_RS21965 | 8033.14 | 6954 | 50S ribosomal protein L15 (E. coli O157:H7 str. Sakai) (E. coli) |
| GO Category | GO Term ID | NR Annotation | GeneRatio | BgRatio | p-Value |
|---|---|---|---|---|---|
| Biological Process | GO:0006811 | Ion transport | 39 (8.44%) | 58 (5.88%) | 0.000997 |
| GO:0044282 | Small molecule catabolic process | 12 (2.6%) | 14 (1.42%) | 0.003079 | |
| GO:0015711 | Organic anion transport | 20 (4.33%) | 27 (2.74%) | 0.003370 | |
| Cellular Component | GO:0071944 | Cell periphery | 9 (3.64%) | 10 (2.03%) | 0.010471 |
| GO:0005618 | Cell wall | 8 (3.24%) | 9 (1.83%) | 0.019240 | |
| GO:0030312 | External encapsulating structure | 8 (3.24%) | 9 (1.83%) | 0.019240 | |
| Molecular Function | GO:0008514 | Organic anion transmembrane transporter activity | 14 (3.28%) | 18 (1.92%) | 0.005266 |
| GO:0022891 | Substrate-specific transmembrane transporter activity | 43 (10.07%) | 71 (7.58%) | 0.006012 | |
| GO:0015075 | Ion transmembrane transporter activity | 28 (6.56%) | 43 (4.59%) | 0.006591 |
| Number of Repetitive Times | Differences Expression Gene | NR Annotation |
|---|---|---|
| 5 | dcuA, Z_RS24500, Z_RS23645, Z_RS21565, Z_RS21275, Z_RS18545, tyrP, Z_RS12305, proY, metN, glpT | IT, OAT, OATTA, SSTTA, ITTA |
| 4 | mgtA | IT, OAT, SSTTA, ITTA |
| 3 | Z_RS26985, Z_RS26485, Z_RS25225, Z_RS24665, Z_RS22875, Z_RS22705, Z_RS22635, Z_RS22415, Z_RS21515, Z_RS17370, Z_RS17145, Z_RS13740, Z_RS09350, Z_RS09310, Z_RS06530, Z_RS05705, Z_RS02760, Z_RS02175, Z_RS24940, nikE, msbB, kdpC | IT, CP, CW, EES, SSTTA, ITTA |
| 2 | Z_RS22695, Z_RS16125, Z_RS13335, Z_RS12460, Z_RS10735, Z_RS03375, ugpC, rhtB, kdgT, brnQ | IT, OAT, SSTTA, ITTA |
| 1 | Z_RS26780, Z_RS26335, Z_RS25895, Z_RS25370, Z_RS24525, Z_RS24370, Z_RS23610, Z_RS23465, Z_RS22905, Z_RS21350, Z_RS21150, Z_RS21140, Z_RS21025, Z_RS19885, Z_RS18625, Z_RS16005, Z_RS12610, Z_RS12490, Z_RS09475, Z_RS09120, Z_RS04060, Z_RS03890, Z_RS02910, Z_RS02490, Z_RS01960, secF, pstS, pstA, nepI, lamB, gldA, gcvT, gcvH, fucI, fghA, fadE | IT, SMCP, SSTTA |
| Pathway ID | Pathway | DEGs Genes with Pathway Annotation (681) | All Genes with Pathway Annotation (1480) | p-Value | q-Value |
|---|---|---|---|---|---|
| ko03010 | Ribosome | 45 (6.61%) | 58 (3.92%) | 0.000001 | 0.000059 |
| ko00920 | Sulfur metabolism | 26 (3.82%) | 36 (2.43%) | 0.001160 | 0.059179 |
| ko00520 | Amino sugar and nucleotide sugar metabolism | 32 (4.7%) | 48 (3.24%) | 0.002740 | 0.093167 |
| ko00020 | Citrate cycle (TCA cycle) | 20 (2.94%) | 29 (1.96%) | 0.010074 | 0.256884 |
| ko00261 | Monobactam biosynthesis | 7 (1.03%) | 8 (0.54%) | 0.020602 | 0.420276 |
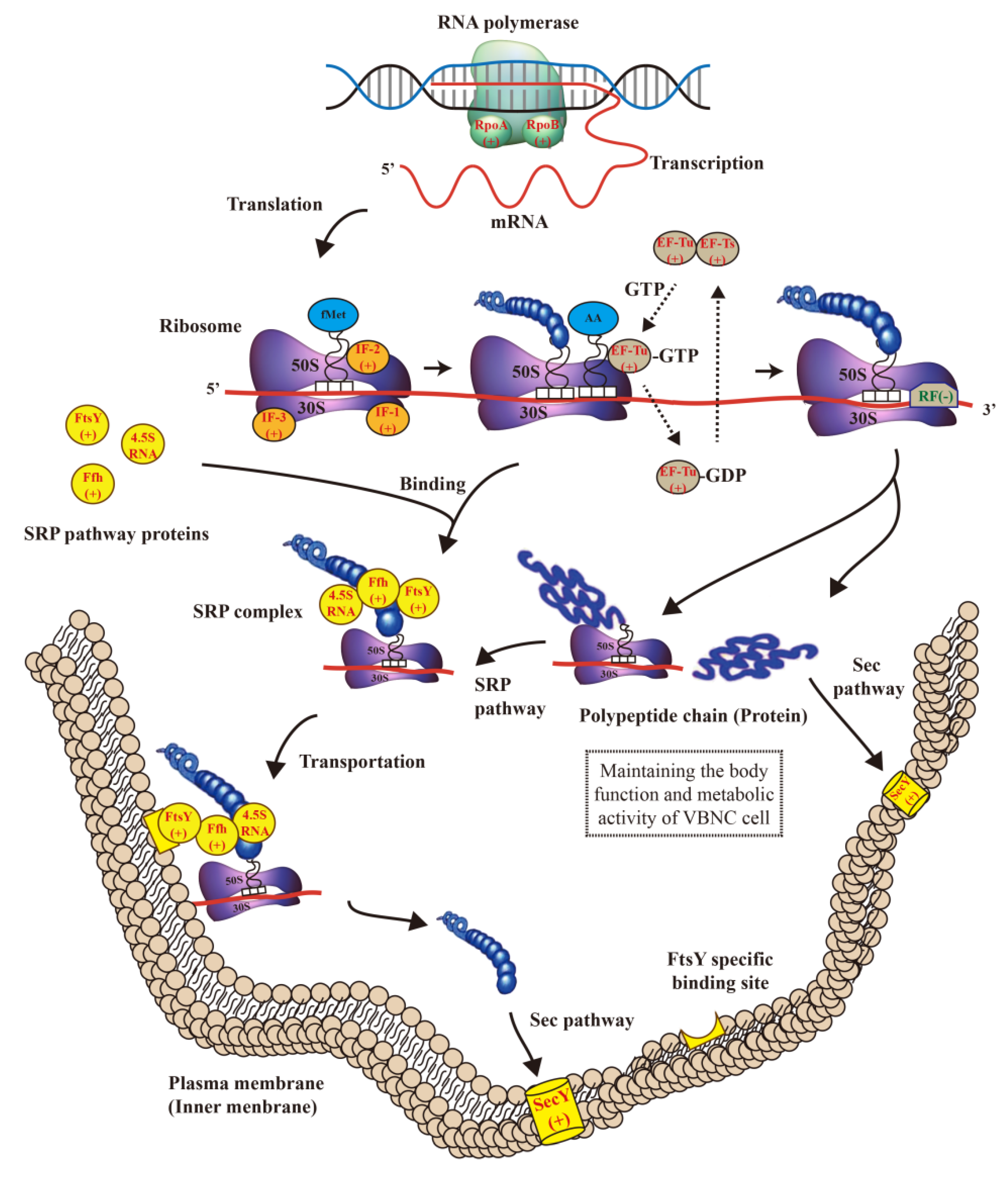
| Pathway ID | Signal Pathway | Differentially Expressed Genes in the Signal Pathway |
| Ko03010 | Ribosome | rpsO, rplS, rpsM, rplF, Z_RS23800, rplT, rpsQ, rpsR, rpsD, rpsB, rplU, Z_RS08020, rplB, rplR, rplD, rplV, Z_RS16205, rpsC, rplC, rplW, rpmC, Z_RS25730, rplX, Z_RS25530, rpsI, rplM, rpsE, rpmA, rplP, rpsK, rpmG, rplE, rpmI, rpsG, rpsS, rpmD, Z_RS20780, rpsJ, Z_RS27210, rplJ, rplQ, Z_RS27195, rplO, Z_RS00125, Z_RS30885 |
| Related protein annotations | Differentially expressed genes of significant protein annotation in the signal pathway | |
| RNA Polymerase Subunit α and β (+) | Subunit α: rplQ, rplM, rpsI; Subunit β: rplJ | |
| Initiation factor (+) | IF-1: rpsM, rpsK, rpsD; IF-2: rpsO; IF3: rpmI, rplT | |
| Elongation factor Tu (+) | EF-Tu: rpsJ, rplC, rplD, rplW, rplB, rpsS, rplV, rpsC, rplP, rpmC; EF-Ts: rpsB; EF-G: rpsG | |
| Preprotein Translocase Subunit SecY (+) | rpsQ, rplX, rplE, rplF, rplR, rpsE, rpmD, rplO | |
| FtsY and Ffh (+) | rplS | |
| release factor RF-1 (−) | rpmE | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, J.; Zhao, X. Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature. Microorganisms 2019, 7, 634. https://doi.org/10.3390/microorganisms7120634
Zhong J, Zhao X. Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature. Microorganisms. 2019; 7(12):634. https://doi.org/10.3390/microorganisms7120634
Chicago/Turabian StyleZhong, Junliang, and Xihong Zhao. 2019. "Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature" Microorganisms 7, no. 12: 634. https://doi.org/10.3390/microorganisms7120634
APA StyleZhong, J., & Zhao, X. (2019). Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature. Microorganisms, 7(12), 634. https://doi.org/10.3390/microorganisms7120634





