Bacteriophage and the Innate Immune System: Access and Signaling
Abstract
1. Introduction
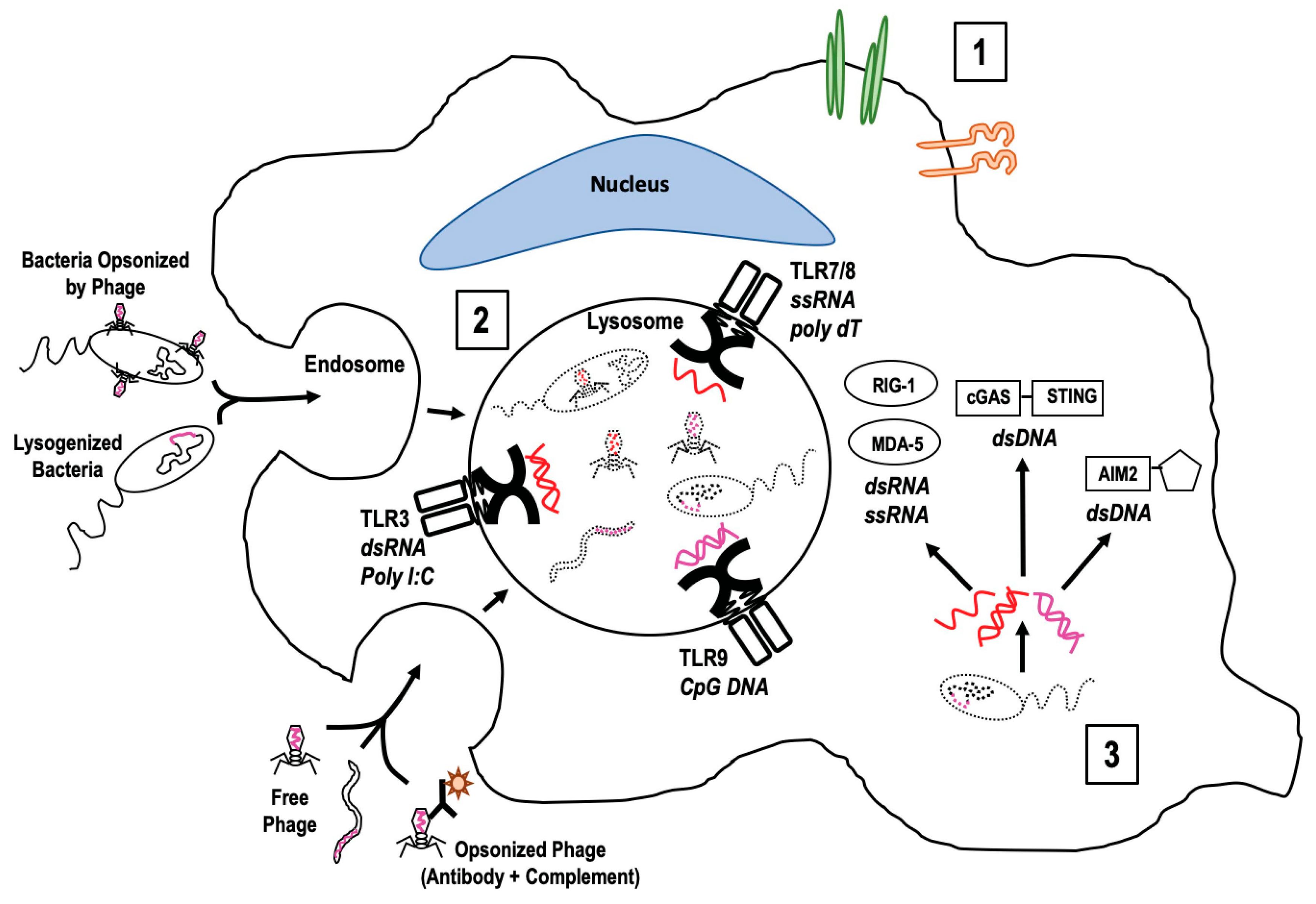
2. Accessing Innate Immune Cells
3. Signaling Innate Immune Cells
4. Summary and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Andrew, M.Q.; King, M.J.A.; Eric, B.C.; Elliot, J.L. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; p. 1338. [Google Scholar]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobián-Güemes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Lytic to temperate switching of viral communities. Nature 2016, 531, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lewis, D.E.A.; Adhya, S. The Developmental Switch in Bacteriophage lambda: A Critical Role of the Cro Protein. J. Mol. Biol. 2018, 430, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Nanda, A.M.; Thormann, K.; Frunzke, J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 2015, 197, 410–419. [Google Scholar] [CrossRef]
- Abeles, S.R.; Ly, M.; Santiago-Rodriguez, T.M.; Pride, D.T. Effects of Long Term Antibiotic Therapy on Human Oral and Fecal Viromes. PLoS ONE 2015, 10, e0134941. [Google Scholar] [CrossRef]
- Brussow, H.; Canchaya, C.; Hardt, W.D. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef]
- Enault, F.; Briet, A.; Bouteille, L.; Roux, S.; Sullivan, M.B.; Petit, M.A. Phages rarely encode antibiotic resistance genes: A cautionary tale for virome analyses. ISME J. 2017, 11, 237–247. [Google Scholar] [CrossRef]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.A.; Winter, S.E.; Rivera-Chávez, F.; Xavier, M.N.; Poon, V.; Nuccio, S.P.; Tsolis, R.M.; Bäumler, A.J. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 2012, 3, e00143-12. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.R.; Lee, H.H.; Spina, C.S.; Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013, 499, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [PubMed]
- Bergh, O.; Børsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef]
- Paul, J.H. Prophages in marine bacteria: Dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008, 2, 579–589. [Google Scholar] [CrossRef]
- Minot, S.; Sinha, R.; Chen, J.; Li, H.; Keilbaugh, S.A.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011, 21, 1616–1625. [Google Scholar] [CrossRef]
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338. [Google Scholar] [CrossRef]
- Keen, E.C.; Dantas, G. Close Encounters of Three Kinds: Bacteriophages, commensal Bacteria, and Host Immunity. Trends Microbiol. 2018, 26, 943–954. [Google Scholar] [CrossRef]
- Kim, M.S.; Bae, J.W. Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 2018, 12, 1127–1141. [Google Scholar] [CrossRef]
- Mills, S.; Shanahan, F.; Stanton, C.; Hill, C.; Coffey, A.; Ross, R.P. Movers and shakers: Influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 2013, 4, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Van Belleghem, J.D.; Dąbrowska, K.; Vaneechoutte, M.; Barr, J.J.; Bollyky, P.L. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses 2018, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776. [Google Scholar] [CrossRef] [PubMed]
- Gorski, A.; Wazna, E.; Dabrowska, B.W.; Dabrowska, K.; Switała-Jeleń, K.; Miedzybrodzki, R. Bacteriophage translocation. FEMS Immunol. Med. Microbiol. 2006, 46, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.; Xie, C.; Kirkness, E.; Biggs, W.; Wong, E.; Turpaz, Y.; Bloom, K.; Delwart, E.; Nelson, K.E.; Venter, J.C.; et al. The blood DNA virome in 8000 humans. PLoS Pathog. 2017, 13, 1006292. [Google Scholar]
- Holers, V.M. Complement and its receptors: New insights into human disease. Annu. Rev. Immunol. 2014, 32, 433–459. [Google Scholar] [CrossRef]
- Blasius, A.L.; Beutler, B. Intracellular toll-like receptors. Immunity 2010, 32, 305–315. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Ori, D.; Murase, M.; Kawai, T. Cytosolic nucleic acid sensors and innate immune regulation. Int. Rev. Immunol. 2017, 36, 74–88. [Google Scholar] [CrossRef]
- Xiao, T.S. The nucleic acid-sensing inflammasomes. Immunol. Rev. 2015, 265, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Kieser, K.J.; Kagan, J.C. Multi-receptor detection of individual bacterial products by the innate immune system. Nat. Rev. Immunol. 2017, 17, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Bieghs, V.; Trautwein, C. Innate immune signaling and gut-liver interactions in non-alcoholic fatty liver disease. Hepatobiliary Surg. Nutr. 2014, 3, 377–385. [Google Scholar]
- Gordon, S. Pattern recognition receptors: Doubling up for the innate immune response. Cell 2002, 111, 927–930. [Google Scholar] [CrossRef]
- Opitz, B.; Hippenstiel, S.; Eitel, J.; Suttorp, N. Extra- and intracellular innate immune recognition in endothelial cells. Thromb. Haemost. 2007, 98, 319–326. [Google Scholar]
- Lusiak-Szelachowska, M.; Żaczek, M.; Weber-Dąbrowska, B.; Międzybrodzki, R.; Letkiewicz, S.; Fortuna, W.; Rogóż, P.; Szufnarowski, K.; Jończyk-Matysiak, E.; Olchawa, E.; et al. Antiphage activity of sera during phage therapy in relation to its outcome. Future Microbiol. 2017, 12, 109–117. [Google Scholar] [CrossRef]
- Dabrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 2014, 88, 12551–12557. [Google Scholar] [CrossRef]
- Dabrowska, K.; Switała-Jelen, K.; Opolski, A.; Weber-Dabrowska, B.; Gorski, A. Bacteriophage penetration in vertebrates. J. Appl. Microbiol. 2005, 98, 7–13. [Google Scholar] [CrossRef]
- Jerne, N.K. The presence in normal serum of specific antibody against bacteriophage T4 and its increase during the earliest stages of immunization. J. Immunol. 1956, 76, 209–216. [Google Scholar]
- Tuma, P.; Hubbard, A.L. Transcytosis: Crossing cellular barriers. Physiol. Rev. 2003, 83, 871–932. [Google Scholar] [CrossRef] [PubMed]
- Aronow, R.; Danon, D.; Shahar, A.; Aronson, M. Electron Microscopy of in Vitro Endocytosis of T2 Phage by Cells from Rabbit Peritoneal Exudate. J. Exp. Med. 1964, 120, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J. A bacteriophages journey through the human body. Immunol. Rev. 2017, 279, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Duerkop, B.A.; Hooper, L.V. Resident viruses and their interactions with the immune system. Nat. Immunol. 2013, 14, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wu, M.; Liu, X.; Liu, Z.; Zhou, Q.; Niu, Z.; Huang, Y. Probing the endocytic pathways of the filamentous bacteriophage in live cells using ratiometric pH fluorescent indicator. Adv. Healthc. Mater. 2015, 4, 413–419. [Google Scholar] [CrossRef]
- Hsia, R.; Ohayon, H.; Gounon, P.; Dautry-Varsat, A.; Bavoil, P.M. Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2000, 2, 761–772. [Google Scholar] [CrossRef]
- Claesson-Welsh, L. Vascular permeability—The essentials. Upsala J. Med. Sci. 2015, 120, 135–143. [Google Scholar] [CrossRef]
- Huh, H.; Wong, S.; St Jean, J.; Slavcev, R. Bacteriophage interactions with mammalian tissue: Therapeutic applications. Adv. Drug Deliv. Rev 2019, 145, 4–17. [Google Scholar] [CrossRef]
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers. MBio 2017, 8, e01874-17. [Google Scholar] [CrossRef]
- Ivanenkov, V.; Felici, F.; Menon, A.G. Uptake and intracellular fate of phage display vectors in mammalian cells. Biochim. Biophys. Acta 1999, 1448, 450–462. [Google Scholar] [CrossRef]
- Shan, J.; Ramachandran, A.; Thanki, A.M.; Vukusic, F.B.I.; Barylski, J.; Clokie, M.R.J. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 2018, 8, 5091. [Google Scholar] [CrossRef] [PubMed]
- Kucharzik, T.; Lügering, N.; Rautenberg, K.; Lügering, A.; Schmidt, M.A.; Stoll, R.; Domschke, W. Role of M cells in intestinal barrier function. Annu. N. Y. Acad. Sci. 2000, 915, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Petris, C.K.; Golomb, M.; Phillips, T.E. Bacterial transcytosis across conjunctival M cells. Investig. Ophthalmol. Vis. Sci. 2007, 48, 2172–2177. [Google Scholar] [CrossRef]
- Bille, E.; Meyer, J.; Jamet, A.; Euphrasie, D.; Barnier, J.P.; Brissac, T.; Larsen, A.; Pelissier, P.; Nassif, X. A virulence-associated filamentous bacteriophage of Neisseria meningitidis increases host-cell colonisation. PLoS Pathog. 2017, 13, e1006495. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F. Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv. Virus Res. 2012, 82, 91–118. [Google Scholar] [PubMed]
- Wagner, P.L.; Waldor, M.K. Bacteriophage control of bacterial virulence. Infect. Immun. 2002, 70, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Farache, J.; Koren, I.; Milo, I.; Gurevich, I.; Kim, K.W.; Zigmond, E.; Furtado, G.C.; Lira, S.A.; Shakhar, G. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 2013, 38, 581–595. [Google Scholar] [CrossRef]
- Lelouard, H.; Fallet, M.; de Bovis, B.; Méresse, S.; Gorvel, J.P. Peyer’s patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology 2012, 142, 592–601. [Google Scholar] [CrossRef]
- Barfoot, R.; Denham, S.; Gyure, L.A.; Hall, J.G.; Hobbs, S.M.; Jackson, L.E.; Robertson, D. Some properties of dendritic macrophages from peripheral lymph. Immunology 1989, 68, 233–239. [Google Scholar]
- Roxas, J.L.; Viswanathan, V.K. Modulation of Intestinal Paracellular Transport by Bacterial Pathogens. Compr. Physiol. 2018, 8, 823–842. [Google Scholar]
- Tetz, G.; Tetz, V. Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gut Pathog. 2016, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Przerwa, A.; Zimecki, M.; Switała-Jeleń, K.; Dabrowska, K.; Krawczyk, E.; Łuczak, M.; Weber-Dabrowska, B.; Syper, D.; Miedzybrodzki, R.; Górski, A. Effects of bacteriophages on free radical production and phagocytic functions. Med. Microbiol. Immunol. 2006, 195, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Miernikiewicz, P.; Kłopot, A.; Soluch, R.; Szkuta, P.; Kęska, W.; Hodyra-Stefaniak, K.; Konopka, A.; Nowak, M.; Lecion, D.; Kaźmierczak, Z.; et al. T4 Phage Tail Adhesin Gp12 Counteracts LPS-Induced Inflammation In vivo. Front. Microbiol. 2016, 7, 1112. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Zhang, H.M.; Wu, T.T.; Xu, L.; Gan, Y.L.; Jiang, L.S.; Zhang, L.; Guo, S.L. Titer dynamic analysis of D29 within MTB-infected macrophages and effect on immune function of macrophages. Exp. Lung Res. 2014, 40, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Gorski, A.; Dabrowska, K.; Switala-Jeleń, K.; Nowaczyk, M.; Weber-Dabrowska, B.; Boratynski, J.; Wietrzyk, J.; Opolski, A. New insights into the possible role of bacteriophages in host defense and disease. Med. Immunol. 2003, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Wenger, S.L.; Turner, J.H.; Petricciani, J.C. The cytogenetic, proliferative and viability effects of four bacteriophages on human lymphocytes. In Vitro 1978, 14, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Nykvist, P.; Tasanen, K.; Viitasalo, T.; Kapyla, J.; Jokinen, J.; Bruckner-Tuderman, L.; Heino, J. The cell adhesion domain of type XVII collagen promotes integrin-mediated cell spreading by a novel mechanism. J. Biol. Chem. 2001, 276, 38673–38679. [Google Scholar] [CrossRef]
- Prasad, K.S.; Andre, P.; He, M.; Bao, M.; Manganello, J.; Phillips, D.R. Soluble CD40 ligand induces beta3 integrin tyrosine phosphorylation and triggers platelet activation by outside-in signaling. Proc. Natl. Acad. Sci. USA 2003, 100, 12367–12371. [Google Scholar] [CrossRef]
- Fraser, J.S.; Yu, Z.; Maxwell, K.L.; Davidson, A.R. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J. Mol. Biol. 2006, 359, 496–507. [Google Scholar] [CrossRef]
- Sathaliyawala, T.; Islam, M.Z.; Li, Q.; Fokine, A.; Rossmann, M.G.; Rao, V.B. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 2010, 77, 444–455. [Google Scholar] [CrossRef]
- Bork, P.; Holm, L.; Sander, C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 1994, 242, 309–320. [Google Scholar] [CrossRef]
- Halaby, D.M.; Mornon, J.P. The immunoglobulin superfamily: An insight on its tissular, species, and functional diversity. J. Mol. Evol. 1998, 46, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Hodyra-Stefaniak, K.; Miernikiewicz, P.; Drapała, J.; Drab, M.; Jończyk-Matysiak, E.; Lecion, D.; Kaźmierczak, Z.; Beta, W.; Majewska, J.; Harhala, M.; et al. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci. Rep. 2015, 5, 14802. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, Z.; Piotrowicz, A.; Owczarek, B.; Hodyra, K.; Miernikiewicz, P.; Lecion, D.; Harhala, M.; Górski, A.; Dąbrowska, K. Molecular imaging of T4 phage in mammalian tissues and cells. Bacteriophage 2014, 4, e28364. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef]
- Noppert, S.J.; Fitzgerald, K.A.; Hertzog, P.J. The role of type I interferons in TLR responses. Immunol. Cell Biol. 2007, 85, 446–457. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 285–299. [Google Scholar] [CrossRef]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, eaat9691. [Google Scholar] [CrossRef]
- Pjanova, D.; Mandrika, L.; Petrovska, R.; Vaivode, K.; Donina, S. Comparison of the effects of bacteriophage-derived dsRNA and poly(I:C) on ex vivo cultivated peripheral blood mononuclear cells. Immunol. Lett. 2019, 212, 114–119. [Google Scholar] [CrossRef]
- Diner, E.J.; Burdette, D.L.; Wilson, S.C.; Monroe, K.M.; Kellenberger, C.A.; Hyodo, M.; Hayakawa, Y.; Hammond, M.C.; Vance, R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013, 3, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- McCartney, S.A.; Colonna, M. Viral sensors: Diversity in pathogen recognition. Immunol. Rev. 2009, 227, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Thomsen, A.R. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef]
- Rosenwald, A.G.; Murray, B.; Toth, T.; Madupu, R.; Kyrillos, A.; Arora, G. Evidence for horizontal gene transfer between Chlamydophila pneumoniae and Chlamydia phage. Bacteriophage 2014, 4, e965076. [Google Scholar] [CrossRef]
- Metcalf, J.A.; Funkhouser-Jones, L.J.; Brileya, K.; Reysenbach, A.L.; Bordenstein, S.R. Antibacterial gene transfer across the tree of life. Elife 2014, 3, e04266. [Google Scholar] [CrossRef]
- Moran, Y.; Fredman, D.; Szczesny, P.; Grynberg, M.; Technau, U. Recurrent horizontal transfer of bacterial toxin genes to eukaryotes. Mol. Biol. Evol. 2012, 29, 2223–2230. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Bordenstein, S.R. Eukaryotic association module in phage WO genomes from Wolbachia. Nat. Commun. 2016, 7, 13155. [Google Scholar] [CrossRef]
- Chatterjee, A.; Duerkop, B.A. Beyond Bacteria: Bacteriophage-Eukaryotic Host Interactions Reveal Emerging Paradigms of Health and Disease. Front. Microbiol. 2018, 9, 1394. [Google Scholar] [CrossRef]
- Gorski, A.; Międzybrodzki, R.; Jończyk-Matysiak, E.; Żaczek, M.; Borysowski, J. Phage-specific diverse effects of bacterial viruses on the immune system. Future Microbiol. 2019, 14, 1171–1174. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed]
- Sergueev, K.V.; Filippov, A.A.; Farlow, J.; Su, W.; Kvachadze, L.; Balarjishvili, N.; Kutateladze, M.; Nikolich, M.P. Correlation of Host Range Expansion of Therapeutic Bacteriophage Sb-1 with Allele State at a Hypervariable Repeat Locus. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in Vitro Evolution of Therapeutic Bacteriophages: The Appelmans Protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carroll-Portillo, A.; Lin, H.C. Bacteriophage and the Innate Immune System: Access and Signaling. Microorganisms 2019, 7, 625. https://doi.org/10.3390/microorganisms7120625
Carroll-Portillo A, Lin HC. Bacteriophage and the Innate Immune System: Access and Signaling. Microorganisms. 2019; 7(12):625. https://doi.org/10.3390/microorganisms7120625
Chicago/Turabian StyleCarroll-Portillo, Amanda, and Henry C. Lin. 2019. "Bacteriophage and the Innate Immune System: Access and Signaling" Microorganisms 7, no. 12: 625. https://doi.org/10.3390/microorganisms7120625
APA StyleCarroll-Portillo, A., & Lin, H. C. (2019). Bacteriophage and the Innate Immune System: Access and Signaling. Microorganisms, 7(12), 625. https://doi.org/10.3390/microorganisms7120625




