Meta-16S rRNA Gene Phylogenetic Reconstruction Reveals the Astonishing Diversity of Cosmopolitan Myxobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Construction of 16S rRNA Gene Sequences of Myxobacteria
2.2. Sequences Validation and Environmental Categories of Myxobacteria
2.3. Phylogenetic Analysis of 16S rRNA Gene Sequences of Myxobacteria
3. Results
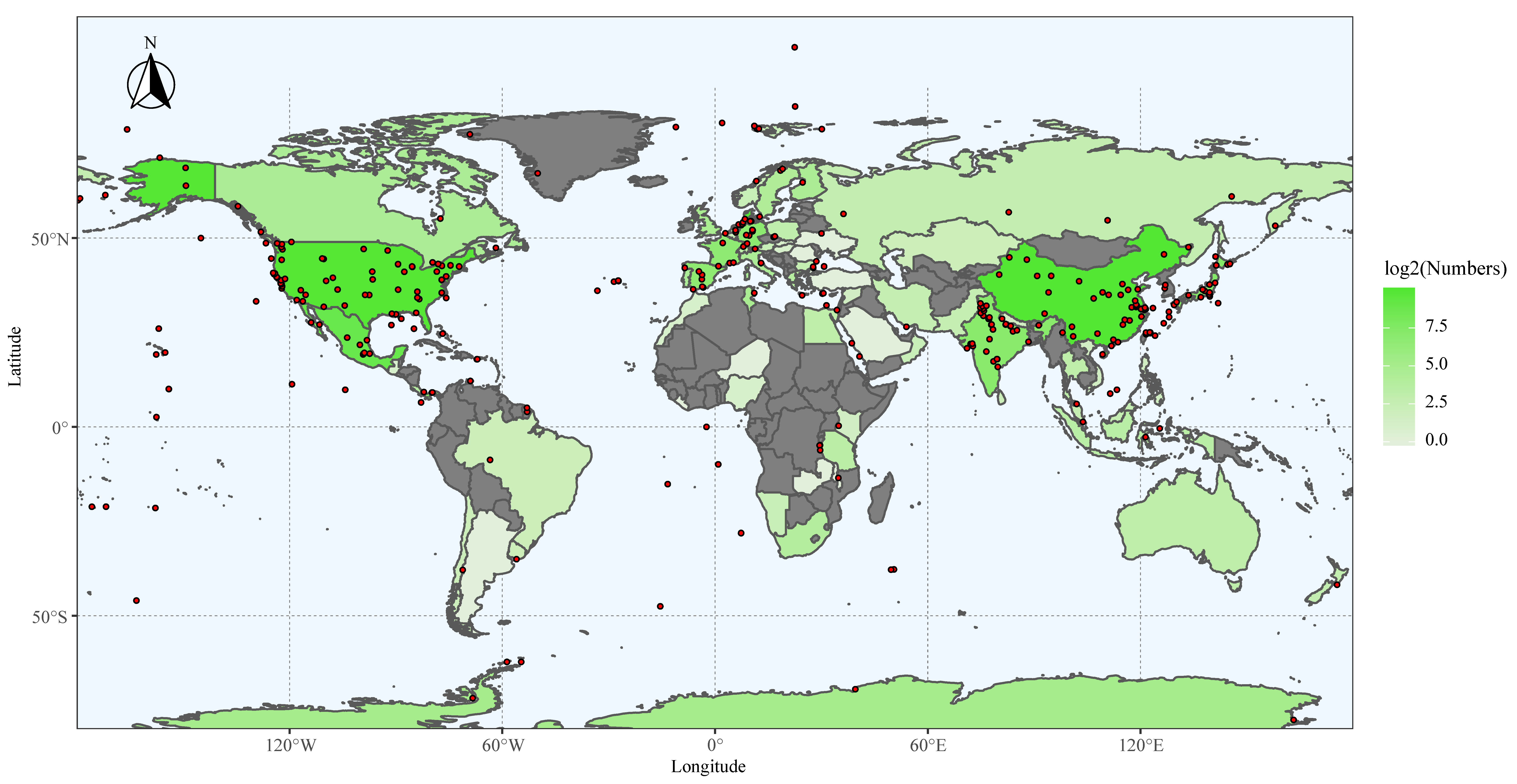
3.1. The Isolation Sources and Locations of 16S rRNA Gene Sequences of Myxobacteria
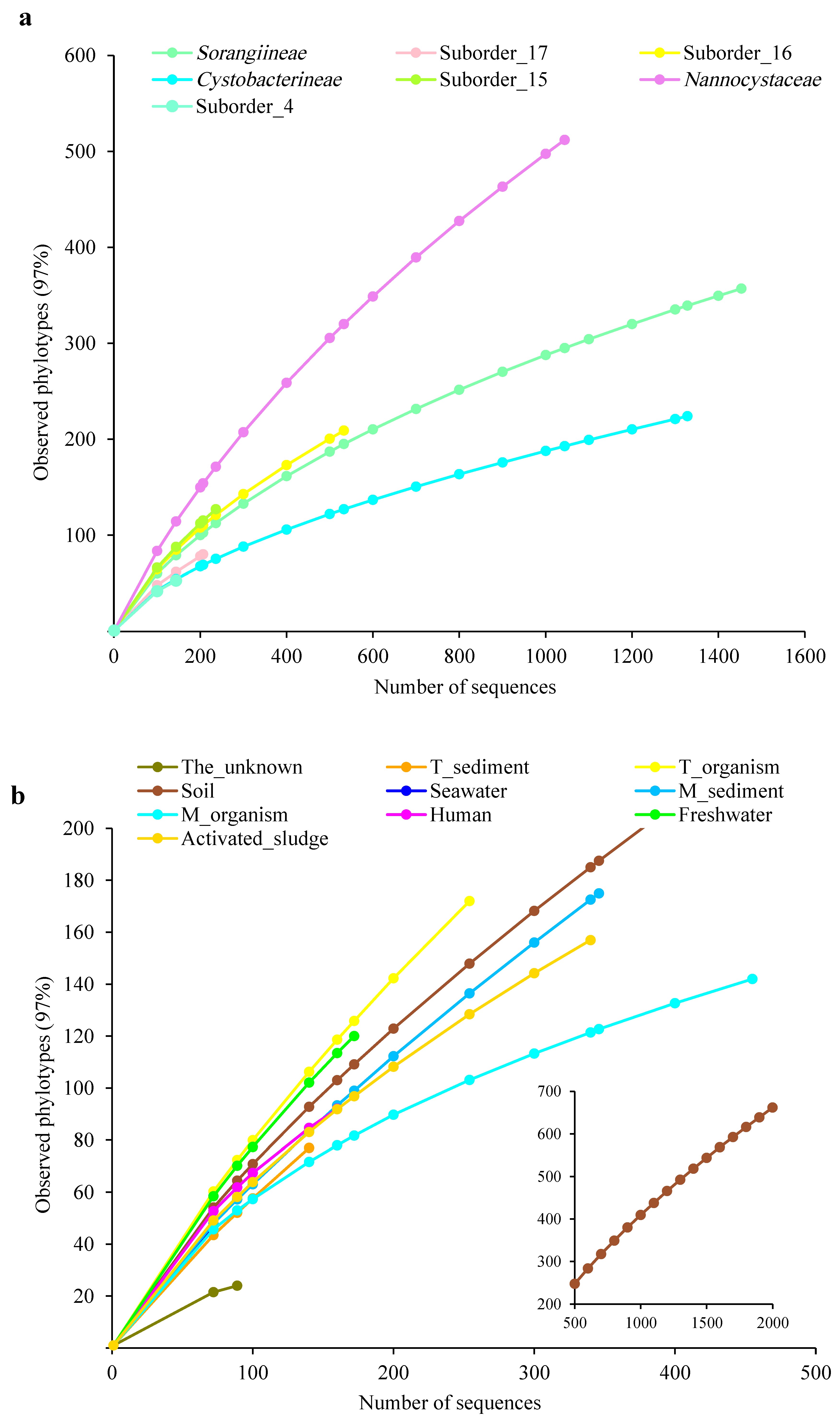
3.2. The Unexpected Diversity of Myxobacteria
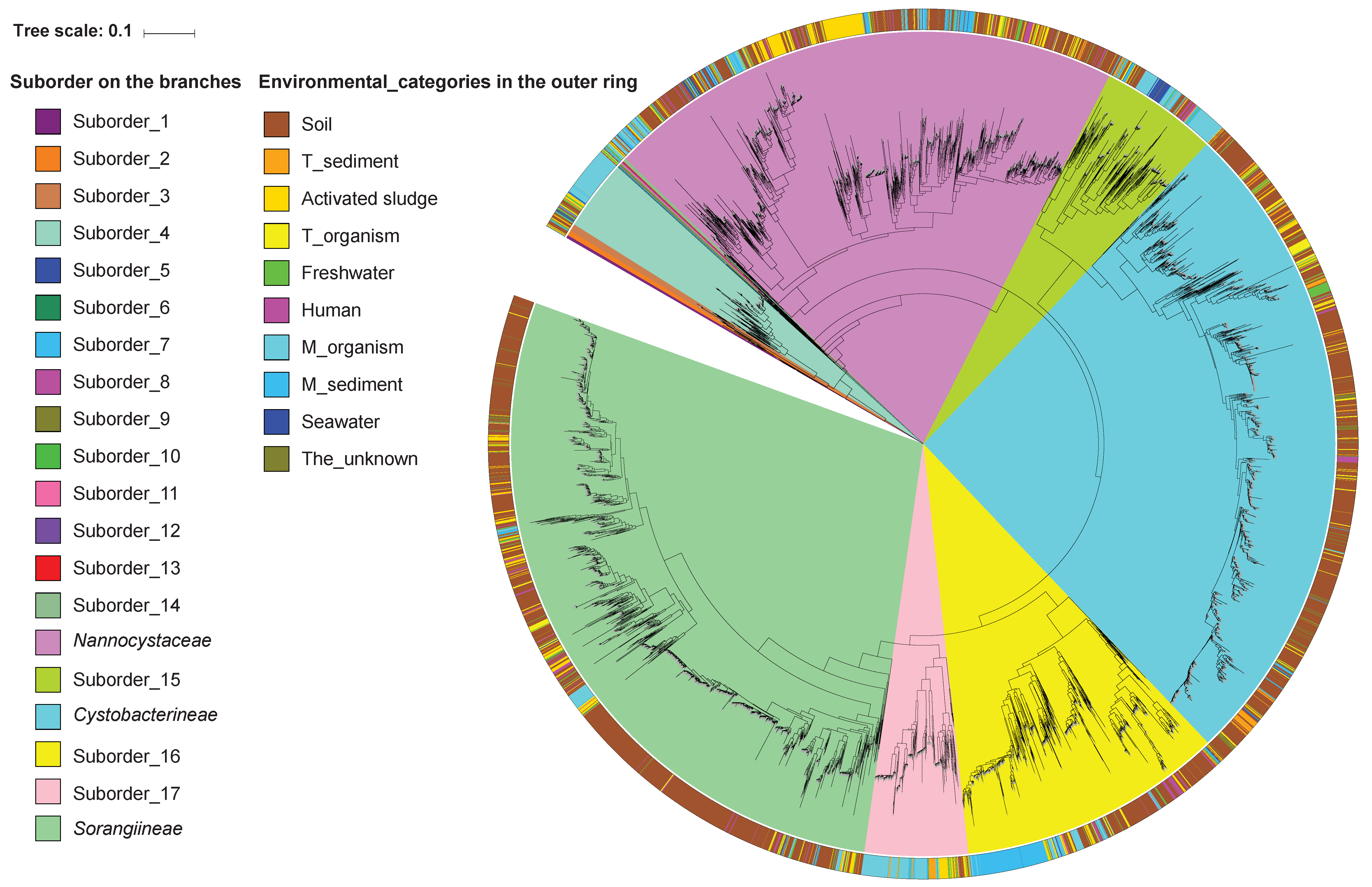
3.3. The Expanded Phylogeny of Myxobacteria
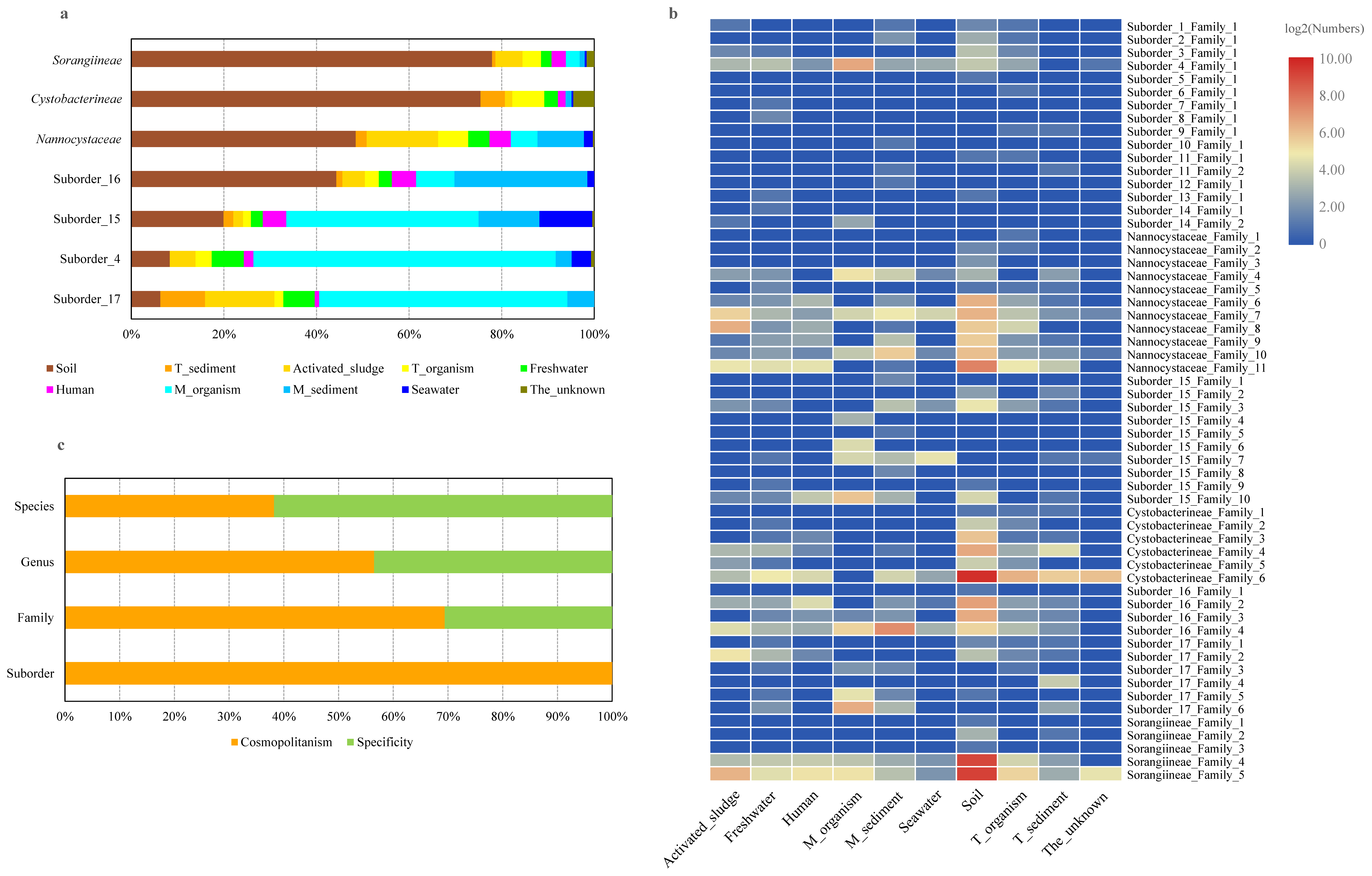
3.4. Global Biogeographic Distribution of Myxobacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shimkets, L.; Woese, C.R. A phylogenetic analysis of the myxobacteria: Basis for their classification. Proc. Natl. Acad. Sci. USA. 1992, 89, 9459–9463. [Google Scholar] [CrossRef] [PubMed]
- Velicer, G.J.; Vos, M. Sociobiology of the myxobacteria. Annu. Rev. Microbiol. 2009, 63, 599–623. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Dorado, J.; Marcos-Torres, F.J.; García-Bravo, E.; Moraleda-Muñoz, A.; Pérez, J. Myxobacteria: Moving, killing, feeding, and surviving together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef] [PubMed]
- Nan, B.; Chen, J.; Neu, J.C.; Berry, R.M.; Oster, G.; Zusman, D.R. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc. Natl. Acad. Sci. USA 2011, 108, 2498–2503. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, P.G.; Morphew, R.M.; Whitworth, D.E. Myxobacteria are able to prey broadly upon clinically-relevant pathogens, exhibiting a prey range which cannot be explained by phylogeny. Front. Microbiol. 2017, 8, 1593. [Google Scholar] [CrossRef] [PubMed]
- Huntley, S.; Hamann, N.; Wegener-Feldbrügge, S.; Treuner-Lange, A.; Kube, M.; Reinhardt, R.; Klages, S.; Müller, R.; Ronning, C.M.; Nierman, W.C.; et al. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol. Biol. Evol. 2011, 28, 1083–1097. [Google Scholar] [CrossRef]
- Yan, Z.C.; Wang, B.; Li, Y.Z.; Gong, X.; Zhang, H.Q.; Gao, P.J. Morphologies and phylogenetic classification of cellulolytic myxobacteria. Syst. Appl. Microbiol. 2003, 26, 104–109. [Google Scholar] [CrossRef]
- Zusman, D.R.; Scott, A.E.; Yang, Z.; Kirby, J.R. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007, 5, 862–872. [Google Scholar] [CrossRef]
- Herrmann, J.; Fayad, A.A.; Müller, R. Natural products from myxobacteria: Novel metabolites and bioactivities. Nat. Prod. Rep. 2017, 34, 135–160. [Google Scholar] [CrossRef]
- Mulwa, L.S.; Stadler, M. Antiviral compounds from myxobacteria. Microorganisms 2018, 6, 73. [Google Scholar] [CrossRef]
- Li, Z.; Ye, X.; Liu, M.; Xia, C.; Zhang, L.; Luo, X.; Wang, T.; Chen, Y.; Zhao, Y.; Qiao, Y.; et al. A novel outer membrane beta-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Moraleda-Mñnoz, A.; Pérez, J.; Fontes, M.; Murillo, F.J.; Muñoz-Dorado, J. Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 2005, 56, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Sanford, R.A.; Cole, J.R.; Tiedje, J.M. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 2002, 68, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sanford, R.A.; Löffler, F.E. Uranium(VI) reduction by Anaeromyxobacter dehalogenans strain 2CP-C. Appl. Environ. Microbiol. 2006, 72, 3608–3614. [Google Scholar] [CrossRef]
- Sproer, C.; Reichenbach, H.; Stackebrandt, E. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Bacteriol. 1999, 49 Pt 3, 1255–1262. [Google Scholar] [CrossRef]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Gerth, K.; Steinmetz, H.; Müller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9, 803. [Google Scholar] [CrossRef]
- Jiang, D.M.; Kato, C.; Zhou, X.W.; Wu, Z.H.; Sato, T.; Li, Y.Z. Phylogeographic separation of marine and soil myxobacteria at high levels of classification. ISME J. 2010, 4, 1520–1530. [Google Scholar] [CrossRef]
- Li, S.G.; Zhou, X.W.; Li, P.F.; Han, K.; Li, W.; Li, Z.F.; Wu, Z.H.; Li, Y.Z. The existence and diversity of myxobacteria in lake mud—A previously unexplored myxobacteria habitat. Environ. Microbiol. Rep. 2012, 4, 587–595. [Google Scholar] [CrossRef]
- Reichenbach, H. The ecology of the myxobacteria. Environ. Microbiol. 1999, 1, 15–21. [Google Scholar] [CrossRef]
- Dawid, W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 2000, 24, 403–427. [Google Scholar] [CrossRef]
- Mohr, K.I.; Zindler, T.; Wink, J.; Wilharm, E.; Stadler, M. Myxobacteria in high moor and fen: An astonishing diversity in a neglected extreme habitat. Microbiologyopen 2017, 6. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Q.; Cai, Z.; Xie, X.; Zhu, H. Isolation and identification of myxobacteria from saline-alkaline soils in Xinjiang, China. PLoS ONE 2013, 8, e70466. [Google Scholar] [CrossRef]
- Mohr, K.I.; Moradi, A.; Glaeser, S.P.; Kämpfer, P.; Gemperlein, K.; Nübel, U.; Schumann, P.; Müller, R.; Wink, J. Nannocystis konarekensis sp. nov., a novel myxobacterium from an Iranian desert. Int. J. Syst. Evol. Microbiol. 2018, 68, 721–729. [Google Scholar] [CrossRef]
- Iizuka, T.; Tokura, M.; Jojima, Y.; Hiraishi, A.; Yamanaka, S.; Fudou, R. Enrichment and phylogenetic analysis of moderately thermophilic myxobacteria from hot springs in Japan. Microbes Environ. 2006, 21, 189–199. [Google Scholar] [CrossRef][Green Version]
- Li, Y.Z.; Hu, W.; Zhang, Y.Q.; Qiu, Z.; Zhang, Y.; Wu, B.H. A simple method to isolate salt-tolerant myxobacteria from marine samples. J. Microbiol. Methods 2002, 50, 205–209. [Google Scholar] [CrossRef]
- Iizuka, T.; Jojima, Y.; Fudou, R.; Yamanaka, S. Isolation of myxobacteria from the marine environment. FEMS Microbiol. Lett. 1998, 169, 317–322. [Google Scholar] [CrossRef]
- Brinkhoff, T.; Fischer, D.; Vollmers, J.; Voget, S.; Beardsley, C.; Thole, S.; Mussmann, M.; Kunze, B.; Wagner-Döbler, I.; Daniel, R.; et al. Biogeography and phylogenetic diversity of a cluster of exclusively marine myxobacteria. ISME J. 2012, 6, 1260–1272. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Chen, I.M.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Tamames, J.; Abellán, J.J.; Pignatelli, M.; Camacho, A.; Moya, A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Bursakov, S.; Liu, M.Y.; Payne, W.J.; LeGall, J.; Moura, I.; Moura, J.J. Isolation and preliminary characterization of a soluble nitrate reductase from the sulfate reducing organism Desulfovibrio desulfuricans ATCC 27774. Anaerobe 1995, 1, 55–60. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Yarza, P.; Richter, M.; Peplies, J.; Euzeby, J.; Amann, R.; Schleifer, K.H.; Ludwig, W.; Glöckner, F.O.; Rosselló-Móra, R. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 2008, 31, 241–250. [Google Scholar] [CrossRef]
- Garcia, R.; Gerth, K.; Stadler, M.; Dogma, I.J., Jr.; Müller, R. Expanded phylogeny of myxobacteria and evidence for cultivation of the ‘unculturables’. Mol. Phylogenet. Evol. 2010, 57, 878–887. [Google Scholar] [CrossRef]
- Pedrós-Alió, C. The rare bacterial biosphere. Ann. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef]
- Wu, Z.H.; Jiang, D.M.; Li, P.; Li, Y.Z. Exploring the diversity of myxobacteria in a soil niche by myxobacteria-specific primers and probes. Environ. Microbiol. 2005, 7, 1602–1610. [Google Scholar] [CrossRef]
- Mohr, K.I.; Stechling, M.; Wink, J.; Wilharm, E.; Stadler, M. Comparison of myxobacterial diversity and evaluation of isolation success in two niches: Kiritimati Island and German compost. Microbiologyopen 2016, 5, 268–278. [Google Scholar] [CrossRef]
- Mohr, K.I. Diversity of myxobacteria-we only see the tip of the iceberg. Microorganisms 2018, 6, 84. [Google Scholar] [CrossRef]
- Albataineh, H.; Stevens, D.C. Marine myxobacteria: A few good halophiles. Mar. Drugs 2018, 16, 209. [Google Scholar] [CrossRef]
- Acosta-González, A.; Rosselló-Móra, R.; Marqués, S. Characterization of the anaerobic microbial community in oil-polluted subtidal sediments: Aromatic biodegradation potential after the Prestige oil spill. Environ. Microbiol. 2013, 15, 77–92. [Google Scholar] [CrossRef]
- Ganesh, S.; Parris, D.J.; DeLong, E.F.; Stewart, F.J. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014, 8, 187–211. [Google Scholar] [CrossRef]
- Karst, S.M.; Dueholm, M.S.; McIlroy, S.J.; Kirkegaard, R.H.; Nielsen, P.H.; Albertsen, M. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat. Biotechnol. 2018, 36, 190–195. [Google Scholar] [CrossRef]
- Wells, G.F.; Park, H.D.; Eggleston, B.; Francis, C.A.; Criddle, C.S. Fine-scale bacterial community dynamics and the taxa-time relationship within a full-scale activated sludge bioreactor. Water Res. 2011, 45, 5476–5488. [Google Scholar] [CrossRef]
- Harris, J.K.; Caporaso, J.G.; Walker, J.J.; Spear, J.R.; Gold, N.J.; Robertson, C.E.; Hugenholtz, P.; Goodrich, J.; McDonald, D.; Knights, D.; et al. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J. 2013, 7, 50–60. [Google Scholar] [CrossRef]
- Wang, X.; Hu, M.; Xia, Y.; Wen, X.; Ding, K. Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl. Environ. Microbiol. 2012, 78, 7042–7047. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012, 4, 77. [Google Scholar] [CrossRef]
- Shi, J.; Xu, C.; Han, Y.; Han, H. Enhanced anaerobic biodegradation efficiency and mechanism of quinoline, pyridine, and indole in coal gasification wastewater. Chem. Eng. J. 2019, 361, 1019–1029. [Google Scholar] [CrossRef]
| Abundant/Rare | Suborder | Family | Genus | Species | Numbers (%) |
|---|---|---|---|---|---|
| Rare | Suborder_1 | 1 | 5 | 5 | 5 (0.10) |
| Rare | Suborder_2 | 1 | 3 | 6 | 10 (0.20) |
| Rare | Suborder_3 | 1 | 3 | 8 | 15 (0.30) |
| Abundant | Suborder_4 | 1 | 28 | 44 | 144 (2.88) |
| Rare | Suborder_5 | 1 | 1 | 1 | 1 (0.02) |
| Rare | Suborder_6 | 1 | 1 | 1 | 1 (0.02) |
| Rare | Suborder_7 | 1 | 1 | 1 | 1 (0.02) |
| Rare | Suborder_8 | 1 | 1 | 1 | 2 (0.04) |
| Rare | Suborder_9 | 1 | 2 | 2 | 2 (0.04) |
| Rare | Suborder_10 | 1 | 1 | 1 | 1 (0.02) |
| Rare | Suborder_11 | 2 | 3 | 3 | 4 (0.08) |
| Rare | Suborder_12 | 1 | 1 | 1 | 1 (0.02) |
| Rare | Suborder_13 | 1 | 2 | 2 | 2 (0.04) |
| Rare | Suborder_14 | 2 | 3 | 3 | 7 (0.14) |
| Abundant | Nannocystaceae | 11 | 157 | 309 | 1044 (20.89) |
| Abundant | Suborder_15 | 10 | 48 | 108 | 236 (4.72) |
| Abundant | Cystobacterineae | 6 | 50 | 131 | 1328 (26.58) |
| Abundant | Suborder_16 | 4 | 54 | 116 | 533 (10.67) |
| Abundant | Suborder_17 | 6 | 32 | 60 | 207 (4.14) |
| Abundant | Sorangiineae | 5 | 49 | 195 | 1453 (29.08) |
| Total | 20 | 58 | 445 | 998 | 4997 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Yao, Q.; Zhu, H. Meta-16S rRNA Gene Phylogenetic Reconstruction Reveals the Astonishing Diversity of Cosmopolitan Myxobacteria. Microorganisms 2019, 7, 551. https://doi.org/10.3390/microorganisms7110551
Liu Y, Yao Q, Zhu H. Meta-16S rRNA Gene Phylogenetic Reconstruction Reveals the Astonishing Diversity of Cosmopolitan Myxobacteria. Microorganisms. 2019; 7(11):551. https://doi.org/10.3390/microorganisms7110551
Chicago/Turabian StyleLiu, Yang, Qing Yao, and Honghui Zhu. 2019. "Meta-16S rRNA Gene Phylogenetic Reconstruction Reveals the Astonishing Diversity of Cosmopolitan Myxobacteria" Microorganisms 7, no. 11: 551. https://doi.org/10.3390/microorganisms7110551
APA StyleLiu, Y., Yao, Q., & Zhu, H. (2019). Meta-16S rRNA Gene Phylogenetic Reconstruction Reveals the Astonishing Diversity of Cosmopolitan Myxobacteria. Microorganisms, 7(11), 551. https://doi.org/10.3390/microorganisms7110551





