A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
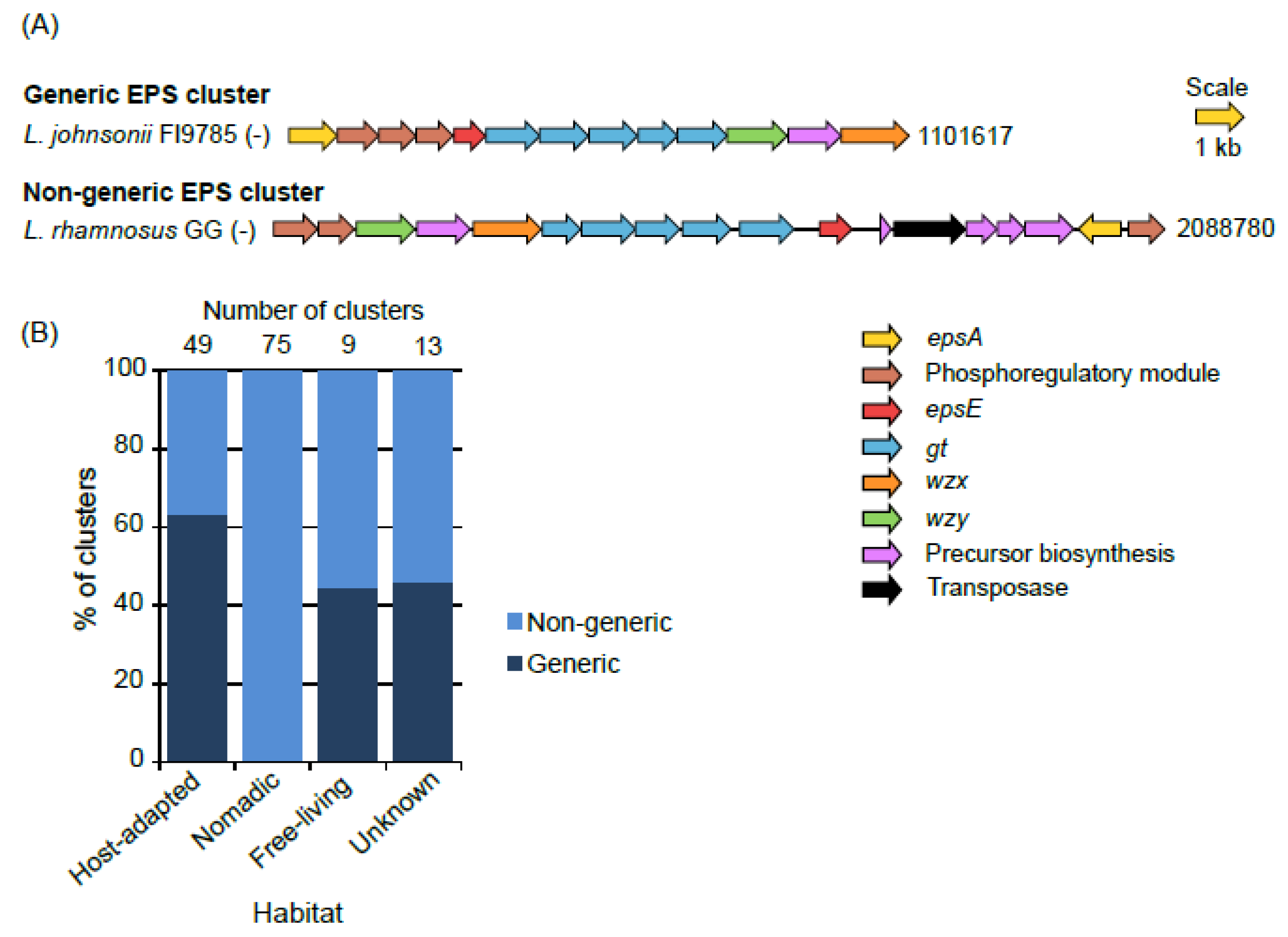
3.1. Number of Clusters and Gene Composition
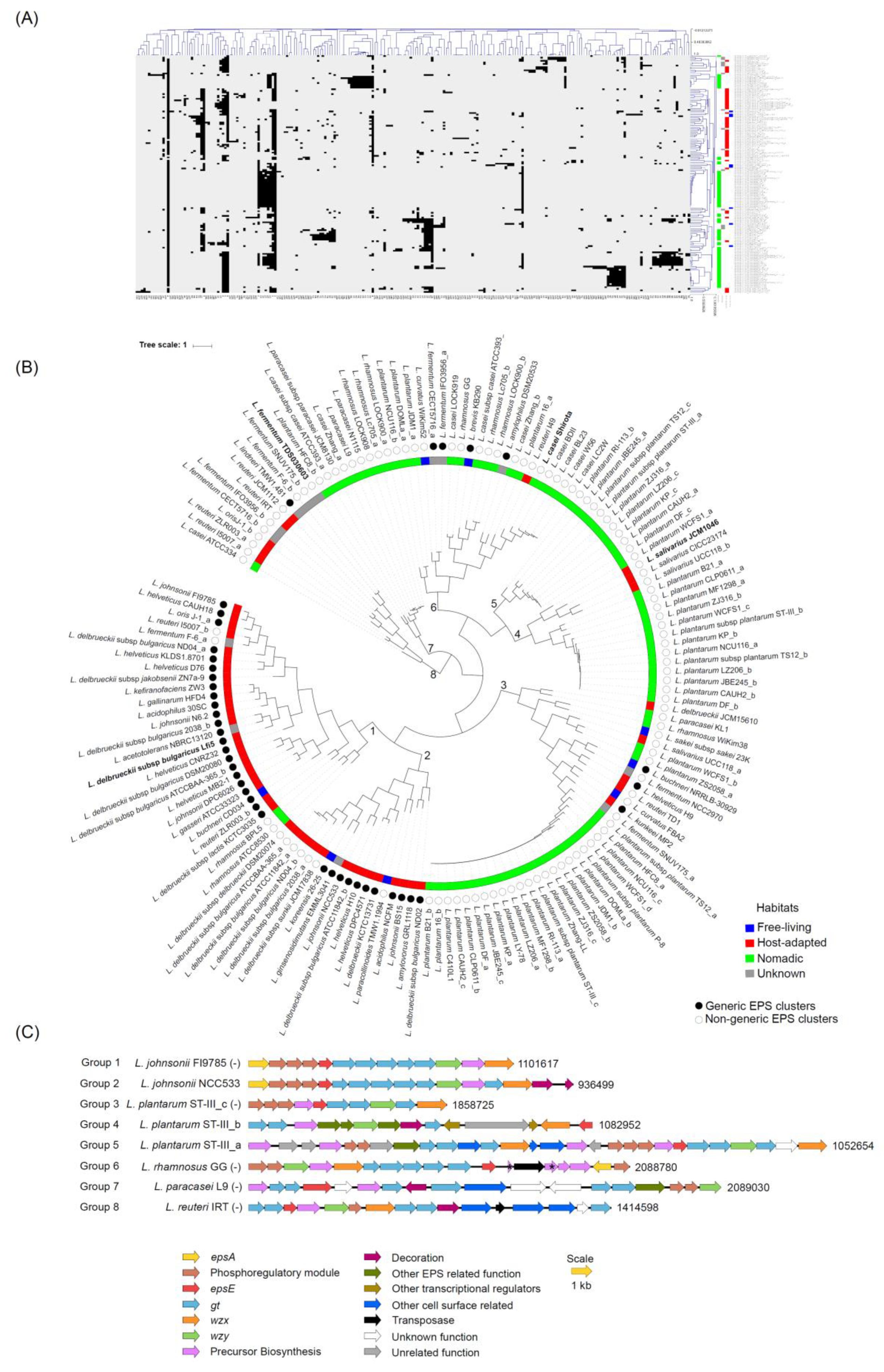
3.2. Organization of Genes in the Clusters
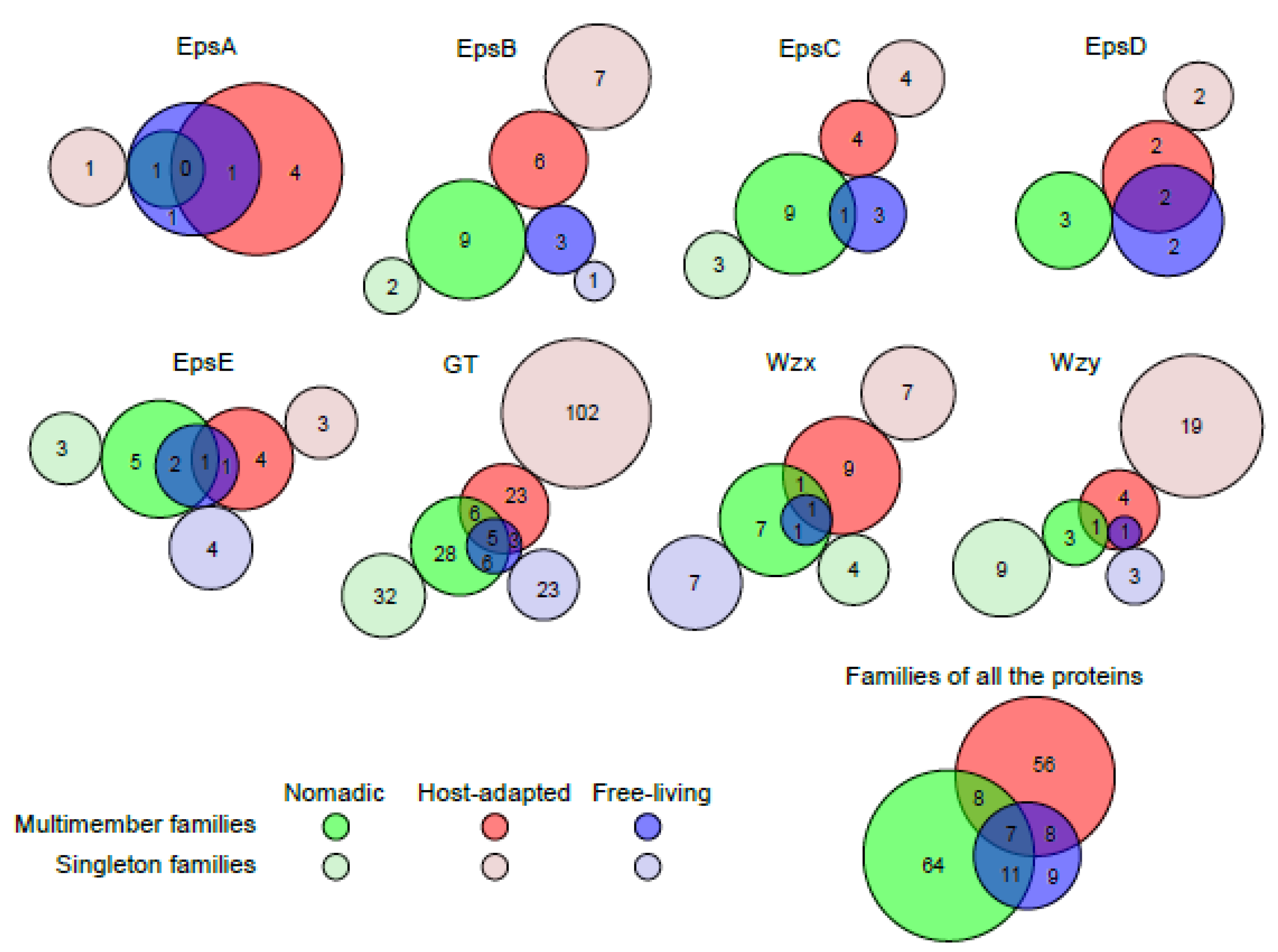
3.3. Variation in the Number of Protein Families across Various Gene Functionalities
3.4. EpsA
3.5. Phosphoregulatory Module: EpsB, C and D
3.6. EpsE
3.7. GTs
3.8. Wzx and Wzy
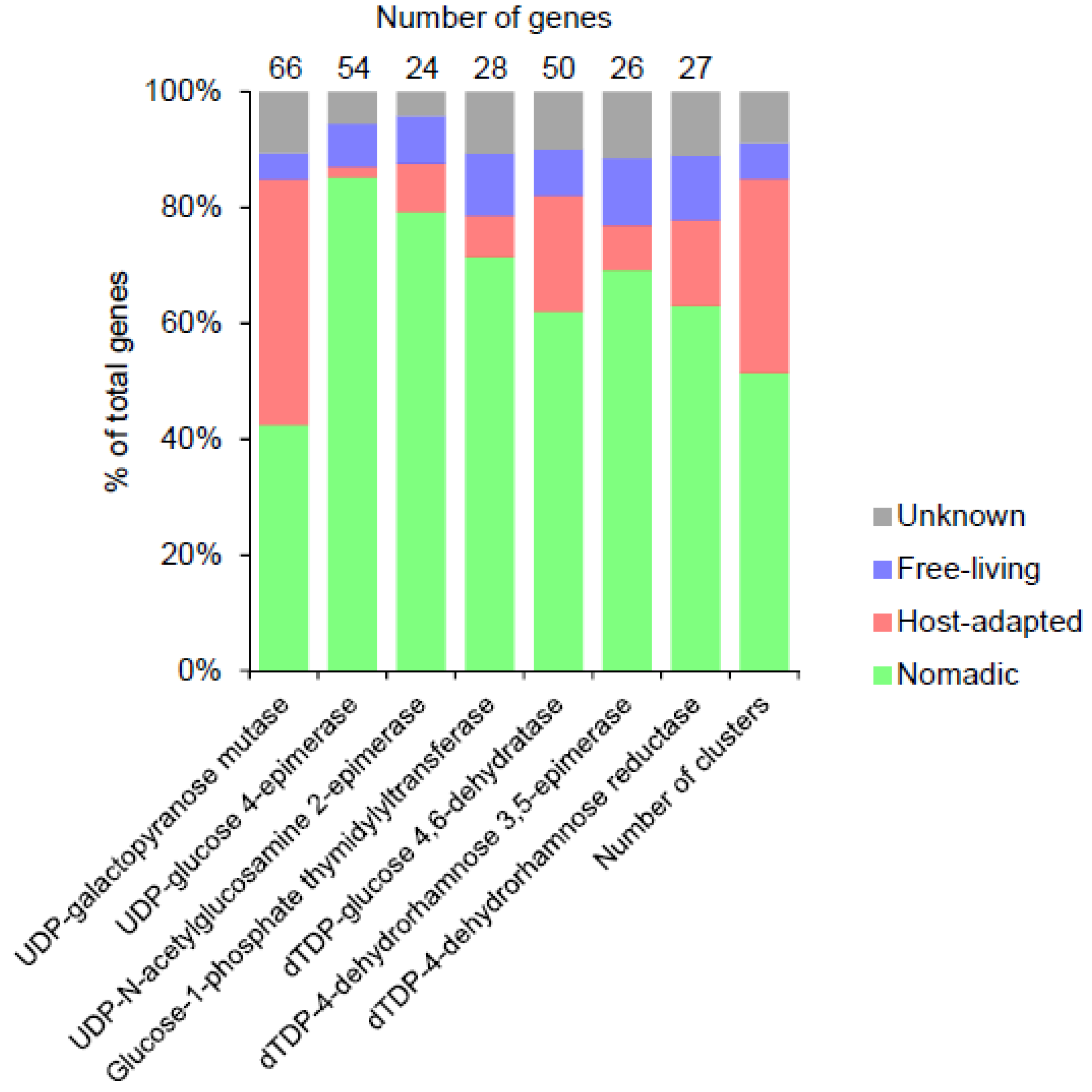
3.9. Precursor Biosynthesis
3.10. Other Genes
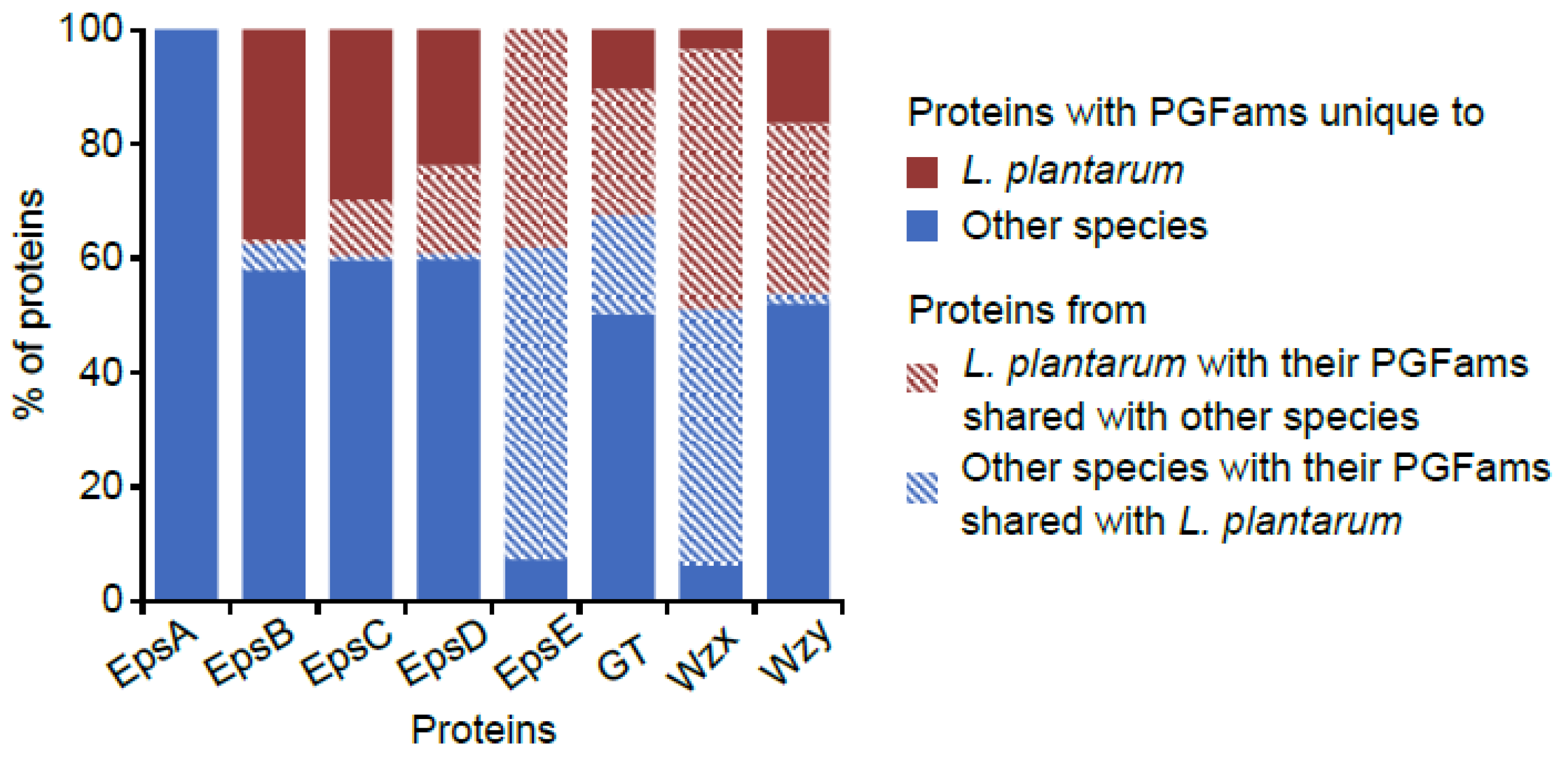
3.11. Sharing of Protein Families Across Various Habitats
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turpin, W.; Humblot, C.; Thomas, M.; Guyot, J.-P. Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int. J. Food Microbiol. 2010, 143, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Hama, S.; Mizuno, S.; Kihara, M.; Tanaka, T.; Ogino, C.; Noda, H.; Kondo, A. Production of d-lactic acid from hardwood pulp by mechanical milling followed by simultaneous saccharification and fermentation using metabolically engineered Lactobacillus plantarum. Bioresour. Technol. 2015, 187, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 2008, 72, 728–764. [Google Scholar] [CrossRef] [PubMed]
- Lamothe, G.; Jolly, L.; Mollet, B.; Stingele, F. Genetic and biochemical characterization of exopolysaccharide biosynthesis by Lactobacillus delbrueckii subsp. bulgaricus. Arch. Microbiol. 2002, 178, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Pridmore, R.D.; Berger, B.; Desiere, F.; Vilanova, D.; Barretto, C.; Pittet, A.-C.; Zwahlen, M.-C.; Rouvet, M.; Altermann, E.; Barrangou, R.; et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 2004, 101, 2512–2517. [Google Scholar] [CrossRef]
- Lebeer, S.; Verhoeven, T.L.A.; Francius, G.; Schoofs, G.; Lambrichts, I.; Dufrêne, Y.; Vanderleyden, J.; De Keersmaecker, S.C.J. Identification of a Gene Cluster for the Biosynthesis of a Long, Galactose-Rich Exopolysaccharide in Lactobacillus rhamnosus GG and Functional Analysis of the Priming Glycosyltransferase. Appl. Environ. Microbiol. 2009, 75, 3554–3563. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Ahmed, Z.; Bai, X.; Wang, J. Complete genome sequence of Lactobacillus kefiranofaciens ZW3. J. Bacteriol. 2011, 193, 4280–4281. [Google Scholar] [CrossRef]
- Dertli, E.; Colquhoun, I.J.; Gunning, A.P.; Bongaerts, R.J.; Le Gall, G.; Bonev, B.B.; Mayer, M.J.; Narbad, A. Structure and biosynthesis of two exopolysaccharides produced by Lactobacillus johnsonii FI9785. J. Biol. Chem. 2013, 288, 31938–31951. [Google Scholar] [CrossRef]
- Song, X.; Xiong, Z.; Kong, L.; Wang, G.; Ai, L. Relationship Between Putative eps Genes and Production of Exopolysaccharide in Lactobacillus casei LC2W. Front. Microbiol. 2018, 9, 1882. [Google Scholar] [CrossRef]
- Dertli, E.; Mayer, M.J.; Narbad, A. Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol. 2015, 15, 8. [Google Scholar] [CrossRef]
- Donot, F.; Fontana, A.; Baccou, J.C.; Schorr-Galindo, S. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 2012, 87, 951–962. [Google Scholar] [CrossRef]
- Ozturk, S.; Aslim, B.; Suludere, Z. Evaluation of chromium(VI) removal behaviour by two isolates of Synechocystis sp. in terms of exopolysaccharide (EPS) production and monomer composition. Bioresour. Technol. 2009, 100, 5588–5593. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; De Vin, F. Exopolysaccharides from Lactic Acid Bacteria. In Comprehensive Glycosciences; Kamerling, H., Ed.; Elsevier Ltd.: Oxford, UK, 2007; pp. 477–519. [Google Scholar]
- Ruas-Madiedo, P.; Gueimonde, M.; Margolles, A.; De Los Reyes-Gavilán, C.G.; Salminen, S. Exopolysaccharides Produced by Probiotic Strains Modify the Adhesion of Probiotics and Enteropathogens to Human Intestinal Mucus. J. Food Prot. 2006, 69, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; López, P.; Capozzi, V.; de Palencia, P.F.; Dueñas, M.T.; Spano, G.; Fiocco, D.; Russo, P.; López, P.; Capozzi, V.; et al. Beta-Glucans Improve Growth, Viability and Colonization of Probiotic Microorganisms. Int. J. Mol. Sci. 2012, 13, 6026–6039. [Google Scholar] [CrossRef] [PubMed]
- Živković, M.; Miljković, M.S.; Ruas-Madiedo, P.; Markelić, M.B.; Veljović, K.; Tolinački, M.; Soković, S.; Korać, A.; Golić, N. EPS-SJ Exopolisaccharide Produced by the Strain Lactobacillus paracasei subsp. paracasei BGSJ2-8 Is Involved in Adhesion to Epithelial Intestinal Cells and Decrease on E. coli Association to Caco-2 Cells. Front. Microbiol. 2016, 7, 286. [Google Scholar]
- Juvonen, R.; Honkapää, K.; Maina, N.H.; Shi, Q.; Viljanen, K.; Maaheimo, H.; Virkki, L.; Tenkanen, M.; Lantto, R. The impact of fermentation with exopolysaccharide producing lactic acid bacteria on rheological, chemical and sensory properties of pureed carrots (Daucus carota L.). Int. J. Food Microbiol. 2015, 207, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Hamet, M.F.; Piermaria, J.A.; Abraham, A.G. Selection of EPS-producing Lactobacillus strains isolated from kefir grains and rheological characterization of the fermented milks. LWT Food Sci. Technol. 2015, 63, 129–135. [Google Scholar] [CrossRef]
- Wang, K.; Li, W.; Rui, X.; Chen, X.; Jiang, M.; Dong, M. Characterization of a novel exopolysaccharide with antitumor activity from Lactobacillus plantarum 70810. Int. J. Biol. Macromol. 2014, 63, 133–139. [Google Scholar] [CrossRef]
- Tang, Y.; Dong, W.; Wan, K.; Zhang, L.; Li, C.; Zhang, L.; Liu, N. Exopolysaccharide Produced by Lactobacillus plantarum Induces Maturation of Dendritic Cells in BALB/c Mice. PLoS ONE 2015, 10, e0143743. [Google Scholar] [CrossRef]
- Wang, Y.; Ahmed, Z.; Feng, W.; Li, C.; Song, S. Physicochemical properties of exopolysaccharide produced by Lactobacillus kefiranofaciens ZW3 isolated from Tibet kefir. Int. J. Biol. Macromol. 2008, 43, 283–288. [Google Scholar] [CrossRef]
- Li, S.; Huang, R.; Shah, N.P.; Tao, X.; Xiong, Y.; Wei, H. Antioxidant and antibacterial activities of exopolysaccharides from Bifidobacterium bifidum WBIN03 and Lactobacillus plantarum R315. J. Dairy Sci. 2014, 97, 7334–7343. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, A.; Nácher-Vázquez, M.; Notararigo, S.; López, P.; Mohedano, M.L. Chapter 22—Current and Future Applications of Bacterial Extracellular Polysaccharides. In Probiotics, Prebiotics, and Synbiotics; Watson, R.R., Preedy, V.R., Eds.; Elsevier Ltd.: Oxford, UK, 2016; pp. 329–344. [Google Scholar]
- Zeidan, A.A.; Poulsen, V.K.; Janzen, T.; Buldo, P.; Derkx, P.M.F.; Øregaard, G.; Neves, A.R. Polysaccharide production by lactic acid bacteria: From genes to industrial applications. FEMS Microbiol. Rev. 2017, 41, S168–S200. [Google Scholar] [CrossRef]
- Schmid, J.; Sieber, V.; Rehm, B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015, 6, 496. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.; Newell, J.; Porcelli, I.; Vincent, S.J.F.; Stingele, F. Lactobacillus helveticus glycosyltransferases: From genes to carbohydrate synthesis. Glycobiology 2002, 12, 319–327. [Google Scholar] [CrossRef]
- Horn, N.; Wegmann, U.; Dertli, E.; Mulholland, F.; Collins, S.R.A.; Waldron, K.W.; Bongaerts, R.J.; Mayer, M.J.; Narbad, A. Spontaneous Mutation Reveals Influence of Exopolysaccharide on Lactobacillus johnsonii Surface Characteristics. PLoS ONE 2013, 8, e59957. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, M.; Miljkovic, M.; Ruas-Madiedo, P.; Strahinic, I.; Tolinacki, M.; Golic, N.; Kojic, M. Exopolysaccharide production and ropy phenotype are determined by two gene clusters in putative probiotic strain Lactobacillus paraplantarum BGCG11. Appl. Environ. Microbiol. 2015, 81, 1387–1396. [Google Scholar] [CrossRef] [PubMed]
- I-Chiao, L.; Graziano, C.; Iris, I.; Nico, T.; Marjolein, M.; Peter, A.; Giuseppe, S.; Michiel, K. Strain-specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host interactions. Appl. Environ. Microbiol. 2016, 82, 3959–3970. [Google Scholar]
- Vastano, V.; Perrone, F.; Marasco, R.; Sacco, M.; Muscariello, L. Transcriptional analysis of exopolysaccharides biosynthesis gene clusters in Lactobacillus plantarum. Arch. Microbiol. 2016, 198, 295–300. [Google Scholar] [CrossRef]
- Remus, D.M.; van Kranenburg, R.; van Swam, I.I.; Taverne, N.; Bongers, R.S.; Wels, M.; Wells, J.M.; Bron, P.A.; Kleerebezem, M. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb. Cell Fact. 2012, 11, 149. [Google Scholar] [CrossRef]
- Peant, B.; LaPointe, G.; Gilbert, C.; Atlan, D.; Ward, P.; Roy, D. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 2005, 151, 1839–1851. [Google Scholar] [CrossRef] [PubMed]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.F.T.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Springer: Boston, MA, USA, 2010; pp. 267–277. [Google Scholar]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Aanensen, D.M.; Mavroidi, A.; Saunders, D.; Rabbinowitsch, E.; Collins, M.; Donohoe, K.; Harris, D.; Murphy, L.; Quail, M.A.; et al. Genetic Analysis of the Capsular Biosynthetic Locus from All 90 Pneumococcal Serotypes. PLoS Genet. 2006, 2, e31. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Siezen, R.J.; Nauta, A. In silico prediction of horizontal gene transfer events in Lactobacillus bulgaricus and Streptococcus thermophilus reveals protocooperation in yogurt manufacturing. Appl. Environ. Microbiol. 2009, 75, 4120–4129. [Google Scholar] [CrossRef] [PubMed]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.; Stingele, F. Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int. Dairy J. 2001, 11, 733–745. [Google Scholar] [CrossRef]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Jr, A.F.; Folador, E.L.; Gomide, A.C.P.; Goes-Neto, A.; Azevedo, V.A.C.; Wattam, A.R.; Oliveira, A.F., Jr.; Folador, E.L.; Gomide, A.C.P.; Goes-Neto, A.; et al. Cell Division in genus Corynebacterium: Protein-protein interaction and molecular docking of SepF and FtsZ in the understanding of cytokinesis in pathogenic species. An. Acad. Bras. Cienc. 2018, 90, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.V.C.; Figueiredo, H.; Ramos, R.; Guimarães, L.C.; Pereira, F.L.; Dorella, F.A.; Selim, S.A.K.; Salaheldean, M.; Silva, A.; Wattam, A.R.; et al. Comparative genomic analysis between Corynebacterium pseudotuberculosis strains isolated from buffalo. PLoS ONE 2017, 12, e0176347. [Google Scholar] [CrossRef] [PubMed]
- Satti, S.M.; Shah, A.A.; Auras, R.; Marsh, T.L. Genome annotation of Poly(lactic acid) degrading Pseudomonas aeruginosa and Sphingobacterium sp. bioRxiv 2019, 609883. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar, P.; Das, P.; Kapur, M.K. Draft genome of Streptomyces sp. strain 130 and functional analysis of extracellular enzyme producing genes. Mol. Biol. Rep. 2019, 46, 5063–5071. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulou, M.; Vuillemin, M.; Campbell-Sills, H.; Lucas, P.M.; Ballestra, P.; Miot-Sertier, C.; Favier, M.; Coulon, J.; Moine, V.; Doco, T.; et al. Exopolysaccharide (EPS) Synthesis by Oenococcus oeni: From Genes to Phenotypes. PLoS ONE 2014, 9, e98898. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, M.A.; Reeves, P.R. Progress in Our Understanding of wzx Flippase for Translocation of Bacterial Membrane Lipid-Linked Oligosaccharide. J. Bacteriol. 2018, 200, e00154-17. [Google Scholar] [CrossRef]
- Eberhardt, A.; Hoyland, C.N.; Vollmer, D.; Bisle, S.; Cleverley, R.M.; Johnsborg, O.; Håvarstein, L.S.; Lewis, R.J.; Vollmer, W. Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae. Microb. Drug Resist. 2012, 18, 240–255. [Google Scholar] [CrossRef]
- Chan, Y.G.-Y.; Kim, H.K.; Schneewind, O.; Missiakas, D. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-cpsA-Psr (LCP) family of enzymes. J. Biol. Chem. 2014, 289, 15680–15690. [Google Scholar] [CrossRef]
- Cieslewicz, M.J.; Kasper, D.L.; Wang, Y.; Wessels, M.R. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 2001, 276, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Morona, J.K.; Paton, J.C.; Miller, D.C.; Morona, R. Tyrosine phosphorylation of cpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 2002, 35, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, J.R.; McMahon, D.J.; Welker, D.L.; Oberg, C.J.; Moineau, S. Biochemistry, Genetics, and Applications of Exopolysaccharide Production in Streptococcus thermophilus: A Review. J. Dairy Sci. 2003, 86, 407–423. [Google Scholar] [CrossRef]
- Lazarevic, V.; Margot, P.; Soldo, B.; Karamata, D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: A regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J. Gen. Microbiol. 1992, 138, 1949–1961. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dertli, E.; Mayer, M.J.; Colquhoun, I.J.; Narbad, A. epsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785. Microb. Biotechnol. 2016, 9, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Bender, M.H.; Cartee, R.T.; Yother, J. Positive correlation between tyrosine phosphorylation of cpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 2003, 185, 6057–6066. [Google Scholar] [CrossRef] [PubMed]
- Muscariello, L.; Marino, C.; Capri, U.; Vastano, V.; Marasco, R.; Sacco, M. CcpA and three newly identified proteins are involved in biofilm development in Lactobacillus plantarum. J. Basic Microbiol. 2013, 53, 62–71. [Google Scholar] [CrossRef] [PubMed]
- D’Abrosca, G.; Paladino, A.; Cuoco, E.; Marasco, R.; Pacifico, S.; Piccolella, S.; Vastano, V.; Sacco, M.; Isernia, C.; Muscariello, L.; et al. Structural Characterization of the Lactobacillus plantarum FlmC Protein Involved in Biofilm Formation. Molecules 2018, 23, 2252. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Marie, C.; Delorme, C.; Faurie, J.-M.; Mercier, G.; Ehrlich, D.; Renault, P. Control of epsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by epsD tyrosine kinase. J. Bacteriol. 2007, 189, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Elsholz, A.K.W.; Wacker, S.A.; Losick, R. Self-regulation of exopolysaccharide production in Bacillus subtilis by a tyrosine kinase. Genes Dev. 2014, 28, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Toniolo, C.; Balducci, E.; Romano, M.R.; Proietti, D.; Ferlenghi, I.; Grandi, G.; Berti, F.; Ros, I.M.Y.; Janulczyk, R. Streptococcus agalactiae capsule polymer length and attachment is determined by the proteins cpsABCD. J. Biol. Chem. 2015, 290, 9521–9532. [Google Scholar] [CrossRef] [PubMed]
- Almirón-Roig, E.; Gasson, M.J.; Mulholland, F.; Griffin, A.M. The complete cps gene cluster from Streptococcus thermophilus NCFB 2393 involved in the biosynthesis of a new exopolysaccharide. Microbiology 2000, 146, 2793–2802. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.B.; Jones, C.; Larson, T.R.; Calix, J.J.; Zartler, E.R.; Yother, J.; Nahm, M.H. Streptococcus pneumoniae serotype 11D has a bispecific glycosyltransferase and expresses two different capsular polysaccharide repeating units. J. Biol. Chem. 2013, 288, 21945–21954. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.T.; Taylor, V.L.; Qi, M.; Lam, J.S. Membrane Topology Mapping of the O-Antigen Flippase (wzx), Polymerase (wzy), and Ligase (WaaL) from Pseudomonas aeruginosa PAO1 Reveals Novel Domain Architectures. MBio 2010, 1, e00189-10. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; Salazar, N.; Clara, G. Biosynthesis and Chemical Composition of Exopolysaccharides; Caister Academic Press: Norfolk, UK, 2009. [Google Scholar]
- Ishiyama, N.; Creuzenet, C.; Lam, J.S.; Berghuis, A.M. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: Substrate specificity in udp-hexose 4-epimerases. J. Biol. Chem. 2004, 279, 22635–22642. [Google Scholar] [CrossRef] [PubMed]
- Giraud, M.-F.; Naismith, J.H. The rhamnose pathway. Curr. Opin. Struct. Biol. 2000, 10, 687–696. [Google Scholar] [CrossRef]
- Pretzer, G.; Snel, J.; Molenaar, D.; Wiersma, A.; Bron, P.A.; Lambert, J.; de Vos, W.M.; van der Meer, R.; Smits, M.A.; Kleerebezem, M. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 2005, 187, 6128–6136. [Google Scholar] [CrossRef]
- Gao, M.; Coggin, A.; Yagnik, K.; Teplitski, M. Role of Specific Quorum-Sensing Signals in the Regulation of Exopolysaccharide II Production within Sinorhizobium meliloti Spreading Colonies. PLoS ONE 2012, 7, e42611. [Google Scholar] [CrossRef]
- Schäper, S.; Steinchen, W.; Krol, E.; Altegoer, F.; Skotnicka, D.; Søgaard-Andersen, L.; Bange, G.; Becker, A. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. Proc. Natl. Acad. Sci. USA 2017, 114, E4822–E4831. [Google Scholar] [CrossRef]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Walter, J. Ecological role of lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl. Environ. Microbiol. 2008, 74, 4985–4996. [Google Scholar] [CrossRef] [PubMed]


| Genes | Abbreviation | Total Number of Genes | Number of PLFams | Number of Clusters not Having the Gene | Number of PGFams | Proteins: PLFams | PLFams: PGFams | % of Singleton Families # | Number of Clusters Having Multicopy Genes | For Clusters Having >2 Copies of Gene, Average Number of Those | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Families # | |||||||||||
| 1 | LytR-transcriptional regulator | epsA | 78 | 8 | 74 | 2 | 9.8 | 4 | 12.5 | 5 | 2.2 | 1 |
| 2 | Tyrosine kinase modulator | epsB | 130 | 29 | 25 | 15 | 4.4 | 1.9 | 37.9 | 9 | 2 | 2 |
| 3 | Tyrosine kinase | epsC | 125 | 25 | 30 | 10 | 5 | 2.5 | 28 | 8 | 2 | 2 |
| 4 | Phosphotyrosine phosphatase | epsD | 97 | 11 | 49 | 1 | 8.8 | 11 | 9.1 | 0 | - | - |
| 5 | Priming glycosyltransferase | epsE | 140 | 24 | 10 | 8 | 5.8 | 3 | 33.3 | 4 | 2 | 1.8 |
| 6 | Glycosyltransferase | gt | 670 | 343 | 0 | 246 | 1.9 | 1.4 | 66.6 | 140 | 4.6 | 4.3 |
| 7 | Flippase | wzx | 147 | 39 | 18 | 16 | 3.8 | 2.4 | 46.2 | 17 | 2.17 | 2 |
| 8 | Polysaccharide polymerase | wzy | 103 | 50 | 42 | 42 | 2 | 1.2 | 73.8 | 2 | 2 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deo, D.; Davray, D.; Kulkarni, R. A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes. Microorganisms 2019, 7, 444. https://doi.org/10.3390/microorganisms7100444
Deo D, Davray D, Kulkarni R. A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes. Microorganisms. 2019; 7(10):444. https://doi.org/10.3390/microorganisms7100444
Chicago/Turabian StyleDeo, Dipti, Dimple Davray, and Ram Kulkarni. 2019. "A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes" Microorganisms 7, no. 10: 444. https://doi.org/10.3390/microorganisms7100444
APA StyleDeo, D., Davray, D., & Kulkarni, R. (2019). A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes. Microorganisms, 7(10), 444. https://doi.org/10.3390/microorganisms7100444





